Integrative Genomics and Bioactivity-Guided Isolation of Novel Antimicrobial Compounds from Streptomyces sp. KN37 in Agricultural Applications
Abstract
1. Introduction
2. Results
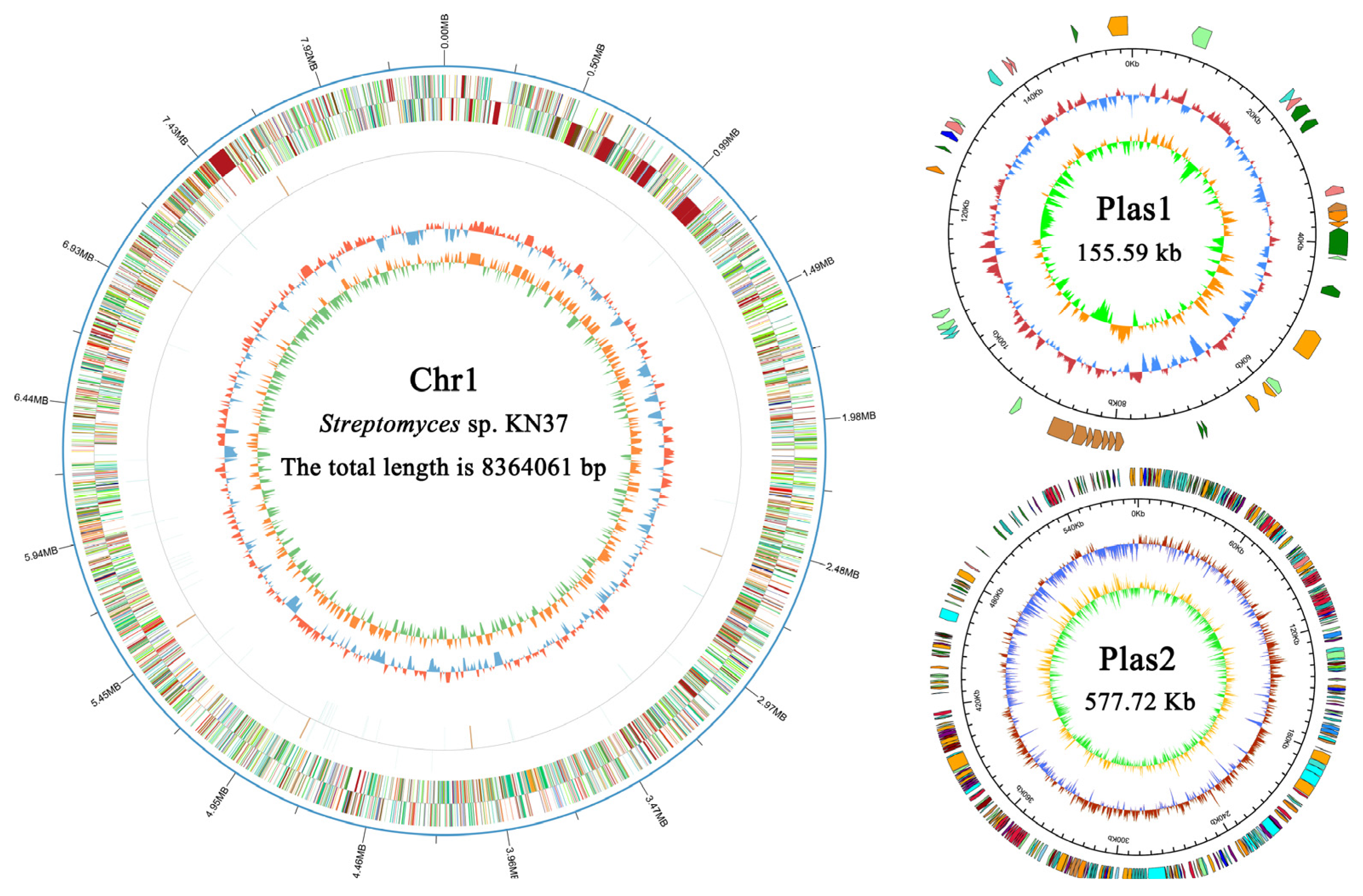
2.1. General Properties of the Streptomyces sp. KN37 Genome
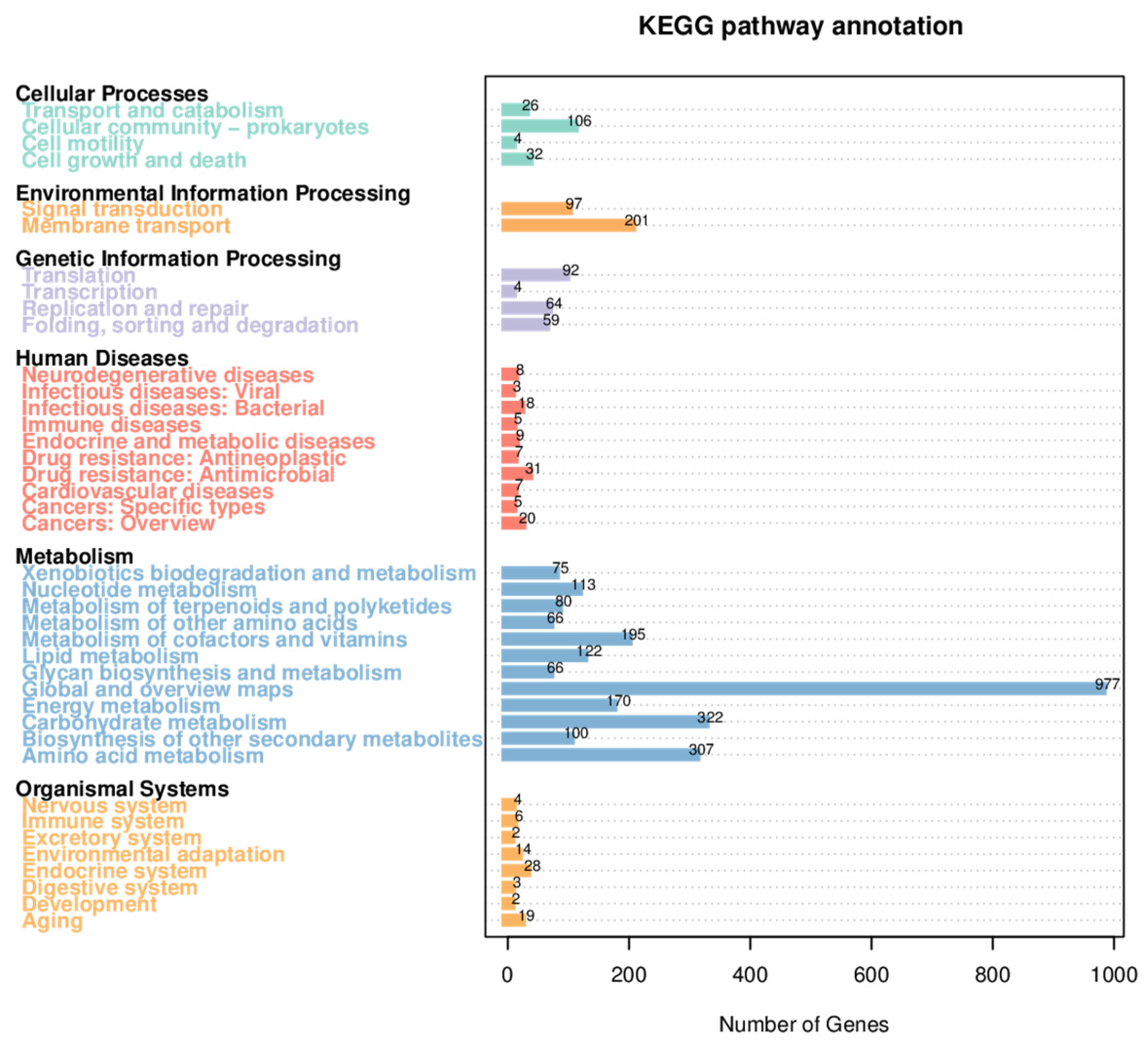
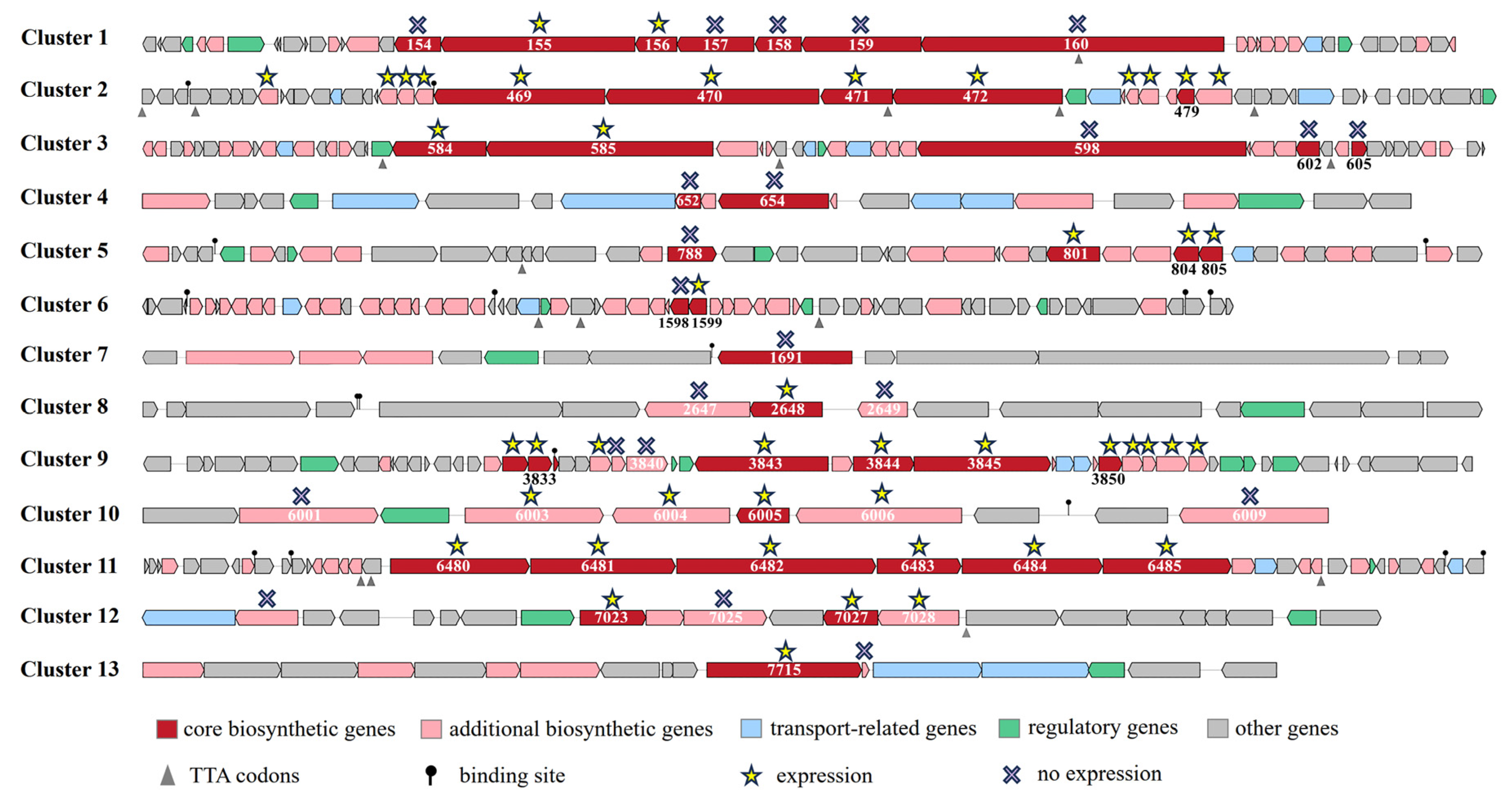
2.2. Analysis of Secondary Metabolism Gene Clusters
2.3. Isolation of Active Substances by Traditional Methods
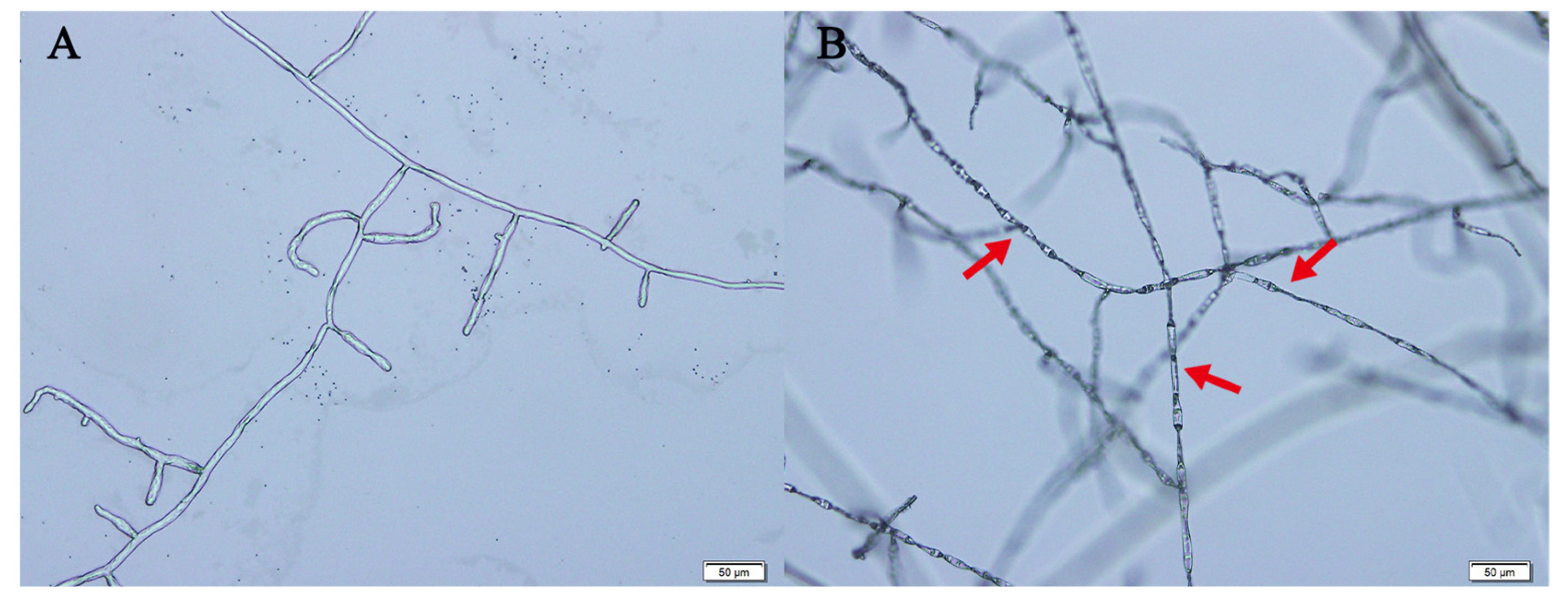
2.4. Preliminary Bioactivity Assays
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Cultures Conditions
4.2. Genome Analysis
4.3. Nucleotide Sequence Accession Number
4.4. Isolation and Characterization of Secondary Metabolites
4.5. Reverse Transcription PCR
4.6. In Vitro Antimicrobial Activity Test
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tudi, M.; Daniel Ruan, H.; Wang, L.; Lyu, J.; Sadler, R.; Connell, D.; Chu, C.; Phung, D.T. Agriculture development, pesticide application and its impact on the environment. Int. J. Environ. Res. Public Health 2021, 18, 1112. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Razek, A.S.; El-Naggar, M.E.; Allam, A.; Morsy, O.M.; Othman, S.I. Microbial natural products in drug discovery. Processes 2020, 8, 470. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.K.; Zhang, S.Y.; Qi, H.; Xiang, W.S.; Li, J.S.; Wang, J.D. A novel spiro-heterocycle milbemycin metabolite from a genetically engineered strain of Streptomyces bingchenggensis. Nat. Prod. Res. 2023, 37, 449–454. [Google Scholar] [CrossRef]
- Lan, Y.; Yan, Z.; Duan, T. Luobuma Leaf Spot Disease Caused by Alternaria tenuissima in China. J. Fungi 2023, 9, 1062. [Google Scholar] [CrossRef] [PubMed]
