Enhancing the Pharmacological Properties of Triterpenes Through Acetylation: An Anticancer and Antioxidant Perspective
Abstract
1. Introduction
2. Results
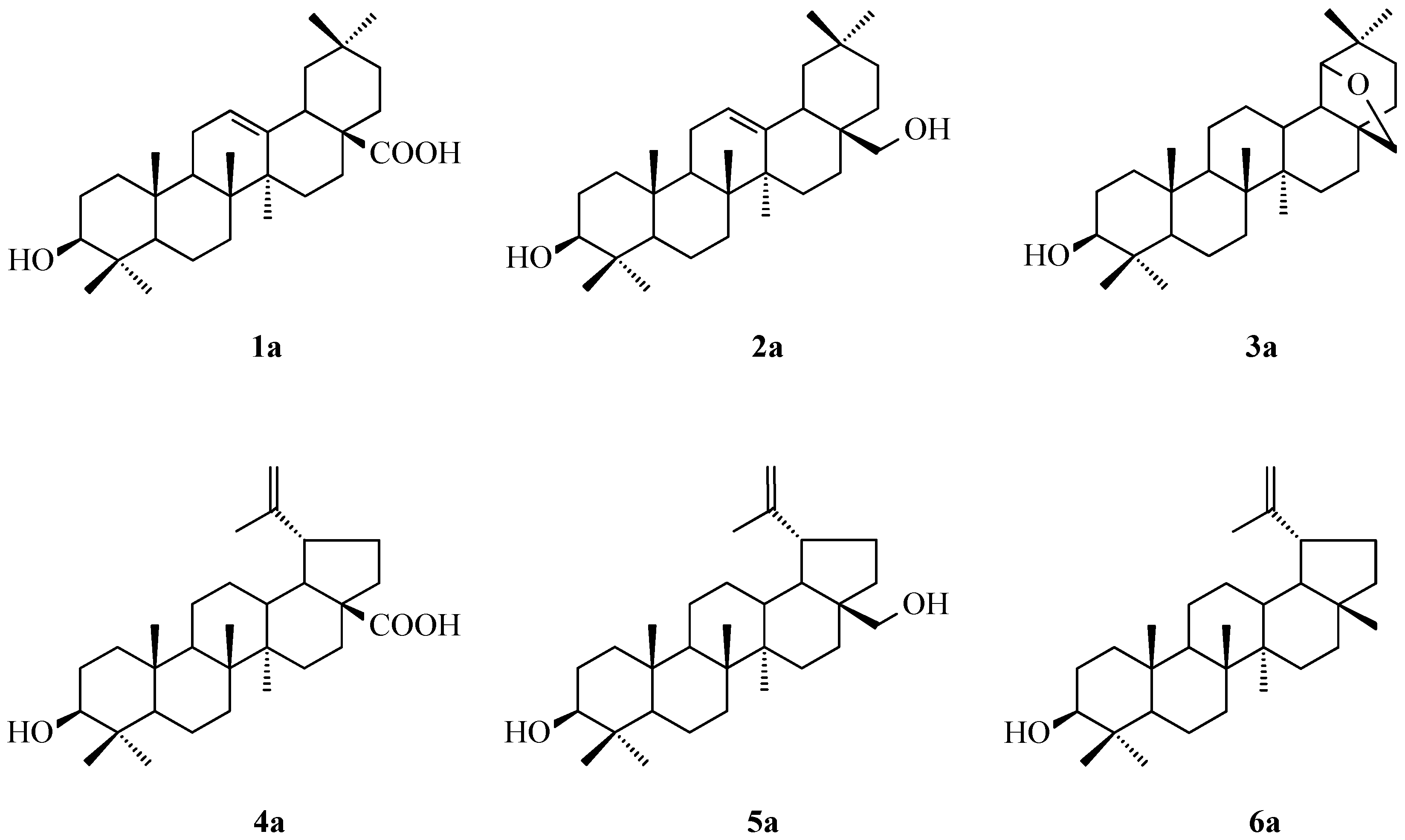
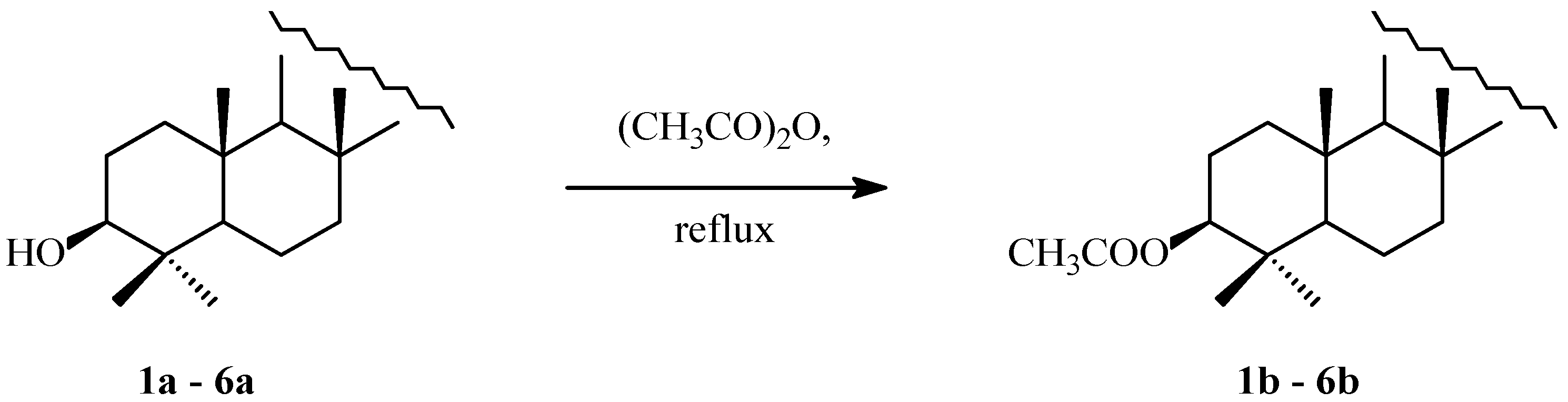
2.1. Acylation of Triterpenes
2.2. SAR Analysis
2.3. In Vivo Assay
2.3.1. MTT Results
2.3.2. Selectivity Index
2.3.3. The Apoptosis Assay
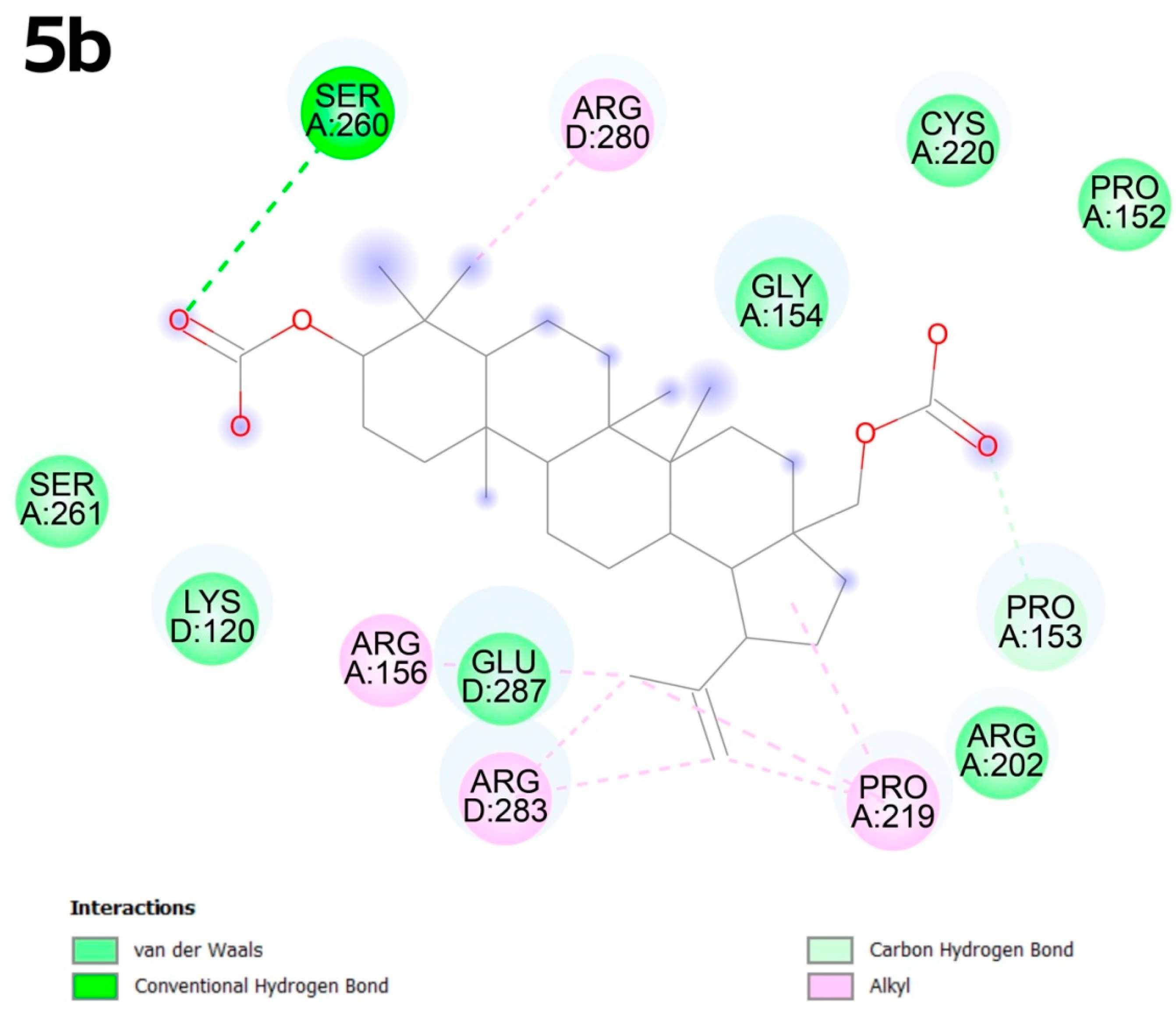
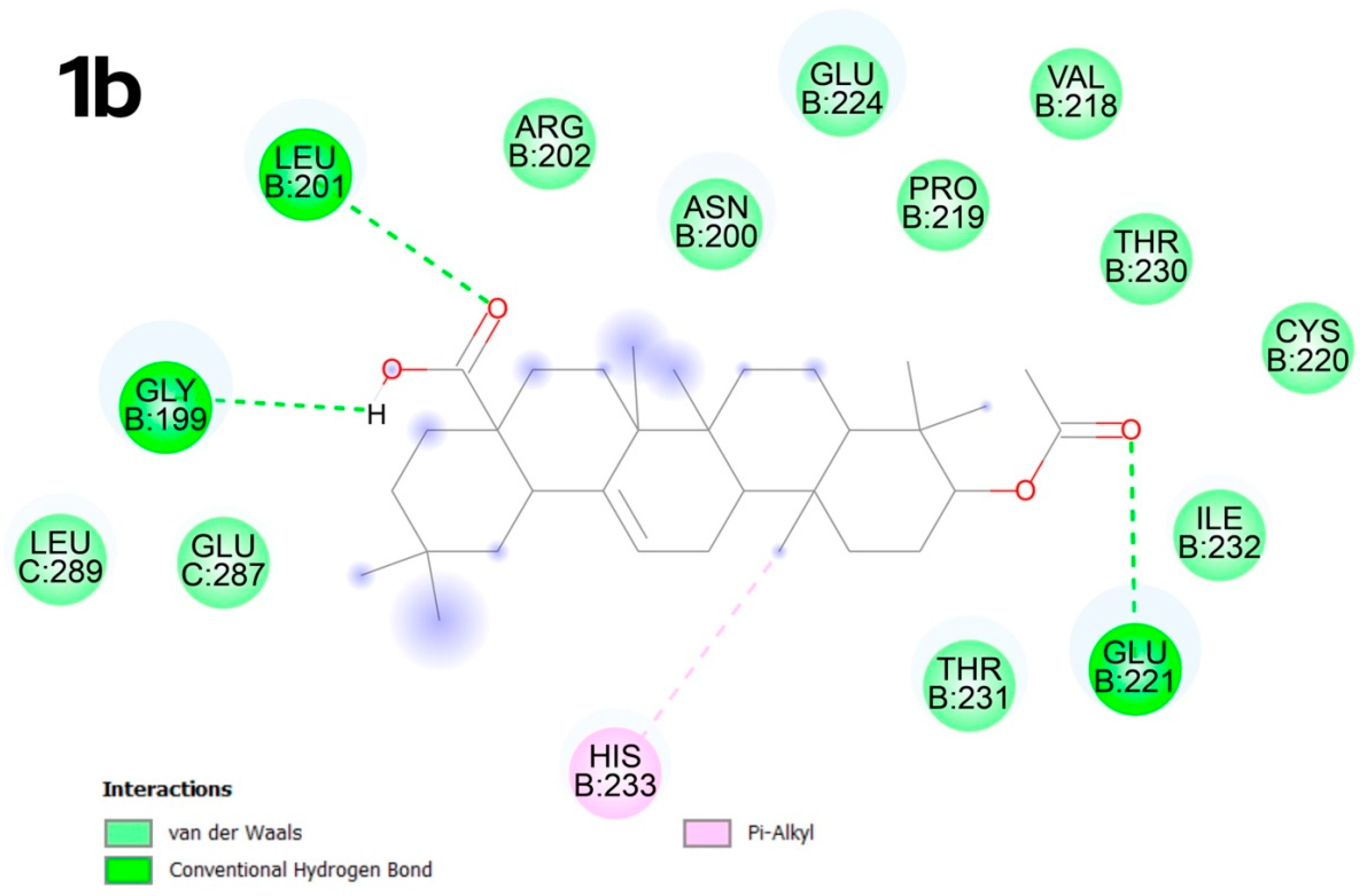
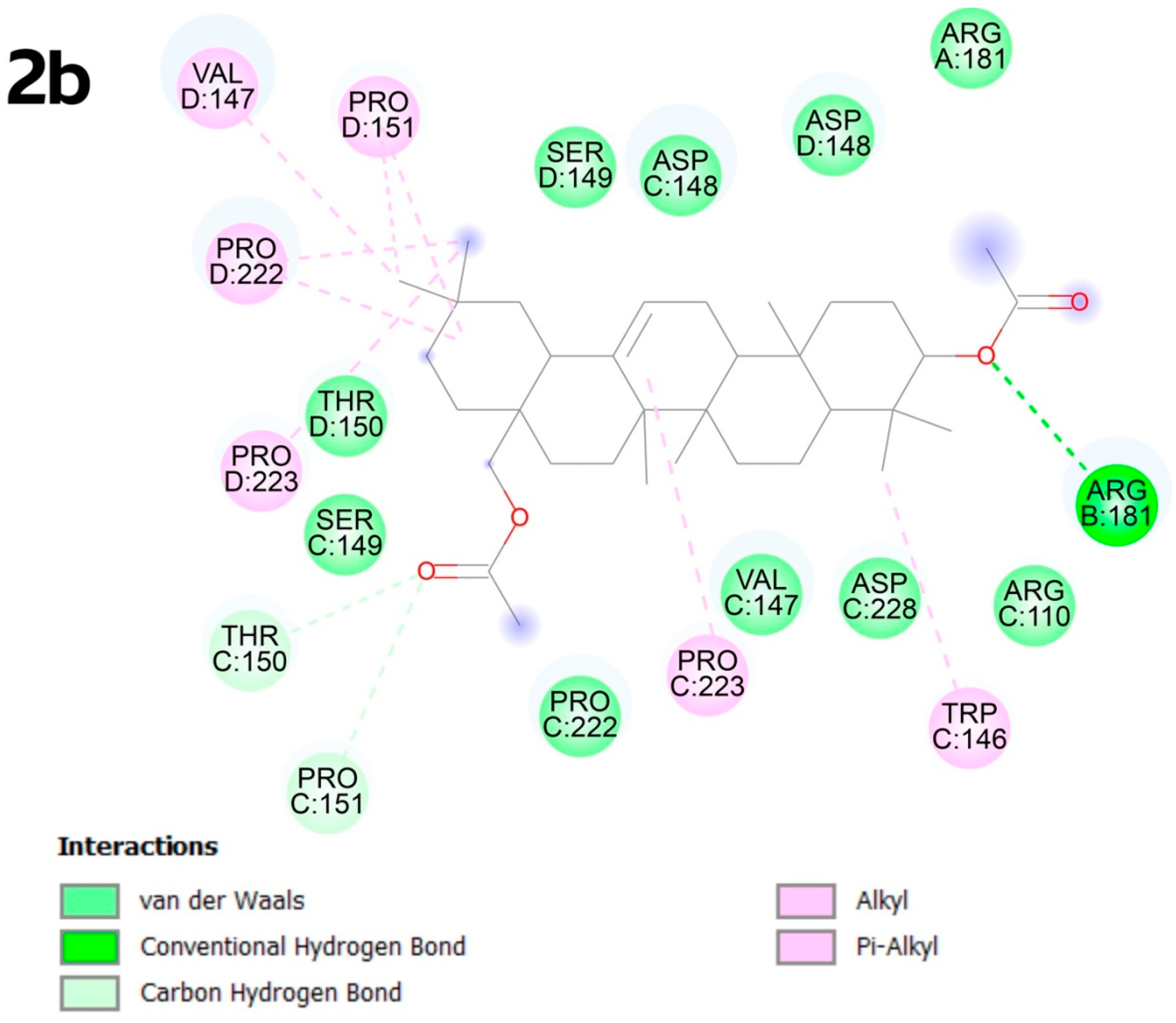
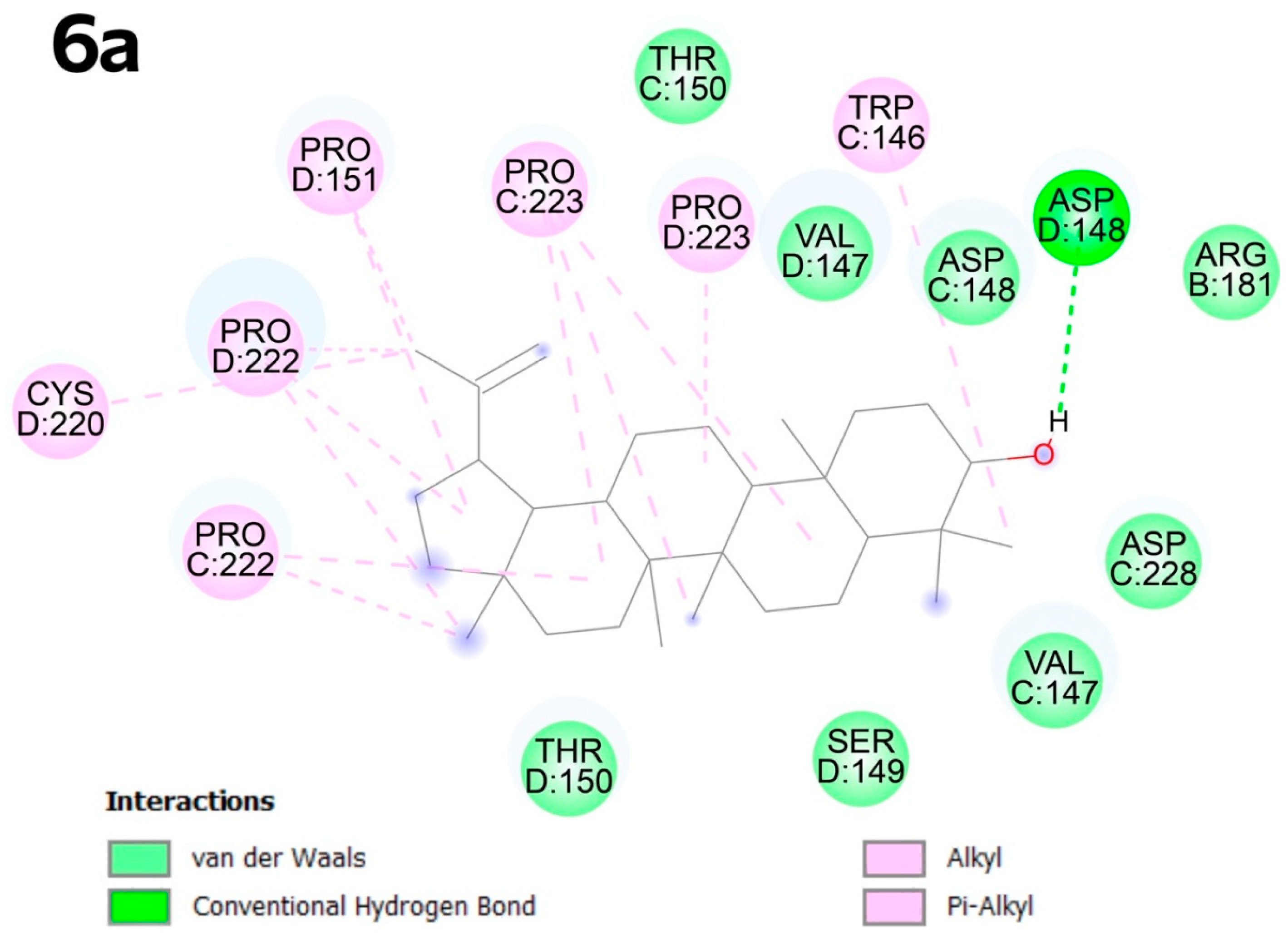
2.4. Molecular Docking
2.4.1. Detecting Cavities
2.4.2. Molecular Docking
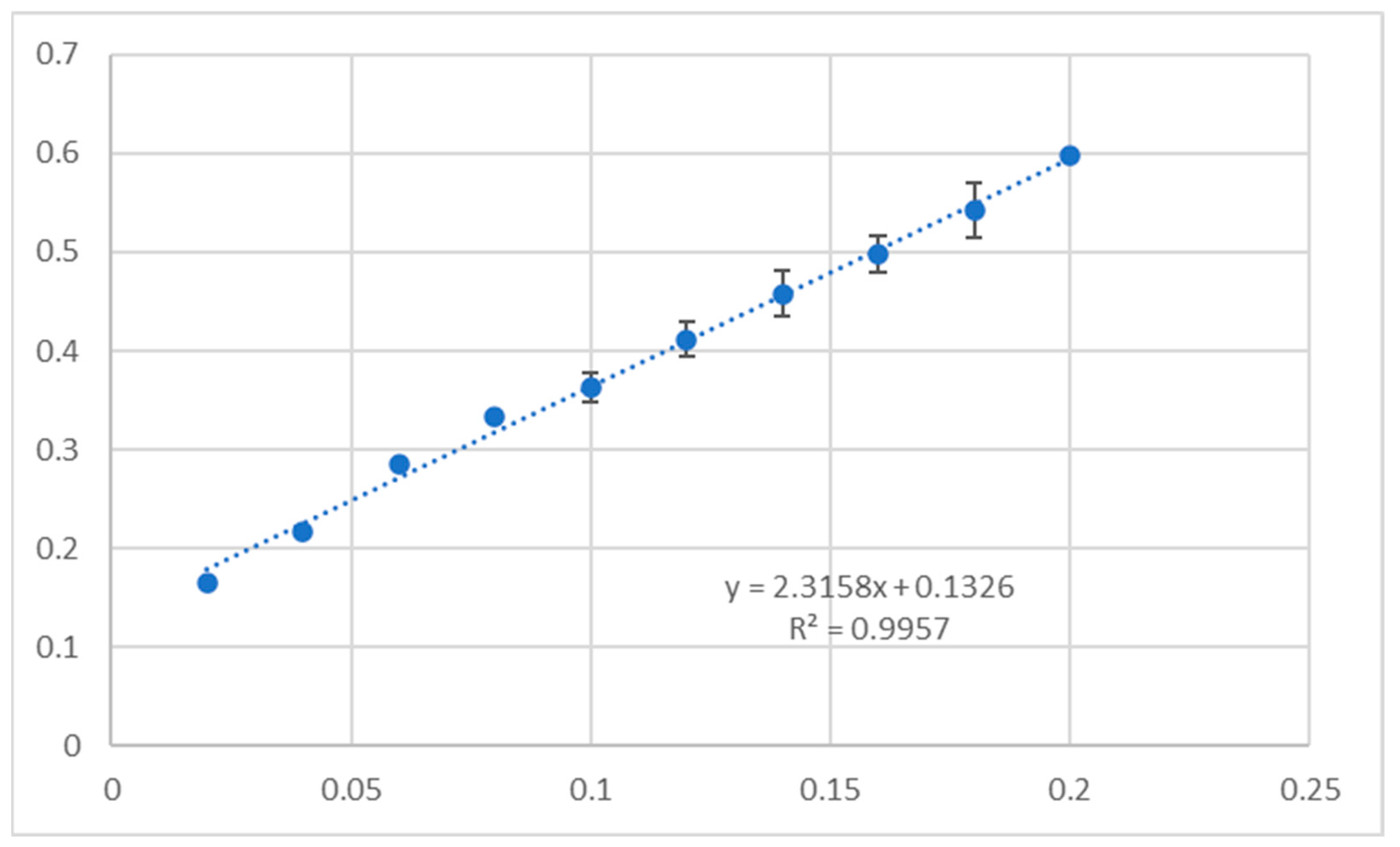
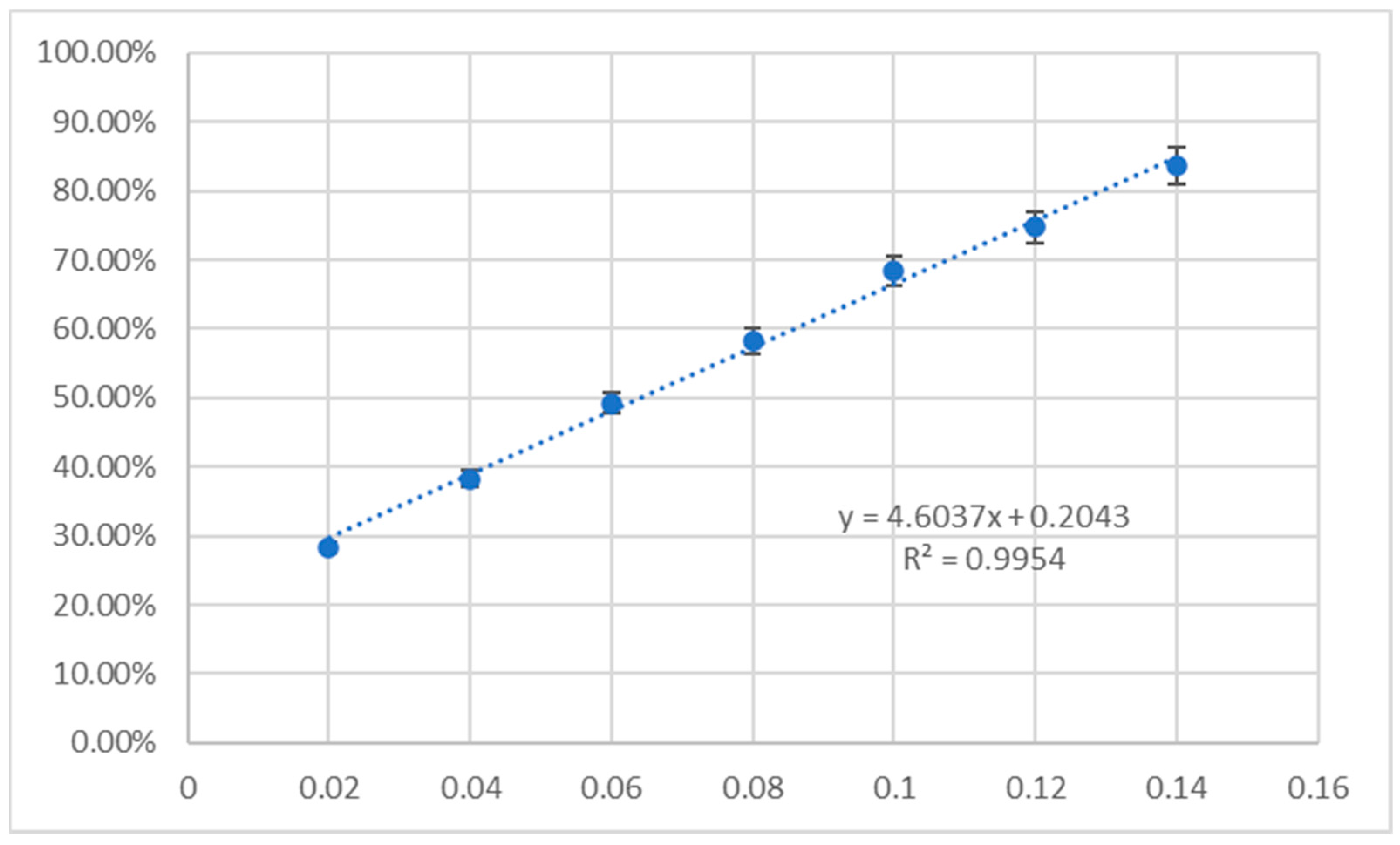
2.5. Antioxidant Assay
2.6. ADMETox Analysis
3. Discussion
3.1. Acylation of Triterpenes 1a–6a
3.2. Structure–Activity Analysis
3.3. Cytotoxic Activity of Triterpenes 1a–6a and 1b–6b
3.3.1. In Vivo Assay
3.3.2. Selectivity Index
3.3.3. Apoptosis
3.4. Molecular Docking
3.5. Antioxidant Activity
3.6. ADMETox Analysis
4. Materials and Methods
4.1. NMR
4.2. Acetylation of Triterpenes 1a–6a
4.2.1. General Information
4.2.2. Syntheses
- 3-O-Acetyloleanolic acid (1b): m.p. 267–268 °C (lit. m.p. 268–269 °C [96]).
- 3,28-di-O-Acetylerythrodiol (2b): m.p. 185–187 °C (lit. m.p. 185–185.5 °C [95]).
- 3-O-Acetylbetulinic acid (4b): m.p. 290–291 °C (lit. m.p. 285–290 °C [99]).
- 3,28-di-O-Acetylbetulin (5b): m.p. 215–217 °C (lit. m.p. 213–218 °C [98]).
- 3-O-Acetylolupeol (6b): m.p. 216–218 °C (lit. m.p. 217–218 °C [98]).
4.3. SAR Analysis
4.4. MTT Assay
4.5. Apoptosis
4.6. Molecular Docking
4.7. Antioxidant Activity
4.8. ADMETox Profile
5. Conclusions
6. Future Research Directions
- First comparative evaluation: Our study is the first to juxtapose acetylated and non-acetylated triterpenes across six cancer cell lines (HeLa, KB, MCF-7, A-549, PC-3, SKOV-3), including rarely investigated derivatives like 3b and 6b. This broad-spectrum analysis reveals stark contrasts in cytotoxicity, with acetylated derivatives 1b and 4b exhibiting submicromolar IC50 values, underscoring the transformative impact of C-3 acylation.
- Systematic acylation effects: By systematically modifying six triterpenes, we identify structure–activity relationships (SARs) that highlight the critical role of the C-17 carboxyl group. For instance, acetylation of oleanolic acid (1a) and betulinic acid (4a) enhanced activity by 40–100-fold, whereas analogous modifications in erythrodiol (2a) or lupeol (6a) reduced efficacy, emphasizing the necessity of tailored functionalization.
- Mechanistic insights via novel docking: Unlike prior studies, we explore interactions with the p53 Y220C mutant (PDB: 8DC4), a high-priority oncogenic target. Molecular docking using CB-Dock2—a machine learning–enhanced tool—revealed unique binding modes, such as 2b’s strong affinity for the C3 pocket (−10.1 kcal × mol−1), suggesting potential inhibition of p53-driven tumorigenesis.
- CB-Dock2 advancements: Our use of CB-Dock2, which incorporates ligand and protein flexibility, outperformed traditional rigid docking methods. This approach provided unprecedented accuracy in predicting binding poses, as evidenced by 5b’s alkyl interactions with arginine residues in the C1 pocket, validating its utility in drug discovery.
- Integrated methodology: By combining cytotoxicity, apoptosis, antioxidant assays and ADMETox profiling with computational analyses, we provide a holistic pharmacological evaluation. For example, 1b’s high CUPRAC activity (0.21986 mg × mL−1 Trolox equivalent) contrasted with its low DPPH response, highlighting electron transfer as its primary antioxidant mechanism—a distinction critical for therapeutic applications.
- Structural modifications: An example of planned chemical transformations is the introduction of additional functional groups (e.g., nitro, halogens) to increase molecular interactions with target proteins. More branched acyl groups will be introduced into the triterpene molecule to optimize lipophilicity and penetration of cell membranes.
- Synergistic studies: The combination of unsubstituted/acetylated triterpenes with other active substances (e.g., kinase inhibitors) may lead to synergistic anticancer activity.
- Exploration of mechanisms of action: the moderate activity of some triterpene derivatives may result from mechanisms of internal cellular resistance (e.g., expression of MDR pumps). Studying these mechanisms may indicate ways to improve pharmacological activity.
- Application of nanocarriers: Research on encapsulating compounds in lipid or polymer nanoparticles may increase their bioavailability and specificity towards cancer cells.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yeung, M.F.; Che, C.T. A review of presence of oleanolic acid in natural products. Nat. Proda Med. 2009, 2, 77–290. [Google Scholar]
- Smina, T.P.; Mathew, J.; Janardhanan, K.K.; Devasagayam, T.P.A. Antioxidant activity and toxicity profile of total triterpenes isolated from Ganoderma lucidum (Fr.) P. Karst occurring in South India. Environ. Toxicol. Pharmacol. 2011, 32, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Cui, Y.; Ding, H. Optimization of betulin extraction process from Inonotus obliguus with pulsed electric fields. Innov. Food Sci. Emerg. Technol. 2008, 9, 306–310. [Google Scholar] [CrossRef]
- Chen, J.; Liu, J.; Zhang, L.; Wu, G.; Hua, W.; Wu, X.; Sun, H. Pentacyclic triterpenes. Part 3: Synthesis and biological evaluation of oleanolic acid derivatives as novel inhibitors of glycogen phosphorylase. Bioorg. Med. Chem. Lett. 2006, 16, 2915–2919. [Google Scholar] [CrossRef]
- Kvasnica, M.; Sarek, J.; Klinotova, E.; Dzubak, P.; Hajduch, M. Synthesis of phthalates of betulinic acid and betulin with cytotoxic activity. Bioorg. Med. Chem. 2005, 13, 3447–3454. [Google Scholar] [CrossRef]
- Kazakova, O.B.; Khusnutdinova, E.F.; Tolstikov, G.A.; Suponitsky, K.Y. Synthesis of new olean-18(19)-ene derivatives from allobetulin. Russ. J. Bioorg. Chem. 2010, 36, 512–515. [Google Scholar] [CrossRef]
- Kazakova, O.B.; Giniyatullina, G.V.; Yamansarov, E.Y.; Tolstikov, G.A. Betulin and ursolic acid synthetic derivatives as inhibitors of Papilloma virus. Bioorg. Med. Chem. Lett. 2010, 20, 4088–4090. [Google Scholar] [CrossRef]
- Errichiello, F.; D’Amato, M.; Gambuti, A.; Moio, L.; Pastore, A.; AL-Hmadi, H.; Stornaiuolo, M.; Serino, E.; Taglialatela-Scafati, O.; Forino, M. Oleanolic acid: A promising antidiabetic metabolite detected in Aglianico grape pomace. J. Funct. Foods 2023, 104, 105548. [Google Scholar] [CrossRef]
- Castellano, J.M.; Garcia-Rodriguez, S.; Espinosa, J.M.; Millan-Linares, M.C.; Rada, M.; Perona, J.S. Oleanolic acid exerts a neuroprotective effect against microglial cell activation by modulating cytokine release and antioxidant defense systems. Biomolecules 2019, 9, 683. [Google Scholar] [CrossRef]
- Hong, W.; Fu, W.; Zhao, Q.; Xue, C.; Cai, W.; Dong, N.; Shan, A. Effects of oleanolic acid on acute liver injury triggered by lipopolysaccharide in broiler chickens. Br. Poult. Sci. 2023, 64, 697–709. [Google Scholar] [CrossRef]
- Bai, X.; Lai, T.; Zhou, T.; Li, Y.; Li, X.; Zhang, H. In vitro antioxidant activities of phenols and oleanolic acid from mango peel and their cytotoxic effect on A549 cell line. Molecules 2018, 23, 1395. [Google Scholar] [CrossRef] [PubMed]
- Salman, I.; Fakhoury, M.; Fouani, M.; Lawand, N. Peripheral anti-nociceptive and anti-inflammatory effect of oleanolic acid in a rat model of osteoarthritis. Antiinflamm. Antiallergy Agents Med. Chem. 2021, 20, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.H.; Ma, H.Y.; Yi, Y.L.; Zhu, S.J.; Yu, Z.W.; Zhu, J.; Mei, S.; Bahetibike, S.; Lu, Y.Q.; Huang, L.T.; et al. Oleanolic acid derivative alleviates cardiac fibrosis through inhibiting PTP1B activity and regulating AMPK/TGF-β/Smads pathway. Eur. J. Pharmacol. 2023, 960, 176116. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, Y.; Wang, X.; Tian, Z.; Qi, D.; Li, Y.; Jiang, H. Antihypertensive activity of oleanolic acid is mediated via downregulation of secretory phospholipase A2 and fatty acid synthase in spontaneously hypertensive rats. Int. J. Mol. Med. 2020, 46, 2019–2034. [Google Scholar] [CrossRef] [PubMed]
- Shan, T.; Ye, J.; Jia, J.; Wang, Z.; Jiang, Y.; Wang, Y.; Wang, Y.; Zheng, K.; Ren, Z. Viral UL8 is involved in the antiviral activity of oleanolic acid against HSV-1 infection. Front. Microbiol. 2021, 12, 689607. [Google Scholar] [CrossRef]
- Hichri, F.; Ben Jannet, H.; Cheriaa, J.; Jegham, S.; Mighri, Z. Antibacterial activities of a few prepared derivatives of oleanolic acid and of other natural triterpenic compounds. Comptes Rendus Chim. 2003, 6, 473–483. [Google Scholar] [CrossRef]
- Pan, Y.; Zhou, F.; Song, Z.; Huang, H.; Chen, Y.; Shen, Y.; Jia, Y.; Chen, J. Oleanolic acid protects against pathogenesis of atherosclerosis, possibly via FXR-mediated angiotensin (Ang)-(1-7) upregulation. Biomed. Pharmacother. 2018, 97, 1694–1700. [Google Scholar] [CrossRef]
- Bednarczyk-Cwynar, B.; Zaprutko, L.; Ruszkowski, P.; Hładoń. Synthesis and in vitro cytotoxic activity of A- or/and C-ring modified oleanolic acid derivatives. Org. Biomol. Chem. 2012, 10, 2201–2205. [Google Scholar] [CrossRef]
- Bednarczyk-Cwynar, B.; Ruszkowski, P.; Bobkiewicz-Kozlowska, T.; Zaprutko, L. Oleanolic Acid A-lactams inhibit the growth of HeLa, KB, MCF-7 and Hep-G2 cancer cell lines at micromolar concentrations. Anticancer Agents Med. Chem. 2016, 16, 579–592. [Google Scholar] [CrossRef]
- Sasikumar, K.; Dubey, V.; Ghosh, A.R. Oleanolic acid from black raisins, Vitis vinifera with antioxidant and antiproliferative potentials on HCT 116 colon cancer cell line. Braz. J. Pharm. Sci. 2020, 56, 17158. [Google Scholar] [CrossRef]
- Allouche, Y.; Warleta, F.; Campos, M.; Sánchez-Quesada, C.; Uceda, M.; Beltrán, G.; Gaforio, J.J. Antioxidant, antiproliferative, and pro-apoptotic capacities of pentacyclic triterpenes found in the skin of olives on MCF-7 human breast cancer cells and their effects on DNA damage. J. Agric. Food Chem. 2011, 59, 121–130. [Google Scholar] [CrossRef]
- Allouche, Y.; Beltrán, G.; Gaforio, J.J.; Uceda, M.; Mesa, M.D. Antioxidant and antiatherogenic activities of pentacyclic triterpenic diols and acids. Food Chem. Toxicol. 2010, 48, 2885–2890. [Google Scholar] [CrossRef]
- Martín, R.; Miana, M.; Jurado-López, R.; Martínez-Martínez, E.; Gómez-Hurtado, N.; Delgado, C.; Bartolomé, M.V.; San Román, J.A.; Cordova, C.; Lahera, V.; et al. Diol triterpenes block profibrotic effects of angiotensin II and protect from cardiac hypertrophy. PLoS ONE 2012, 7, e41545. [Google Scholar] [CrossRef][Green Version]
- Peñas-Fuentes, J.L.; Siles, E.; Rufino-Palomares, E.E.; Pérez-Jiménez, A.; Reyes-Zurita, F.J.; Lupiáñez, J.A.; Fuentes-Almagro, C.; Peragón-Sánchez, J. Effects of erythrodiol on the antioxidant response and proteome of HepG2 cells. Antioxidants 2021, 11, 73. [Google Scholar] [CrossRef]
- Juan, M.E.; Wenzel, U.; Daniel, H.; Planas, J.M. Erythrodiol, a natural triterpenoid from olives, has antiproliferative and apoptotic activity in HT-29 human adenocarcinoma cells. Mol. Nutr. Food Res. 2008, 52, 595–599. [Google Scholar] [CrossRef]