- Kamjam, M.; Sivalingam, P.; Deng, Z.; Hong, K. Deep sea actinomycetes and their secondary metabolites. Front. Microbiol. 2017, 8, 760. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wu, Z.; Zhang, Y.; Zhang, Z.; Fang, W.; Wang, Y.; Wan, Z.; Wang, K.; Ke, S. Herbicidal secondary metabolites from actinomycetes: Structure diversity, modes of action, and their roles in the development of herbicides. J. Agric. Food Chem. 2020, 68, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Kato, W.; Ikeda, H.; Katsuyama, Y.; Ohnishi, Y.; Imoto, M. Discovery of “heat shock metabolites” produced by thermotolerant actinomycetes in high-temperature culture. J. Antibiot. 2020, 73, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Nithya, K.; Muthukumar, C.; Biswas, B.; Alharbi, N.S.; Kadaikunnan, S.; Khaled, J.M.; Dhanasekaran, D. Desert actinobacteria as a source of bioactive compounds production with a special emphases on pyridine-2,5-diacetamide a new pyridine alkaloid produced by Streptomyces sp. Da3-7. Microbiol. Res. 2018, 207, 116–133. [Google Scholar] [CrossRef]
- Xie, F.; Pathom-aree, W. Actinobacteria from desert: Diversity and biotechnological applications. Front. Microbiol. 2021, 12, 765531. [Google Scholar] [CrossRef]
- Hu, D.; Sun, C.; Jin, T.; Fan, G.; Mok, K.M.; Li, K.; Lee, S.M. Exploring the potential of antibiotic production from rare actinobacteria by whole-genome sequencing and guided MS/MS analysis. Front. Microbiol. 2020, 11, 1540. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Wang, B.; Zhao, H. Breaking the silence: New strategies for discovering novel natural products. Curr. Opin. Biotechnol. 2017, 48, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.J.; Lai, H.; Li, J.; Freemont, P.S. Streptomyces cell-free systems for natural product discovery and engineering. Nat. Prod. Rep. 2023, 40, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Kang, H. Phylogeny-guided (meta)genome mining approach for the targeted discovery of new microbial natural products. J. Ind. Microbiol. Biotechnol. 2017, 44, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.N.; Zhang, A.Q.; Baerna, K.; Zhang, G.Q. The Control Effect of Actinomycete Strain KN37 against Tomato Gray Mold. J. Trop. Biol. 2019, 10, 258–263. [Google Scholar] [CrossRef]