- Urban, M.; Vlk, M.; Dzubak, P.; Hajduch, M.; Sarek, J. Cytotoxic heterocyclic triterpenoids derived from betulin and betulinic acid. Bioorg. Med. Chem. 2012, 20, 3666–3674. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, X.; Zhang, X.; Wang, J.; Li, K.; Liu, G.; Lu, K.; Zhang, X.; Xie, C.; Zheng, T.; et al. Synthesis and biological evaluation of novel allobetulon/allobetulin–nucleoside conjugates as antitumor agents. Molecules 2021, 27, 4738. [Google Scholar] [CrossRef]
- Skurydina, E.S.; Vasil’eva, N.Y.; Kuznetsova, S.A.; Titova, N.M.; Kuznetsov, B.N. Development of a one-step method for producing allobetulin from birch outer bark and study of its antioxidant activity. Russ. J. Bioorg. Chem. 2024, 7, 2866–2873. [Google Scholar] [CrossRef]
- Machado, D.G.; Cunha, M.P.; Neis, V.B.; Balen, G.O.; Colla, A.; Bettio, L.E.B.; Oliveira, A.; Pazini, F.L.; Dalmarco, J.B.; Simionatto, E.L.; et al. Antidepressant-like effects of fractions, essential oil, carnosol and betulinic acid isolated from Rosmarinus officinalis L. Food Chem. 2013, 136, 999–1005. [Google Scholar] [CrossRef]
- Adesanwo, J.K.; Makinde, O.O.; Obafemi, C.A. Phytochemical analysis and antioxidant activity of methanol extract and betulinic acid isolated from the roots of Tetracera potatoria. J. Pharm. Res. 2013, 6, 903–907. [Google Scholar] [CrossRef]
- Puniani, E.; Cayer, C.; Kent, P.; Mullally, M.; Sánchez-Vindas, P.; Álvarez, L.P.; Cal, V.; Merali, Z.; Arnason, J.T.; Durst, T. Ethnopharmacology of Souroubea sympetala and Souroubea gilgii (Marcgraviaceae) and identification of betulinic acid as an anxiolytic principle. Phytochem 2015, 113, 73–78. [Google Scholar] [CrossRef]
- Lin, C.K.; Tseng, C.K.; Chen, K.H.; Wu, S.H.; Liaw, C.C.; Lee, J.C. Betulinic acid exerts anti-hepatitis C virus activity via the suppression of NF-κB- and MAPK-ERK1/2-mediated COX-2 expression. Br. J. Pharmacol. 2015, 172, 4481–4492. [Google Scholar] [CrossRef]
- Oliveira Costa, J.F.; Barbosa-Filho, J.M.; De Azevedo Maia, G.L.; Guimarães, E.T.; Meira, C.S.; Ribeiro-dos-Santos, R.; Pontes de Carvalho, L.C.; Soares, M.B.P. Potent anti-inflammatory activity of betulinic acid treatment in a model of lethal endotoxemia. Int. Immunopharmacol. 2014, 23, 469–474. [Google Scholar] [CrossRef]
- Ryu, S.Y.; Oak, M.H.; Yoon, S.K.; Cho, D.I.; Yoo, G.S.; Kim, T.S.; Kim, K.M. Anti-allergic and anti-inflammatory triterpenes from the herb of Prunella vulgaris. Planta Med. 2000, 66, 358–360. [Google Scholar] [CrossRef]
- Kim, S.J.; Quan, H.Y.; Jeong, K.J.; Kim, D.Y.; Kim, G.W.; Jo, H.K.; Chung, S.H. Beneficial effect of betulinic acid on hyperglycemia via suppression of hepatic glucose production. J. Agric. Food Chem. 2014, 62, 434–442. [Google Scholar] [CrossRef]
- Xie, R.; Zhang, H.; Wang, X.Z.; Yang, X.Z.; Wu, S.N.; Wang, H.G.; Shen, P.; Ma, T.H. The protective effect of betulinic acid (BA) diabetic nephropathy on streptozotocin (STZ)-induced diabetic rats. Food Funct. 2017, 8, 299–306. [Google Scholar] [CrossRef]
- Jain, M.; Kapadia, R.; Jadeja, R.N.; Thounaojam, M.C.; Devkar, R.V.; Mishra, S.H. Hepatoprotective potential of Tecomella undulata stem bark is partially due to the presence of betulinic acid. J. Ethnopharmacol. 2012, 143, 194–200. [Google Scholar] [CrossRef]
- Wang, S.; Wang, K.; Zhang, C.; Zhang, W.; Xu, Q.; Wang, Y.; Zhang, Y.; Li, Y.; Zhang, Y.; Zhu, H.; et al. Overaccumulation of p53-mediated autophagy protects against betulinic acid-induced apoptotic cell death in colorectal cancer cells. Cell Death Dis. 2017, 8, e3087. [Google Scholar] [CrossRef]
- Wang, H.; Dong, F.; Wang, Y.; Wang, X.; Hong, D.; Liu, Y.; Zhou, J. Betulinic acid induces apoptosis of gallbladder cancer cells via repressing SCD1. Acta Biochim. Biophys. 2020, 52, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lee, S.R.; Kang, K.S.; Ko, Y.; Pang, C.; Yamabe, N.; Kim, K.H. Betulinic acid suppresses ovarian cancer cell proliferation through induction of apoptosis. Biomolecules 2019, 9, 257. [Google Scholar] [CrossRef]
- Oliveira-Costa, J.F.; Meira, C.S.; Gomes das Neves, M.V.; Claro Dos Reis, B.P.Z.; Soares, M.B.P. Anti-inflammatory activities of betulinic acid: A review. Front. Pharmacol. 2022, 13, 883857. [Google Scholar] [CrossRef] [PubMed]
- Pârvănescu Pană, R.D.; Watz, C.G.; Moacă, E.A.; Vlaia, L.; Marcovici, I.; Macașoi, I.G.; Borcan, F.; Olariu, I.; Coneac, G.; Drăghici, G.A.; et al. Oleogel Formulations for the topical delivery of betulin and lupeol in skin injuries-preparation, physicochemical characterization, and pharmaco-toxicological evaluation. Molecules 2021, 26, 4174. [Google Scholar] [CrossRef] [PubMed]
- Pohjala, L.; Alakurtti, S.; Ahola, T.; Yli-Kauhaluoma, J.; Tammela, P. Betulin-derived compounds as inhibitors of alphavirus replication. J. Nat. Prod. 2009, 72, 1917–1926. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Liu, Z.; Liu, W.; Han, X.; Zhao, M. Betulin attenuates lung and liver injuries in sepsis. Int. Immunopharmacol. 2016, 30, 50–56. [Google Scholar] [CrossRef]
- Tang, J.J.; Li, J.G.; Qi, W.; Qiu, W.W.; Li, P.S.; Li, B.L.; Song, B.L. Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metab. 2011, 13, 44–56. [Google Scholar] [CrossRef] [PubMed]
- de Melo, C.L.; Queiroz, M.G.; Filho, A.C.; Rodrigues, A.M.; de Sousa, D.F.; Almeida, J.G.; Pessoa, O.D.; Silveira, E.R.; Menezes, D.B.; Melo, T.S.; et al. Betulinic acid, a natural pentacyclic triterpenoid, prevents abdominal fat accumulation in mice fed a high-fat diet. J. Agric. Food Chem. 2009, 57, 8776–8781. [Google Scholar] [CrossRef]