- Selim, M.S.M.; Abdelhamid, S.A.; Mohamed, S.S. Secondary metabolites and biodiversity of actinomycetes. J. Genet. Eng. Biotechnol. 2021, 19, 72. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. Antismash 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Tietz, J.I.; Schwalen, C.J.; Patel, P.S.; Maxson, T.; Blair, P.M.; Tai, H.; Zakai, U.I.; Mitchell, D.A. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 2017, 13, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Katsuyama, Y.; Onaka, H.; Fujie, M.; Satoh, N.; Shin Ya, K.; Ohnishi, Y. Production of a novel amide-containing polyene by activating a cryptic biosynthetic gene cluster in Streptomyces sp. Msc090213je08. Chembiochem 2016, 17, 1464–1471. [Google Scholar] [CrossRef]
- Reshetnikov, A.S.; Khmelenina, V.N.; Mustakhimov, I.I.; Trotsenko, Y.A. Chapter two—Genes and enzymes of ectoine biosynthesis in halotolerant methanotrophs. Methods Enzymol. 2011, 495, 15–30. [Google Scholar] [CrossRef]
- Li, H.; Li, H.; Chen, S.; Wu, W.; Sun, P. Isolation and identification of pentalenolactone analogs from Streptomyces sp. NRRL S-4. Molecules 2021, 26, 7377. [Google Scholar] [CrossRef]
- Rhodes, A.; Fantes, K.H.; Boothroyd, B.; Mcgonagle, M.P.; Crosse, R. Venturicidin: A new antifungal antibiotic of potential use in agriculture. Nature 1961, 192, 952–954. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Lin, X.; Lei, L.; Lamb, D.C.; Kelly, S.L.; Waterman, M.R.; Cane, D.E. Biosynthesis of the sesquiterpene antibiotic albaflavenone in Streptomyces coelicolor A3(2). J. Biol. Chem. 2008, 283, 8183–8189. [Google Scholar] [CrossRef]
- Bear, I.J.; Thomas, R.G. Nature of argillaceous odour. Nature 1964, 201, 993–995. [Google Scholar] [CrossRef]
- Pham, V.T.T.; Nguyen, H.T.; Nguyen, C.T.; Choi, Y.S.; Dhakal, D.; Kim, T.; Jung, H.J.; Yamaguchi, T.; Sohng, J.K. Identification and enhancing production of a novel macrolide compound in engineered Streptomyces peucetius. RSC Adv. 2021, 11, 3168–3173. [Google Scholar] [CrossRef]
- Silva, S.P.M.; Teixeira, J.A.; Silva, C.C.G. Application of enterocin-whey films to reduce Listeria monocytogenes contamination on ripened cheese. Food Microbiol. 2023, 109, 104134. [Google Scholar] [CrossRef]
- Culp, E.J.; Waglechner, N.; Wang, W.; Fiebig-Comyn, A.A.; Hsu, Y.; Koteva, K.; Sychantha, D.; Coombes, B.K.; Van Nieuwenhze, M.S.; Brun, Y.V.; et al. Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature 2020, 578, 582–587. [Google Scholar] [CrossRef]
- Tyurin, A.P.; Alferova, V.A.; Paramonov, A.S.; Shuvalov, M.V.; Kudryakova, G.K.; Rogozhin, E.A.; Zherebker, A.Y.; Brylev, V.A.; Chistov, A.A.; Baranova, A.A.; et al. Gausemycins a,b: Cyclic lipoglycopeptides from streptomyces sp. Angew. Chem. Int. Ed. 2021, 60, 18694–18703. [Google Scholar] [CrossRef]
- Meindl, K.; Schmiederer, T.; Schneider, K.; Reicke, A.; Butz, D.; Keller, S.; Gühring, H.; Vértesy, L.; Wink, J.; Hoffmann, H.; et al. Labyrinthopeptins: A new class of carbacyclic lantibiotics. Angew. Chem. Int. Ed. 2010, 49, 1151–1154. [Google Scholar] [CrossRef]
- Yang, J.; Shi, R.; Zhou, P.; Qiu, Q.; Li, H. Asymmetric schiff bases derived from diaminomaleonitrile and their metal complexes. J. Mol. Struct. 2016, 1106, 242–258. [Google Scholar] [CrossRef]
- Korta, E.; Bakkali, A.; Berrueta, L.A.; Gallo, B.; Vicente, F.; Kilchenmann, V.; Bogdanov, S. Study of acaricide stability in honey. Characterization of amitraz degradation products in honey and beeswax. J. Agric. Food Chem. 2001, 49, 5835–5842. [Google Scholar] [CrossRef] [PubMed]