- Rzeski, W.; Stepulak, A.; Szymański, M.; Juszczak, M.; Grabarska, A.; Sifringer, M.; Kaczor, J.; Kandefer-Szerszeń, M. Betulin elicits anti-cancer effects in tumour primary cultures and cell lines in vitro. Basic Clin. Pharmacol. Toxicol. 2009, 105, 425–432. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Jiang, D.; Lin, Y.; Wang, Y.; Li, Q.; Liu, L.; Jin, Y.H. Betulin induces reactive oxygen species-dependent apoptosis in human gastric cancer SGC7901 cells. Arch. Pharm. Res. 2016, 39, 1257–1265. [Google Scholar] [CrossRef]
- Boryczka, S.; Bębenek, E.; Wietrzyk, J.; Kempińska, K.; Jastrzębska, M.; Kusz, J.; Nowak, M. Synthesis, structure and cytotoxic activity of new acetylenic derivatives of betulin. Molecules 2013, 18, 4526–4543. [Google Scholar] [CrossRef]
- Fernández, M.A.; de las Heras, B.; García, M.D.; Sáenz, M.T.; Villar, A. New insights into the mechanism of action of the anti-inflammatory triterpene lupeol. J. Pharm. Pharmacol. 2001, 53, 1533–1539. [Google Scholar] [CrossRef]
- Sudhahar, V.; Ashok Kumar, S.; Varalakshmi, P.; Sujatha, V. Protective effect of lupeol and lupeol linoleate in hypercholesterolemia associated renal damage. Mol. Cell Biochem. 2008, 317, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, J.F.; Teixeira, M.M.; Barbosa-Filho, J.M.; Lúcio, A.S.; Almeida, J.R.; de Queiroz, L.P. The triterpenoid lupeol attenuates allergic airway inflammation in a murine model. Int. Immunopharmacol. 2008, 8, 1216–1221. [Google Scholar] [CrossRef]
- Sultana, S.; Saleem, M.; Sharma, S.; Khan, N. Lupeol, a triterpene, prevents free radical mediated macromolecular damage and alleviates benzoyl peroxide induced biochemical alterations in murine skin. Indian J. Exp. Biol. 2003, 41, 827–831. [Google Scholar] [PubMed]
- You, Y.J.; Nam, N.H.; Kim, Y.; Bae, K.H.; Ahn, B.Z. Antiangiogenic activity of lupeol from Bombax ceiba. Phytother. Res. 2003, 17, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Pitchai, D.; Roy, A.; Ignatius, C. In vitro evaluation of anticancer potentials of lupeol isolated from Elephantopus scaber L. on MCF-7 cell line. J. Adv. Pharm. Technol. Res. 2014, 5, 179–184. [Google Scholar] [CrossRef]
- Saini, M.; Khan, M.F.; Sangwan, R.; Khan, M.A.; Kumar, A.; Verma, R.; Ahamad, T.; Jain, S. Design, Synthesis and in-vitro antitumor activity of lupeol derivatives via modification at C-3 and C-30 positions. Chem. Sel. 2019, 4, 1800–1805. [Google Scholar] [CrossRef]
- Lien, A.P.H.; Hua, H.; Chuong, P.H. Free radicals, antioxidants in disease and health. Int. J. Biomed. Sci. 2008, 4, 89–96. [Google Scholar]
- Gülçin, I.; Elias, R.; Gepdiremen, A.; Boyer, L. Antioxidant activity of lignans from fringe tree (Chionanthus virginicus L.). Eur. Food Res. Technol. 2006, 223, 759–767. [Google Scholar] [CrossRef]
- Lobo, V.; Patil, A.; Phatak, A.; Chandra, N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn. Rev. 2010, 4, 118–126. [Google Scholar] [CrossRef]
- Ling, T.; Boyd, L.; Riva, F. Triterpenoids as reactive oxygen species modulators of cell fate. Chem. Res. Toxicol. 2022, 35, 569–584. [Google Scholar] [CrossRef]
- Günther, A.; Makuch, E.; Nowak, A.; Duchnik, W.; Kucharski, Ł.; Pełech, R.; Klimowicz, A. Enhancement of the antioxidant and skin permeation properties of betulin and its derivatives. Molecules 2021, 26, 3435. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.B.H.; Moghaddam, M.G.; Basri, M.; Rahman, M.B.A. Anticancer activity of 3-O-acylated betulinic acid derivatives obtained by enzymatic synthesis. Biosci. Biotechnol. Biochem. 2010, 74, 1025–1029. [Google Scholar] [CrossRef]
- Bednarczyk-Cwynar, B.; Ruszkowski, P.; Atamanyuk, D.; Lesyk, R.; Zaprutko, L. Hybrids of oleanolic acid with norbornene-2,3-dicarboximide-N-carboxylic acids as potential anticancer agents. Acta Pol. Pharm. Drug Res. 2017, 74, 827–835. [Google Scholar]
- Bednarczyk-Cwynar, B.; Więcaszek, T.; Ruszkowski, P. Cytotoxic activity of some lupeol derivatives. Nat. Prod. Commun. 2016, 11, 1237–1238. [Google Scholar] [CrossRef]
- Günther, A.; Zalewski, P.; Sip, S.; Ruszkowski, P.; Bednarczyk-Cwynar, B. Oleanolic acid dimers with potential application in medicine—Design, synthesis, physico-chemical characteristics, cytotoxic and antioxidant activity. Int. J. Mol. Sci. 2024, 25, 6989. [Google Scholar] [CrossRef] [PubMed]
- Bednarczyk-Cwynar, B.; Ruszkowski, P. Acylation of oleanolic acid oximes effectively improves cytotoxic activity in in vitro studies. Pharmaceutics 2024, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Rali, S.; Oyedeji, O.O.; Aremu, O.O.; Oyedeji, A.O.; Nkeh-Chungag, B.N. Semisynthesis of derivatives of oleanolic acid from Syzygium aromaticum and their antinociceptive and anti-inflammatory properties. Mediators Inflamm. 2016, 2016, 8401843. [Google Scholar] [CrossRef]
- Gunther, A.; Zalewski, P.; Sip, S.; Ruszkowski, P.; Bednarczyk-Cwynar, B. Acetylation of oleanolic acid dimers as a method of synthesis of powerful cytotoxic agents. Molecules 2024, 29, 4291. [Google Scholar] [CrossRef]
- Pistritto, G.; Trisciuoglio, D.; Ceci, C.; Garufi, A.; D’Orazi, G. Apoptosis as anticancer mechanism: Function and dysfunction of its modulators and targeted therapeutic strategies. Aging 2016, 8, 603–619. [Google Scholar] [CrossRef]
- Hu, C.; Cao, Y.; Li, P.; Tang, X.; Yang, M.; Gu, S.; Xiong, K.; Li, T.; Xiao, T. Oleanolic acid induces autophagy and apoptosis via the AMPK-mTOR signaling pathway in colon cancer. J. Oncol. 2021, 2021, 8281718. [Google Scholar] [CrossRef]