- Skrzypczak-Jankun, E.; Borbulevych, O.Y.; Jankun, J. Soybean lipoxygenase-3 in complex with 4-nitrocatechol. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 613–615. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tan, Y.; Zhou, S.; Liu, S.; Wang, W.; Jiang, Y.; Long, H.; Liu, J. Neuroprotective methylsuccinic acid and enoic acid derivatives from the fungus Xylaria longipes. Phytochemistry 2023, 210, 113652. [Google Scholar] [CrossRef] [PubMed]
- Dieuleveux, V.; Lemarinier, S.; Guéguen, M. Antimicrobial spectrum and target site of d-3-phenyllactic acid. Int. J. Food Microbiol. 1998, 40, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Paulus, C.; Rebets, Y.; Tokovenko, B.; Nadmid, S.; Terekhova, L.P.; Myronovskyi, M.; Zotchev, S.B.; Rückert, C.; Braig, S.; Zahler, S.; et al. New natural products identified by combined genomics-metabolomics profiling of marine Streptomyces sp. MP131-18. Sci Rep. 2017, 7, 42382. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Chen, L.; Wu, Q.; Jiang, M.; Guo, H.; Hu, Z.; Chen, S.; Liu, L.; Gao, Z. Genome mining of α-pyrone natural products from ascidian-derived fungus Amphichordafelina SYSU-MS7908. Mar. Drugs 2022, 20, 294. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; He, J.; Wei, X.; Ju, J.; Ma, J. Exploration and genome mining of natural products from marine Streptomyces. Appl. Microbiol. Biotechnol. 2020, 104, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Chatzigeorgiou, S.; Thai, Q.D.; Tchoumtchoua, J.; Tallas, K.; Tsakiri, E.N.; Papassideri, I.; Halabalaki, M.; Skaltsounis, A.; Trougakos, I.P. Isolation of natural products with anti-ageing activity from the fruits of Platanus orientalis. Phytomedicine 2017, 33, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Liu, G.; Liu, X.; Zhang, L.; Li, L.; Liu, L. Pycnidiophorones A–D, four new cytochalasans from the wetland derived fungus Pycnidiophora dispersa. RSC Adv. 2020, 10, 40384–40390. [Google Scholar] [CrossRef]
- Gyeltshen, T.; Jordan, G.J.; Smith, J.A.; Bissember, A.C. Natural products isolation studies of the paleoendemic plant species Nothofagus gunnii and Nothofagus cunninghamii. Fitoterapia 2022, 156, 105088. [Google Scholar] [CrossRef]
- Vásquez, R.; Rios, N.; Solano, G.; Cubilla-Rios, L. Lentinoids A–D, new natural products isolated from Lentinus strigellus. Molecules 2018, 23, 773. [Google Scholar] [CrossRef]
- Molina, L.; Williams, D.E.; Andersen, R.J.; Golsteyn, R.M. Isolation of a natural product with anti-mitotic activity from a toxic canadian prairie plant. Heliyon 2021, 7, e7131. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Tang, L.; Xie, Y.; Xie, L. Secondary metabolites from Hericium erinaceus and their anti-inflammatory activities. Molecules 2022, 27, 2157. [Google Scholar] [CrossRef] [PubMed]
- Kianfé, B.Y.; Kühlborn, J.; Tchuenguem, R.T.; Tchegnitegni, B.T.; Ponou, B.K.; Groß, J.; Teponno, R.B.; Dzoyem, J.P.; Opatz, T.; Tapondjou, L.A. Antimicrobial secondary metabolites from the medicinal plant Crinum glaucum A. Chev. (Amaryllidaceae). S. Afr. J. Bot. 2020, 133, 161–166. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, J.; Zhang, P.; Xie, S.; Yuan, X.; Hou, X.; Yan, N.; Fang, Y.; Du, Y. In vitro and in vivo antifungal activity and preliminary mechanism of cembratrien-diols against botrytis cinerea. Ind. Crop. Prod. 2020, 154, 112745. [Google Scholar] [CrossRef]
- Xiang, Y.P.; Lu, J.Y.; Li, F.F.; Huang, J.; Yang, C.F.; Fu, Z.B.; Gao, L.H. Bactericidal effect of ozonated camellia oil on Staphylococcus aureus in vitro. J. Centr. South Univ Med. Sci. 2018, 43, 139–142. [Google Scholar]
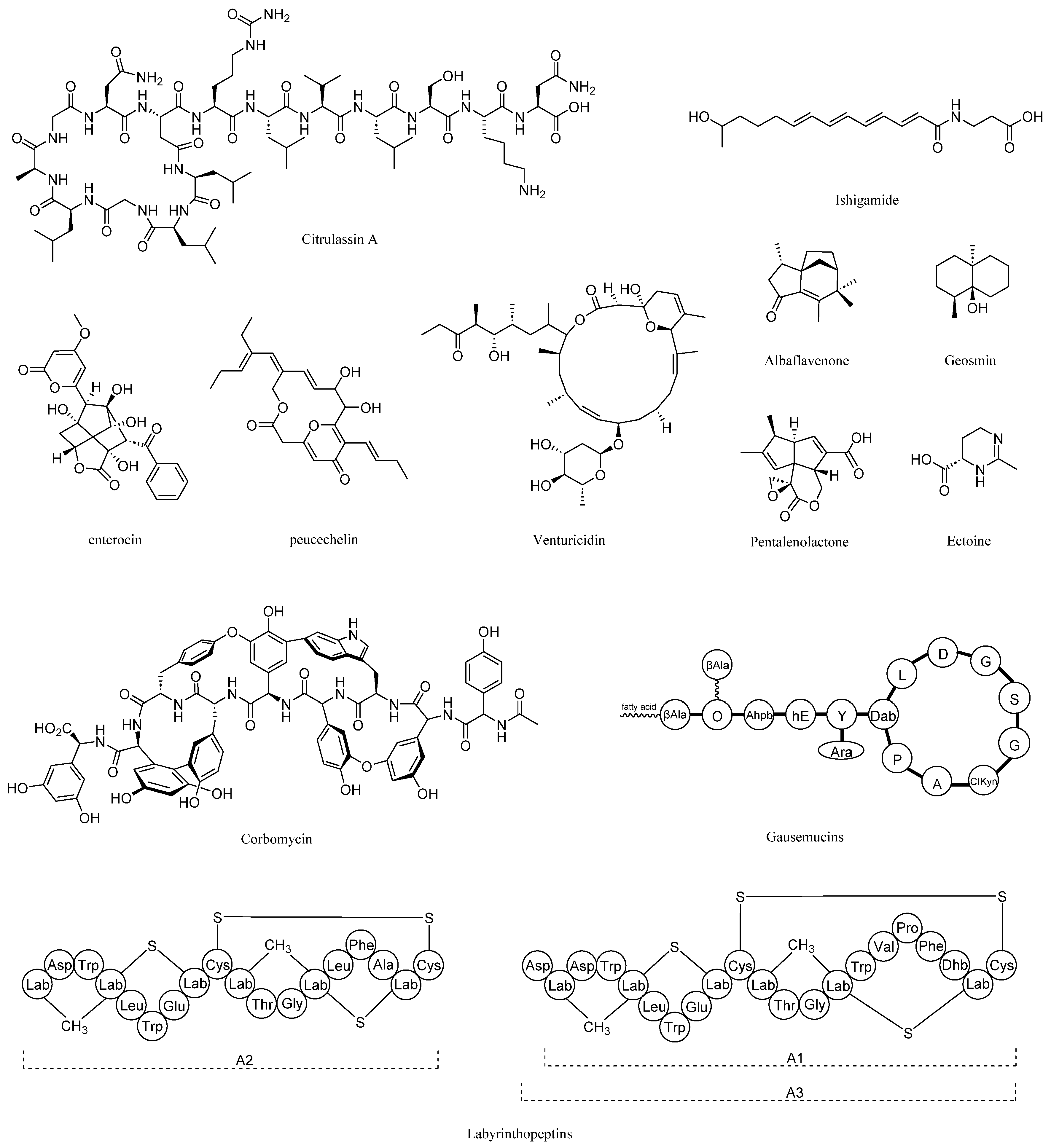

| Gene Cluster | Type | Size (kb) | Predicted Product | Most Similar Known Cluster | Similarity (%) | Reference Strain | Accession Number |
|---|---|---|---|---|---|---|---|
| 1 | NRPS-like transAT-PKS NRPS | 103.717 | NRP + Polyketide | oxalomycin B | 75 | Streptomyces albus | BGC0001106 |
| 2 | NRPS T3PKS | 88.789 | NRP | corbomycin | 96 | Streptomyces sp. WAC 01529 | BGC0002314 |