- Thornberry, N.A.; Lazebnik, Y. Caspases: Enemies within. Science 1998, 281, 1312–1316. [Google Scholar] [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.C.; Cullen, S.P.; Martin, S.J. Apoptosis: Controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 2008, 9, 231–241. [Google Scholar] [CrossRef]
- Kim, G.J.; Jo, H.J.; Lee, K.J.; Choi, J.W.; An, J.H. Oleanolic acid induces p53-dependent apoptosis via the ERK/JNK/AKT pathway in cancer cell lines in prostatic cancer xenografts in mice. Oncotarget 2018, 9, 26370–26386. [Google Scholar] [CrossRef]
- Fulda, S.; Friesen, C.; Los, M.; Scaffidi, C.; Mier, W.; Benedict, M.; Nuñez, G.; Krammer, P.H.; Peter, M.E.; Debatin, K.M. Betulinic acid triggers CD95 (APO-1/Fas)- and p53-independent apoptosis via activation of caspases in neuroectodermal tumors. Cancer Res. 1997, 57, 4956–4964. [Google Scholar]
- Selzer, E.; Pimentel, E.; Wacheck, V.; Schlegel, W.; Pehamberger, H.; Jansen, B.; Kodym, R. Effects of betulinic acid alone and in combination with irradiation in human melanoma cells. J. Investig. Dermatol. 2000, 114, 935–940. [Google Scholar] [CrossRef]
- Fulda, S.; Scaffidi, C.; Susin, S.A.; Krammer, P.H.; Kroemer, G.; Peter, M.E.; Debatin, K. Activation of mitochondria and release of mitochondrial apoptogenic factors by btulinic acid. J. Biol. Chem. 1998, 273, 33942–33948. [Google Scholar] [CrossRef]
- Karin, M. Nuclear factor-κB in cancer development and progression. Nature 2006, 441, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Baud, V.; Karin, M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discov. 2009, 8, 33–40. [Google Scholar] [CrossRef]
- DiDonato, J.A.; Mercurio, F.; Karin, M. NF-κB and the link between inflammation and cancer. Immunol. Rev. 2012, 246, 379–400. [Google Scholar] [CrossRef]
- Yadav, V.R.; Prasad, S.; Sung, B.; Kannappan, R.; Aggarwal, B.B. Targeting inflammatory pathways by triterpenoids for prevention and treatment of cancer. Toxins 2010, 2, 2428–2466. [Google Scholar] [CrossRef] [PubMed]
- Chasov, V.; Davletshin, D.; Gilyazova, E.; Mirgayazova, R.; Kudriaeva, A.; Khadiullina, R.; Yuan, Y.; Bulatov, E. Anticancer therapeutic strategies for targeting mutant p53-Y220C. J. Biomed. Res. 2024, 38, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Sukandar, E.R.; Kaennakam, S.; Raab, P.; Nost, X.; Rassamee, K.; Bauer, R.; Siripong, P.; Ersam, T.; Tip-pyang, S.; Chavasiri, W. Cytotoxic and anti-inflammatory Activities of dihydroisocoumarin and xanthone derivatives from Garcinia picrorhiza. Molecules 2021, 26, 6626. [Google Scholar] [CrossRef]
- Peńa-Morán, O.A.; Villarreal, M.L.; Álvarez-Berber, L.; Meneses-Acosta, A.; Rodríguez-López, V. Cytotoxicity, post-treatment recovery, and selectivity analysis of naturally occurring podophyllotoxins from Bursera fagaroides var. fagaroides on breast cancer cell lines. Molecules 2016, 21, 1013. [Google Scholar] [CrossRef]
- Valderrama, J.A.; Delgado, V.; Sepúlveda, S.; Benites, J.; Theoduloz, C.; Calderon, P.B.; Muccioli, G.G. Synthesis and cytotoxic activity on human cancer cells of novel isoquinolinequinone-amino acid derivatives. Molecules 2016, 21, 1199. [Google Scholar] [CrossRef]
- Pagadala, N.S.; Syed, K.; Tuszynski, J. Software for molecular docking: A review. Biophys. Rev. 2017, 9, 91–102. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Nguyen, T.H.; Pham, T.N.H.; Huy, N.T.; Bay, M.V.; Pham, M.Q.; Nam, P.C.; Vu, V.V.; Ngo, S.T. Autodock Vina adopts more accurate binding poses but autodock4 forms better binding affinity. J. Chem. Inf. Model. 2020, 60, 204–211. [Google Scholar] [CrossRef]
- Pinzi, L.; Rastelli, G. Molecular docking: Shifting paradigms in drug discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.X.; Cao, Y. CB-Dock2: Improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Basse, N.; Kaar, J.L.; Settanni, G.; Joerger, A.C.; Rutherford, T.J.; Fersht, A.R. Toward the rational design of p53-stabilizing drugs: Probing the surface of the oncogenic Y220C mutant. Chem. Biol. 2010, 17, 46–56. [Google Scholar] [CrossRef]
- Strano, S.; Dell’Orso, S.; Mongiovi, A.M.; Monti, O.; Lapi, E.; Agostino, S.D.; Fontemaggi, G.; Blandino, G. Mutant p53 proteins: Between loss and gain of function. Head Neck 2006, 29, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Cadwell, C.; Zambetti, G.P. The effects of wild-type p53 tumor suppressor activity and mutant p53 gain-of-function on cell growth. Gene 2002, 77, 15–30. [Google Scholar] [CrossRef]
- Wu, B.Y.; Parks, L.M. Reduction of triterpene acids with lithium aluminum hydride. J. Am. Pharm. Assoc. 1950, 39, 475–476. [Google Scholar] [CrossRef]
- Wrzeciono, U.; Malecki, I.; Budzianowski, J.; Kierylowicz, H.; Zaprutko, L.; Beimcik, E.; Kostępska, H. Nitrogenous triterpene derivatives. 10. Hemisuccinates of some oleanolic acid derivatives and their antiulcer effect. Pharmazie 1985, 40, 542–544. [Google Scholar] [PubMed]
- Klinot, J.; Vystrčil, A. Nebenprodukte bei der umsetzung von allobetulin zu heterobetulin. Collect. Czechoslov. Chem. Commun. 1964, 29, 516–530. [Google Scholar] [CrossRef]
- Yagishita, K. Isolation and identification of betulin, lupeol, and β-amyrin from the bird-lime of Trochodendron aralioides Siebold et Zuccarini. J. Agric. Chem. Soc. Jap. 1957, 21, 77–81. [Google Scholar] [CrossRef][Green Version]
- Kozubek, M.; Höhlich, L.; Hoenke, S.; Deigner, H.; Al-Harrasi, A.; Csuk, R. Apoptotic activity of substituted 3-O-acetyl-betulinic acid benzylamides. Eur. J. Med. Chem. Rep. 2021, 3, 100016. [Google Scholar] [CrossRef]
- Available online: www.way2drug.com (accessed on 2 October 2024).