| 3 | NRPS betalactone | 104.296 | NRP + Saccharide | gausemycin A, B | 69 | Streptomyces kanamyceticus | BGC0002430 |
| 4 | lassopeptide | 22.591 | Ripp | citrulassin E | 100 | Streptomyces glaucescens | BGC0001551 |
| 5 | NRPS-like terpene | 50.493 | Terpene | hopene | 84 | Streptomyces coelicolor A3(2) | BGC0000663 |
| 6 | T2PKS | 72.416 | Polyketide: Type II polyketide | enterocin | 90 | Streptomyces maritimus | BGC0000220 |
| 7 | terpene | 21.082 | Terpene | geosmin | 100 | Streptomyces coelicolor A3(2) | BGC0001181 |
| 8 | terpene | 17.556 | terpene | albaflavenone | 100 | Streptomyces coelicolor A3(2) | BGC0000660 |
| 9 | thioamide-NRP NRPS ladderane | 73.74 | NRP + Polyketide | ishigamide | 100 | Streptomyces sp. MSC090213JE08 | BGC0001623 |
| 10 | ectoine | 10.404 | Other: Ectoine | ectoine | 100 | Streptomyces sp. | BGC0002052 |
| 11 | T1PKS | 105.514 | Polyketide | venturicidin D, E, F, A | 76 | Streptomyces sp. NRRL S-4 | BGC0002454 |
| 12 | terpene | 25.638 | Terpene | isorenieratene | 85 | Streptomyces griseus subsp. griseus NBRC 13350 | BGC0000664 |
| 13 | lanthipeptide-class-iii | 22.582 | RiPP: Lanthipeptide | labyrinthopeptin A1, A2, A3 | 60 | Actinomadura namibiensis | BGC0000519 |
| Compound | Toxicity Curve | R2 | EC50 (mg/L) | 95% Confidence Interval (mg/L) |
|---|---|---|---|---|
| 4-(Diethylamino)salicylaldehyde | y = 3.560x − 4.133 | 0.967 | 14.487 | 12.014–20.694 |
| 4-Nitrosodiphenylamine | y = 1.499x − 2.414 | 0.903 | 40.785 | 33.702–50.768 |
| N-(2,4-Dimethylphenyl)formamide | y = 2.098x − 4.839 | 0.961 | 202.584 | 173.304–230.679 |
| 4-Nitrocatechol | y = 2.579x − 5.865 | 0.921 | 187.966 | 155.768–214.429 |
| Methylsuccinic acid | y = 0.948x − 3.401 | 0.949 | 3868.586 | 2312.070–9543.943 |
| Phenyllactic acid | y = 1.590x − 4.775 | 0.902 | 1009.024 | 833.982–1266.238 |
| 5,6-Dimethylbenzimidazole | y = 2.081x − 5.070 | 0.989 | 272.795 | 239.417–311.149 |