- Available online: https://cadd.labshare.cn/cb-dock2/index.php (accessed on 8 October 2024).
- Garbiec, E.; Rosiak, N.; Tykarska, E.; Zalewski, P.; Cielecka-Piontek, J. Sinapic acid co-amorphous systems with amino acids for improved solubility and antioxidant activity. Int. J. Mol. Sci. 2023, 24, 5533. [Google Scholar] [CrossRef]
- Available online: https://admetmesh.scbdd.com (accessed on 2 October 2024).
- Chen, Z.; Huang, K.Y.; Ling, Y.; Goto, M.; Duan, H.Q.; Tong, X.H.; Liu, Y.L.; Cheng, Y.Y.; Morris-Natschke, S.L.; Yang, P.C.; et al. Discovery of an oleanolic acid/hederagenin-nitric oxide donor hybrid as an EGFR tyrosine kinase inhibitor for non-small-cell lung cancer. J. Nat. Prod. 2019, 82, 3065–3073. [Google Scholar] [CrossRef] [PubMed]
- Medina-O’Donnell, M.; Rivas, F.; Reyes-Zurita, F.J.; Martinez, A.; Martin-Fonseca, S.; Garcia-Granados, A.; Ferrer-Martín, R.M.; Lupiañez, J.A.; Parra, A. Semi-synthesis and antiproliferative evaluation of PEGylated pentacyclic triterpenes. Eur. J. Med. Chem. 2016, 118, 64–78. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, P.; Patel, N.K.; Gangwal, R.P.; Sangamwar, A.T.; Bhutani, K.K. Oleanolic acid analogs as NO, TNF-α and IL-1β inhibitors: Synthesis, biological evaluation and docking studies. Bioorg. Med. Chem. Lett. 2014, 24, 4114–4119. [Google Scholar] [CrossRef] [PubMed]
- Parra, A.; Martin-Fonseca, S.; Rivas, F.; Reyes-Zurita, F.J.; Medina-O’Donnell, M.; Martinez, A.; Garcia-Granados, A.; Lupiañez, J.A.; Albericio, F. Semi-synthesis of acylated triterpenes from olive-oil industry wastes for the development of anticancer and anti-HIV agents. Eur. J. Med. Chem. 2014, 74, 278–301. [Google Scholar] [CrossRef]


| Triterpene | Examples of Pharmacological Activity | Ref. |
|---|---|---|
| oleanolic acid | antidiabetic | [8] |
| neuroprotective | [9] | |
| hepatoprotective | [10] | |
| antioxidant | [11] | |
| antinociceptive and anti-inflammatory | [12] | |
| cardioprotective | [13] | |
| antihypertensive | [14] | |
| antiviral: EC50 = 13.07 µM (HSV-1/153), EC50 = 12.89 µM (HSV-1/106), EC50 = 13.09 µM (HSV-1/Blue) | [15] | |
| antibacterial: MBC = 700 µg × mL−1 (Staphylococcus aureus), MIC = 95 µg × mL−1 (S. aureus), MBC = 900 µg × mL−1 (Escherichia coli), MIC = 95 µg × mL−1 (E. coli), MBC = 300 µg × mL−1 (Salmonella typhi), MIC = 65 µg × mL−1 (S. typhi) | [16] | |
| antiatherosclerotic | [17] | |
| anticancer: IC50 = 14.93 µM (KB), IC50 = 13.95 µM (MCF-7), IC50 = 11.82 µM (HeLa), IC50 = 16.20 µM (Hep-G2) | [18,19] | |
| anticancer: IC50 = 40 µg × mL−1 (HCT-116) | [20] | |
| antioxidant, antiproliferative | [21] | |
| erythrodiol | antioxidant, antiproliferative | [21] |
| antioxidant, antiatherogenic | [22] | |
| Anti-inflammatory, vasorelaxing and cardioprotective | [23] | |
| antioxidant and anticancer: IC50 = 27.3 µM (Hep-G2) | [24] | |
| anticancer: EC50 = 48.8 µM (HT-29) | [25] | |
| anticancer: IC50 = 250 µM (CCRF-CEM, CEM) | [26] | |
| anticancer: IC50 = 64.96 µM (SMMC-7721), IC50 = 87.73 µM (Hep-G2), IC50 = 62.96 µM (A-459) | [27] | |
| antioxidant | [28] | |
| betulinic acid | antidepressant | [29] |
| antioxidant | [30] | |
| anxiolytic | [31] | |
| antiviral: EC50 = 11.2 μM (HCV) | [32] | |
| anti-inflammatory | [33] | |
| antiallergic and anti-inflammatory | [34] | |
| antihyperglycemic | [35] | |
| nephroprotective | [36] | |
| hepatoprotective | [37] | |
| anticancer: IC50 = 30 µM (CCRF-CEM, CEM) | [26] | |
| anticancer: IC50 = 125 µM (HT-29); IC50 = 58 µM (SW-480); IC50 = 178 µM (HCT-116) | [38] | |
| anticancer: IC50 = 30 µM (NOZ) | [39] | |
| anticancer: IC50 = 44.47 µM (A-2780) | [40] | |
| betulin | anti-inflammatory | [41] |
| wound healing | [42] | |
| antiviral: IC50 = 45.5 µM (SFV) | [43] | |
| antiseptic | [44] | |
| antihyperlipidemic | [45] | |
| antiobesity | [46] | |
| anticancer: IC50 = 250 µM (CCRF-CEM, CEM) | [26] | |
| anticancer: IC50 = 2.5 µM (SK-N-AS); IC50 = 5.9 µM (C-6); IC50 = 10.3 µM (TE-671); IC50 = 4.3 µM (HT-29); IC50 = 5.2 µM (T-47D); IC50 = 6.8 µM (FTC-238); IC50 = 7.4 µM (A-549); IC50 = 6.4 µM (RPMI-8226); IC50 = 6.7 µM (Jurkat 1E.6); IC50 = 2.8 µM (HPOC); IC50 = 3.4 µM (HPCC); IC50 = 3.4 µM (HPGBM) | [47] | |
| antioxidant; anticancer: IC50 = 29.4 µM (13 µg/mL; SGC7901) | [48] | |
| anticancer: IC50 = 73.2 µM (32.4 µg/mL; T-47D); IC50 = 24.6 µM (10.9 µg/mL; CCRF/CEM); IC50 = 51.7 µM (22.9 µg/mL; SW-707); IC50 = 12.4 µM (5.5 µg/mL; P-388) | [49] | |
| lupeol | wound healing | [42] |
| anti-inflammatory | [50] | |
| nephroprotective | [51] | |
| antiallergic | [52] | |
| antioxidant | [53] | |
| antiangiogenic | [54] | |
| anticancer: IC50 = 80 µM (MCF-7) | [55] | |
| anticancer: IC50 = 46.06 µM (MDA MB-231); IC50 = 31.910 µM (HeLa); IC50 = 64.82 µM (A-549) | [56] |
| Activity | Pa Factor (and Pi Factor) of Compounds 1a–6a and 1b–6b | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 2a | 3a | 4a | 5a | 6a | 1b | 2b | 3b | 4b | 5b | 6b | |
| Antineoplastic | 0.876 (0.005) | 0.920 (0.005) | 0.950 (0.004) | 0.925 (0.005) | 0.948 (0.004) | 0.950 (0.004) | 0.890 (0.005) | 0.923 (0.005) | 0.954 (0.004) | 0.934 (0.004) | 0.952 (0.004) | 0.954 (0.004) |
| Antineoplastic (colon c.) | <0.700 | 0.734 (0.005) | 0.917 (0.003) | 0.789 (0.005) | 0.853 (0.004) | 0.831 (0.004) | <0.700 | 0.790 (0.005) | 0.925 (0.003) | 0.836 (0.004) | 0.876 (0.004) | 0.863 (0.004) |
| Antineoplastic (colorectal c.) | <0.700 | 0.736 (0.005 | 0.920 (0.003) | 0.794 (0.005) | 0.858 (0.004) | 0.836 (0.004) | <0.700 | 0.791 (0.005) | 0.927 (0.003) | 0.840 (0.004) | 0.879 (0.004) | 0.867 (0.004) |
| Antineoplastic (lung c.) | 0.766 (0.005) | 0.802 (0.004) | 0.883 (0.003) | 0.815 (0.004) | 0.833 (0.004) | 0.850 (0.004) | 0.792 (0.004) | 0.823 (0.004) | 0.899 (0.003) | 0.831 (0.004) | 0.859 (0.003) | 0.869 (0.003) |
| Antiprotozoal (Leishmania) | 0.721 (0.008) | <0.700 | <0.700 | 0.923 (0.003) | 0.881 (0.003) | 0.891 (0.003) | 0.821 (0.004) | 0.848 (0.004) | 0.790 (0.005) | 0.954 (0.002) | 0.961 (0.002) | 0.940 (0.002) |
| Apoptosis agonist | 0.901 (0.004) | 0.892 (0.004) | 0.759 (0.010) | 0.822 (0.007) | 0.837 (0.005) | 0.883 (0.005) | 0.891 (0.004) | 0.878 (0.005) | 0.747 (0.011) | 0.850 (0.005) | 0.825 (0.006) | 0.874 (0.005) |
| Caspase 3 stim. | 0.984 (0.002) | 0.971 (0.002) | 0.820 (0.005) | <0.700 | 0.974 (0.002) | 0.978 (0.002) | 0.974 (0.002) | 0.870 (0.004) | 0.720 (0.010) | 0.976 (0.002) | 0.880 (0.004) | 0.954 (0.003) |
| Caspase 8 stim. | 0.914 (0.001) | 0.878 (0.001) | 0.808 (0.002) | <0.700 | 0.869 (0.001) | 0.865 (0.001) | 0.910 (0.001) | 0.846 (0.001) | 0.804 (0.002) | 0.900 (0.001) | 0.835 (0.001) | 0.864 (0.001) |
| Chemopre- ventive | 0.937 (0.002) | 0.852 (0.003) | 0.707 (0.006) | 0.835 (0.003) | 0.733 (0.005) | 0.792 (0.004) | 0.948 (0.002) | 0.912 (0.002) | 0.717 (0.006) | 0.855 (0.003) | 0.802 (0.004) | 0.806 (0.004) |
| Mucomem- branous prot. | 0.894 (0.005) | 0.824 (0.013) | 0.732 (0.042) | 0.786 (0.022) | <0.700 | 0.847 (0.009) | 0.935 (0.004) | 0.892 (0.005) | 0.795 (0.020) | <0.700 | 0.778 (0.025) | 0.895 (0.005) |
| Oxidoreductase inh. | 0.904 (0.002) | 0.888 (0.003) | 0.823 (0.005) | 0.809 (0.006) | 0.745 (0.010) | 0.834 (0.005) | 0.915 (0.002) | 0.885 (0.003) | 0.848 (0.004) | <0.700 | 0.724 (0.013) | 0.855 (0.004) |
| TF NF kappa B stim. | 0.954 (0.001) | 0.931 (0.001) | 0.864 (0.002) | 0.804 (0.003) | 0.935 (0.001) | 0.947 (0.001) | 0.936 (0.001) | 0.893 (0.002) | 0.788 (0.003) | 0.941 (0.001) | 0.904 (0.001) | 0.924 (0.001) |
| TF stim. | 0.954 (0.001) | 0.931 (0.001) | 0.864 (0.002) | 0.804 (0.003) | 0.935 (0.001) | 0.947 (0.001) | 0.936 (0.001) | 0.893 (0.002) | 0.788 (0.003) | 0.941 (0.001) | 0.904 (0.001) | 0.924 (0.001) |
| Comp. No. | Cell Line, IC50 [µM] (±SD) | ||||||
|---|---|---|---|---|---|---|---|
| HeLa | KB | MCF-7 | A-549 | PC-3 | SKOV-3 | HDF | |
| 1a | 11.82 (±0.19) * | 14.93 (±0.07) * | 13.95 (±0.11) * | 8.79 (±0.20) * | 18.63 (±0.05) *** | 18.81 (±0.09) *** | 24.87 (±0.04) *** |
| 2a | 54.32 (±0.02) | 66.04 (±0.44) | 65.52 (±0.18) | 63.67 (±0.12) | 21.09 (±0.18) | 18.33 (±0.05) | 27.99 (±0.66) |
| 3a | 49.44 (±0.21) | 45.33 (±0.03) | 71.06 (±0.13) | 51.74 (±0.18) | 20.17 (±0.04) | 20.93 (±0.02) | 48.22 (±0.19) |
| 4a | 27.50 (±0.12) | 35.21 (±0.06) | 27.89 (±0.12) | 26.23 (±0.19) | 10.61 (±0.09) | 9.52 (±0.21) | 19.92 (±0.49) |
| 5a | 18.32 (±0.02) | 19.74 (±0.13) | 18.09 (±0.02) | 19.56 (±0.17) | 6.22 (±0.06) | 6.95 (±0.08) | 15.81 (±0.66) |
| 6a | 37.74 (±0.12) ** | 51.17 (±1.92) ** | 51.82 (±0.15) ** | 45.70 (±0.12) ** | 14.52 (±0.03) | 14.57 (±0.09) | 29.04 (±0.06) |
| 1b | 0.24 (±0.19) | 0.36 (±0.07) | 1.86 (±0.16) | 0.24 (±0.12) | 0.11 (±0.02) | 0.09 (±0.01) | 0.19 (±0.07) |
| 2b | 70.30 (±0.15) | 49.96 (±0.31) | 61.58 (±0.17) | 51.61 (±0.43) | 25.13 (±0.09) | 25.51 (±0.04) | 53.11 (±0.52) |
| 3b | >100 | >100 | >100 | >100 | 52.22 (±0.14) | 53.09 (±0.55) | 62.02 (±0.18) |
| 4b | 1.62 (±0.18) | 1.50 (±0.07) | 1.50 (±0.11) | 1.23 (±0.13) | 0.93 (±0.02) | 1.03 (±0.01) | 2.88 (±0.09) |
| 5b | >100 | >100 | >100 | >100 | 75.91 (±0.37) | 75.96 (±0.84) | 81.73 (±0.49) |
| 6b | 72.64 (±0.14) | 72.52 (±0.44) | 49.97 (±0.31) | 56.22 (±0.59) | 22.16 (±0.15) | 21.09 (±0.11) | 34.97 (±0.39) |
| Comp. No. | Cell Line, SI | |||||
|---|---|---|---|---|---|---|
| HeLa | KB | MCF-7 | A-549 | PC-3 | SKOV-3 | |
| 1a | 2.10 | 1.66 | 1.78 | 2.83 | 1.33 ** | 1.32 ** |
| 2a | 0.87 | 0.42 | 0.43 | 0.44 | 1.32 | 1.53 |
| 3a | 0.97 | 1.06 | 0.68 | 0.93 | 2.39 | 2.30 |
| 4a | 0.72 | 0.56 | 0.71 | 0.76 | 1.88 | 2.09 |
| 5a | 0.86 | 0.80 | 0.87 | 0.81 | 2.54 | 2.27 |
| 6a | 0.77 | 0.57 | 0.56 | 0.63 | 2.00 | 1.99 |
| 1b | 0.79 | 0.53 | 0.10 | 0.79 | 1.73 | 2.11 |
| 2b | 0.75 | 1.06 | 0.86 | 1.03 | 2.11 | 2.08 |
| 3b | - - - * | - - - * | - - - * | - - - * | 1.19 | 1.17 |
| 4b | 1.77 | 1.92 | 1.92 | 2.34 | 3.10 | 2.80 |
| 5b | - - - * | - - - * | - - - * | - - - * | 1.08 | 1.07 |
| 6b | 0.48 | 0.48 | 0.70 | 0.62 | 1.58 | 1.66 |
| Comp. No. | Cell Line, AI | |
|---|---|---|
| SKOV-3 | PC-3 | |
| 1a | 5.16 (0.01) | 5.27 (0.04) |
| 3a | 5.78 (0.01) | 5.27 (0.01) |
| 5a | 5.26 (0.03) | 5.82 (0.03) |
| 1b | 6.79 (0.16) | 6.90 (0.04) |
| 4b | 7.81 (0.02) | 7.09 (0.01) |
| CurPocket ID | Cavity Volume (Å3) | Center (x, y, z) | Cavity Size (x, y, z) |
|---|---|---|---|
| C1 | 3942 | −16, 49, 61 | 30, 18, 27 |
| C2 | 2320 | −6, 46, 9 | 18, 23, 18 |
| C3 | 828 | −33, 65, 34 | 11, 20, 8 |
| C4 | 430 | −40, 55, 40 | 19, 7, 8 |
| C5 | 370 | −30, 44, 24 | 8, 14, 15 |
| Pocket ID | Compound Number | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 1b | 2a | 2b | 3a | 3b | 4a | 4b | 5a | 5b | 6a | 6b | |
| C1 | −6.4 | −9.0 | −7.6 | −7.1 | −9.3 | −8.3 | −6.0 | −7.8 | −7.1 | −9.1 | −8.5 | −9.1 |
| C2 | −6.3 | −9.0 | −6.5 | −6.4 | −6.5 | −6.4 | −6.2 | −5.7 | −5.8 | −6.0 | −6.0 | −6.1 |
| C3 | −8.6 | −7.4 | −9.3 | −10.1 | −9.9 | −9.3 | −8.6 | −9.3 | −8.2 | −8.1 | −9.3 | −9.6 |
| C4 | −8.2 | −8.5 | −8.0 | −8.8 | −9.4 | −9.1 | −8.6 | −8.4 | −8.0 | −8.2 | −9.6 | −8.6 |
| C5 | −7.8 | −7.8 | −7.5 | −8.9 | −8.3 | −7.9 | −6.8 | −9.0 | −7.1 | −7.5 | −7.5 | −7.2 |
| Comp. No. | Trolox Equivalent [mg/mL] | |
|---|---|---|
| CUPRAC Assay | DPPH Assay | |
| 1a | 0.10312 ± 0.00684 | 0.02633 ± 0.001090 |
| 2a | 0.09414 ± 0.00950 | 0.02092 ± 0.000866 |
| 3a | 0.08248 ± 0.00500 | 0.01677 ± 0.000694 |
| 4a | 0.07632 ± 0.00727 | 0.01526 ± 0.000632 |
| 5a | 0.29900 ± 0.00663 | 0.01077 ± 0.000446 |
| 6a | 0.13602 ± 0.00593 | 0.00065 ± 00.01570 |
| 1b | 0.21986 ± 0.00657 | 0.01430 ± 0.000592 |
| 2b | 0.24016 ± 0.00995 | 0.00313 ± 0.000130 |
| 3b | 0.03212 ± 0.00166 | 0.00182 ± 0.000755 |
| 4b | 0.19986 ± 0.00150 | 0.00376 ± 0.000156 |
| 5b | 0.17609 ± 0.00487 | 0.00786 ± 0.000325 |
| 6b | 0.13490 ± 0.00514 | 0.00348 ± 0.000144 |
| Properties (Optimal Values) | Compound Number | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 2a | 3a | 4a | 5a | 6a | 1b | 2b | 3b | 4b | 5b | 6b | |
| Mol. Weight (100~600) | 456.360 | 442.380 | 442.380 | 456.360 | 442.380 | 426.390 | 498.370 | 526.400 | 484.390 | 498.370 | 526.400 | 468.400 |
| Volume | 505.750 | 499.598 | 493.678 | 505.751 | 499.598 | 490.807 | 546.497 | 581.089 | 534.423 | 546.497 | 581.089 | 531.553 |
| Density | 0.902 | 0.885 | 0.896 | 0.902 | 0.885 | 0.869 | 0.912 | 0.906 | 0.906 | 0.912 | 0.906 | 0.881 |
| NHA (0~12) | 3 | 2 | 2 | 3 | 2 | 1 | 4 | 4 | 3 | 4 | 4 | 2 |
| nHD (0~7) | 2 | 2 | 1 | 2 | 2 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| nRot (0~11) | 1 | 1 | 0 | 2 | 2 | 1 | 3 | 5 | 2 | 4 | 6 | 3 |
| nRing (0~6) | 5 | 5 | 6 | 5 | 5 | 5 | 5 | 5 | 6 | 5 | 5 | 5 |
| maxRing (0~18) | 22 | 22 | 7 | 21 | 21 | 21 | 22 | 22 | 7 | 21 | 21 | 21 |
| nHet (1~15) | 3 | 2 | 2 | 3 | 2 | 1 | 4 | 4 | 3 | 4 | 4 | 2 |
| fChar (−4~+4) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| nRig (0~30) | 27 | 26 | 29 | 27 | 26 | 26 | 28 | 28 | 30 | 28 | 28 | 27 |
| Flexibility (≤2) | 0.037 | 0.038 | 0.000 | 0.074 | 0.077 | 0.038 | 0.107 | 0.179 | 0.067 | 0.143 | 0.214 | 0.111 |
| Stereo Centers (≤2) | 8 | 8 | 10 | 10 | 10 | 10 | 8 | 8 | 10 | 10 | 10 | 10 |
| TPSA (0~140) | 57.530 | 40.460 | 29.460 | 57.530 | 40.460 | 20.230 | 63.600 | 52.600 | 35.530 | 63.600 | 52.600 | 26.300 |
| LogS (−4~0.5) | −5.036 | −5.769 | −6.590 | −5.148 | −5.681 | −6.667 | −5.982 | −6.964 | −7.152 | −5.866 | −6.772 | −7.189 |
| LogP (0~3) | 6.113 | 6.538 | 6.515 | 5.574 | 5.747 | 6.689 | 6.889 | 7.674 | 6.994 | 6.149 | 6.843 | 7.170 |
| LogD (1~3) | 4.843 | 4.705 | 5.145 | 4.873 | 4.682 | 5.313 | 4.873 | 4.975 | 5.214 | 4.945 | 4.971 | 5.417 |
| QED (≥0.67) | 0.409 | 0.424 | 0.428 | 0.436 | 0.452 | 0.421 | 0.311 | 0.274 | 0.356 | 0.320 | 0.274 | 0.299 |
| Sascore (≤6) | 4.589 | 4.702 | 5.524 | 4.689 | 4.761 | 4.663 | 4.624 | 4.754 | 5.542 | 4.725 | 4.823 | 4.692 |
| Fsp3 (≥0.420) | 0.900 | 0.933 | 1.000 | 0.900 | 0.933 | 0.933 | 0.875 | 0.882 | 0.969 | 0.875 | 0.882 | 0.906 |
| MCE-18 (≥45) | 105.368 | 102.207 | 116.200 | 104.000 | 100.793 | 100.793 | 106.667 | 105.000 | 117.397 | 105.300 | 103.594 | 102.164 |
| Npscore (−5~5) | 3.272 | 3.326 | 3.146 | 3.072 | 3.233 | 3.054 | 3.217 | 3.073 | 3.061 | 3.012 | 2.975 | 2.956 |
| Lipinski Rule | A | A | A | A | A | A | A | R | A | A | R | A |
| Pfizer Rule | R | R | R | R | R | R | R | R | R | R | R | R |
| GSK Rule | R | R | R | R | R | R | R | R | R | R | R | R |
| Golden Triangle | A | A | R | A | A | R | A | R | R | A | R | R |
| PAINS (alerts) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ALARM NMR (alerts) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BMS (alerts) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Chelator Rule (alerts) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Caco-2 perm. (≥−5.15) | −5.198 | −4.867 | −5.115 | −6.283 | −4.942 | −5.042 | −5.166 | −4.989 | −5.040 | −5.203 | −5.029 | −4.946 |
| MDCK perm. (≤2 × 10−6 cm/s) | 2.00 × 10−5 | 8.83 × 10−6 | 1.91 × 10−5 | 1.80 × 10−5 | 1.30 × 10−5 | 1.00 × 10−5 | 1.60 × 10−5 | 1.30 × 10−5 | 2.22 × 10−5 | 2.46 × 10−5 | 1.97 × 10−5 | 1.47 × 10−5 |
| Pgp-inh. (≤0.300) | 0.000 | 0.002 | 0.002 | 0.002 | 0.008 | 0.029 | 0.001 | 0.171 | 0.028 | 0.014 | 0.813 | 0.174 |
| Pgp-sub. (≤0.300) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| HIA (≤0.300) | 0.012 | 0.011 | 0.005 | 0.007 | 0.004 | 0.003 | 0.014 | 0.019 | 0.005 | 0.007 | 0.005 | 0.004 |
| F20% (≤0.300) | 0.074 | 0.878 | 0.250 | 0.340 | 0.880 | 0.742 | 0.031 | 0.455 | 0.018 | 0.012 | 0.021 | 0.014 |
| F30% (≤0.300) | 0.756 | 0.952 | 0.956 | 0.898 | 0.939 | 0.891 | 0.828 | 0.929 | 0.865 | 0.899 | 0.962 | 0.833 |
| PPB (≤90%) | 98.130 | 98.230 | 97.480 | 96.515 | 97.762 | 98.998 | 97.013 | 97.247 | 96.056 | 96.841 | 95.967 | 98.268 |