| Compound | Toxicity Curve | R2 | EC50 (mg/L) | 95% Confidence Interval (mg/L) |
|---|---|---|---|---|
| 4-(Diethylamino)salicylaldehyde | y = 1.953x − 3.836 | 0.994 | 92.083 | 83.831–101.509 |
| 4-Nitrosodiphenylamine | y = 3.466x − 2.624 | 0.980 | 5.715 | 5.433–6.015 |
| N-(2,4-Dimethylphenyl)formamide | y = 2.281x − 4.830 | 0.941 | 131.123 | 121.605–141.629 |
| 4-Nitrocatechol | y = 6.033x − 7.833 | 0.956 | 19.871 | 19.092–20.627 |
| Methylsuccinic acid | y = 2.676x − 5.647 | 0.948 | 128.852 | 119.496–138.284 |
| Phenyllactic acid | y = 3.415x − 8.977 | 0.899 | 424.891 | 400.864–450.311 |
| 5,6-Dimethylbenzimidazole | y = 4.768x − 8.960 | 0.968 | 75.716 | 72.100–79.502 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Li, Q.; Zeeshan, M.; Zhang, G.; Wang, C.; Han, X.; Yang, D. Integrative Genomics and Bioactivity-Guided Isolation of Novel Antimicrobial Compounds from Streptomyces sp. KN37 in Agricultural Applications. Molecules 2024, 29, 2040. https://doi.org/10.3390/molecules29092040
Zhao J, Li Q, Zeeshan M, Zhang G, Wang C, Han X, Yang D. Integrative Genomics and Bioactivity-Guided Isolation of Novel Antimicrobial Compounds from Streptomyces sp. KN37 in Agricultural Applications. Molecules. 2024; 29(9):2040. https://doi.org/10.3390/molecules29092040
Chicago/Turabian StyleZhao, Jing, Qinghua Li, Muhammad Zeeshan, Guoqiang Zhang, Chunjuan Wang, Xiaoqiang Han, and Desong Yang. 2024. "Integrative Genomics and Bioactivity-Guided Isolation of Novel Antimicrobial Compounds from Streptomyces sp. KN37 in Agricultural Applications" Molecules 29, no. 9: 2040. https://doi.org/10.3390/molecules29092040
APA StyleZhao, J., Li, Q., Zeeshan, M., Zhang, G., Wang, C., Han, X., & Yang, D. (2024). Integrative Genomics and Bioactivity-Guided Isolation of Novel Antimicrobial Compounds from Streptomyces sp. KN37 in Agricultural Applications. Molecules, 29(9), 2040. https://doi.org/10.3390/molecules29092040