| VD (0.04–20 L/kg) | 0.718 | 1.194 | 1.441 | 0.614 | 1.103 | 1.650 | 0.686 | 1.196 | 1.264 | 0.687 | 1.303 | 1.681 |
| BBB penetr. (≤0.300) | 0.674 | 0.562 | 0.376 | 0.492 | 0.471 | 0.627 | 0.329 | 0.254 | 0.176 | 0.206 | 0.262 | 0.353 |
| Fu (≥5.000%) | 3.524 | 2.365 | 2.072 | 2.579 | 1.625 | 1.713 | 3.461 | 1.927 | 2.151 | 2.123 | 1.714 | 1.646 |
| CYP1A2 inh. (≥0.700) | 0.012 | 0.038 | 0.032 | 0.021 | 0.047 | 0.037 | 0.011 | 0.022 | 0.028 | 0.016 | 0.029 | 0.033 |
| CYP1A2 sub. (≥0.700) | 0.323 | 0.194 | 0.336 | 0.549 | 0.333 | 0.598 | 0.166 | 0.119 | 0.152 | 0.243 | 0.130 | 0.455 |
| CYP2C19 inh. (≥0.700) | 0.028 | 0.054 | 0.063 | 0.027 | 0.054 | 0.073 | 0.029 | 0.070 | 0.068 | 0.026 | 0.065 | 0.080 |
| CYP2C19 sub. (≥0.700) | 0.916 | 0.924 | 0.932 | 0.928 | 0.939 | 0.947 | 0.893 | 0.914 | 0.931 | 0.931 | 0.930 | 0.948 |
| CYP2C9 inh. (≥0.700) | 0.157 | 0.187 | 0.100 | 0.129 | 0.142 | 0.093 | 0.218 | 0.196 | 0.101 | 0.138 | 0.125 | 0.090 |
| CYP2C9 sub. (≥0.700) | 0.813 | 0.124 | 0.175 | 0.703 | 0.160 | 0.569 | 0.723 | 0.080 | 0.143 | 0.678 | 0.110 | 0.596 |
| CYP2D6 inh. (≥0.700) | 0.012 | 0.049 | 0.050 | 0.005 | 0.031 | 0.057 | 0.012 | 0.111 | 0.033 | 0.005 | 0.079 | 0.029 |
| CYP2D6 sub. (≥0.700) | 0.528 | 0.245 | 0.837 | 0.764 | 0.807 | 0.898 | 0.177 | 0.096 | 0.675 | 0.600 | 0.475 | 0.879 |
| CYP3A4 inh. (≥0.700) | 0.172 | 0.717 | 0.252 | 0.127 | 0.519 | 0.223 | 0.246 | 0.605 | 0.278 | 0.155 | 0.530 | 0.250 |
| CYP3A4 sub. (≥0.700) | 0.208 | 0.455 | 0.335 | 0.223 | 0.464 | 0.433 | 0.377 | 0.667 | 0.504 | 0.325 | 0.580 | 0.525 |
| CL (≥15 mL/min/kg) | 3.094 | 10.509 | 4.701 | 3.039 | 6.486 | 5.372 | 2.119 | 3.091 | 3.454 | 2.454 | 3.165 | 3.552 |
| T1/2< 3h (≤0.300) | 0.023 | 0.017 | 0.043 | 0.063 | 0.051 | 0.037 | 0.013 | 0.009 | 0.027 | 0.039 | 0.025 | 6.486 |
| hERG Blockers (≤0.300) | 0.004 | 0.021 | 0.511 | 0.031 | 0.056 | 0.124 | 0.003 | 0.008 | 0.326 | 0.021 | 0.044 | 0.102 |
| H-HT (≤0.300) | 0.296 | 0.402 | 0.254 | 0.275 | 0.262 | 0.085 | 0.278 | 0.313 | 0.271 | 0.298 | 0.258 | 0.126 |
| DILI (≤0.300) | 0.010 | 0.007 | 0.031 | 0.009 | 0.011 | 0.024 | 0.024 | 0.202 | 0.437 | 0.046 | 0.492 | 0.341 |
| AMES Tox. (≤0.300) | 0.008 | 0.003 | 0.005 | 0.003 | 0.002 | 0.002 | 0.005 | 0.002 | 0.003 | 0.002 | 0.002 | 0.002 |
| ROAT (≤0.300) | 0.228 | 0.162 | 0.168 | 0.221 | 0.129 | 0.185 | 0.080 | 0.021 | 0.038 | 0.045 | 0.009 | 0.032 |
| FDAMDD (≤0.300) | 0.909 | 0.925 | 0.939 | 0.928 | 0.928 | 0.919 | 0.623 | 0.753 | 0.727 | 0.628 | 0.726 | 0.515 |
| Skin Sensit. (≤0.300) | 0.028 | 0.076 | 0.893 | 0.329 | 0.624 | 0.727 | 0.036 | 0.035 | 0.532 | 0.195 | 0.129 | 0.680 |
| Carcinogen. (≤0.300) | 0.063 | 0.067 | 0.005 | 0.018 | 0.014 | 0.006 | 0.080 | 0.059 | 0.005 | 0.016 | 0.013 | 0.005 |
| Eye Corrosion (≤0.300) | 0.012 | 0.006 | 0.169 | 0.022 | 0.019 | 0.882 | 0.195 | 0.008 | 0.170 | 0.030 | 0.007 | 0.877 |
| Eye Irritation (≤0.300) | 0.084 | 0.022 | 0.263 | 0.039 | 0.047 | 0.550 | 0.050 | 0.036 | 0.196 | 0.032 | 0.068 | 0.458 |
| Respir. Tox. (≤0.300) | 0.968 | 0.980 | 0.934 | 0.945 | 0.841 | 0.580 | 0.968 | 0.958 | 0.802 | 0.928 | 0.603 | 0.412 |
| Bioconc. Factors | 1.944 | 3.075 | 2.476 | 2.117 | 2.841 | 2.560 | 2.422 | 2.784 | 2.323 | 2.553 | 2.444 | 2.410 |
| IGC50 | 5.021 | 5.176 | 5.726 | 5.176 | 5.357 | 5.710 | 5.048 | 5.311 | 5.789 | 5.240 | 5.491 | 5.773 |
| LC50FM | 5.937 | 6.126 | 6.841 | 6.289 | 6.495 | 6.951 | 6.034 | 6.354 | 6.868 | 6.365 | 8.611 | 6.977 |
| LC50DM | 6.337 | 6.676 | 6.924 | 6.504 | 6.868 | 6.990 | 6.359 | 6.731 | 6.852 | 6.466 | 6.910 | 6.904 |
| NR-AR (≤0.300) | 0.369 | 0.055 | 0.002 | 0.069 | 0.021 | 0.007 | 0.665 | 0.167 | 0.019 | 0.459 | 0.075 | 0.116 |
| NR-AR-LBD (≤0.300) | 0.273 | 0.355 | 0.042 | 0.495 | 0.252 | 0.065 | 0.733 | 0.788 | 0.112 | 0.740 | 0.484 | 0.233 |
| NR-AhR (≤0.300) | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.001 | 0.000 | 0.001 | 0.001 | 0.000 |
| NR-Aromatase (≤0.300) | 0.759 | 0.702 | 0.381 | 0.463 | 0.547 | 0.267 | 0.795 | 0.675 | 0.448 | 0.426 | 0.502 | 0.311 |
| NR-ER (≤0.300) | 0.412 | 0.249 | 0.309 | 0.115 | 0.116 | 0.272 | 0.721 | 0.178 | 0.286 | 0.135 | 0.069 | 0.229 |
| NR-ER-LBD (≤0.300) | 0.593 | 0.765 | 0.696 | 0.432 | 0.237 | 0.546 | 0.785 | 0.633 | 0.805 | 0.657 | 0.737 | 0.786 |
| NR-PPAR γ (≤0.300) | 0.965 | 0.297 | 0.022 | 0.763 | 0.021 | 0.017 | 0.964 | 0.196 | 0.020 | 0.826 | 0.019 | 0.021 |
| SR-ARE (≤0.300) | 0.556 | 0.275 | 0.064 | 0.200 | 0.081 | 0.049 | 0.637 | 0.216 | 0.049 | 0.163 | 0.058 | 0.038 |
| SR-ATAD5 (≤0.300) | 0.052 | 0.118 | 0.008 | 0.078 | 0.040 | 0.009 | 0.428 | 0.385 | 0.015 | 0.190 | 0.148 | 0.016 |
| SR-HSE (≤0.300) | 0.747 | 0.128 | 0.027 | 0.567 | 0.050 | 0.030 | 0.770 | 0.211 | 0.031 | 0.507 | 0.091 | 0.041 |
| SR-MMP (≤0.300) | 0.971 | 0.942 | 0.647 | 0.940 | 0.876 | 0.620 | 0.956 | 0.808 | 0.261 | 0.806 | 0.469 | 0.277 |
| SR-p53 (≤0.300) | 0.271 | 0.130 | 0.010 | 0.443 | 0.112 | 0.012 | 0.647 | 0.432 | 0.011 | 0.537 | 0.353 | 0.015 |
| Acute Toxicity Rule A. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Gen. Carcin. Rule A. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Non Gen. Carcin. Rule A. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Skin Sensit. Rule A. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Aquatic Tox. Rule A. | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Non Biodegr. Rule A. | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| SureChEMBL Rule | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| FAF-Drugs4 Rule | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Toxicophores | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Results | Ref. | |
|---|---|---|
| Oleanolic acid: IC50 = 6.4 µM (A-549) | Acetyloleanolic acid: IC50 = 5.8 µM (A-549) | [104] |
| Oleanolic acid: IC50 > 10 µM (KB) | Acetyloleanolic acid: IC50 = 7.6 µM (KB) | |
| Oleanolic acid: IC50 > 10 µM (KB-VIN) | Acetyloleanolic acid: IC50 = 7.6 µM (KB-VIN) | |
| Oleanolic acid: IC50 = 106.4 µM (B16-F10) | Acetyloleanolic acid: IC50 = 64.7 µM (B16-F10) | [105] |
| Oleanolic acid: IC50 = 429.9 µM (HT-29) | Acetyloleanolic acid: IC50 = 148.5 µM (HT-29) | |
| Oleanolic acid: IC50 = 211.8 µM (Hep-G2) | Acetyloleanolic acid: IC50 = 103.75 µM (Hep-G2) | |
| Oleanolic acid: IC50 = 41.7 µM (RAW 264.7) | Acetyloleanolic acid: IC50 = 13.8 µM (RAW 264.7) | [106] |
| Oleanolic acid: IC50 = 35.2 µM (J774A.1) | Acetyloleanolic acid: IC50 = 16.8 µM (J774A.1) | |
| Oleanolic acid: IC50 = 106.4 µM (B16-F10) | Acetyloleanolic acid: IC50 = 64.7 µM (B16-F10) | [107] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bednarczyk-Cwynar, B.; Ruszkowski, P.; Günther, A.; Sip, S.; Bednarek-Rajewska, K.; Zalewski, P. Enhancing the Pharmacological Properties of Triterpenes Through Acetylation: An Anticancer and Antioxidant Perspective. Molecules 2025, 30, 2661. https://doi.org/10.3390/molecules30122661
Bednarczyk-Cwynar B, Ruszkowski P, Günther A, Sip S, Bednarek-Rajewska K, Zalewski P. Enhancing the Pharmacological Properties of Triterpenes Through Acetylation: An Anticancer and Antioxidant Perspective. Molecules. 2025; 30(12):2661. https://doi.org/10.3390/molecules30122661
Chicago/Turabian StyleBednarczyk-Cwynar, Barbara, Piotr Ruszkowski, Andrzej Günther, Szymon Sip, Katarzyna Bednarek-Rajewska, and Przemysław Zalewski. 2025. "Enhancing the Pharmacological Properties of Triterpenes Through Acetylation: An Anticancer and Antioxidant Perspective" Molecules 30, no. 12: 2661. https://doi.org/10.3390/molecules30122661
APA StyleBednarczyk-Cwynar, B., Ruszkowski, P., Günther, A., Sip, S., Bednarek-Rajewska, K., & Zalewski, P. (2025). Enhancing the Pharmacological Properties of Triterpenes Through Acetylation: An Anticancer and Antioxidant Perspective. Molecules, 30(12), 2661. https://doi.org/10.3390/molecules30122661











