Electropositive Citric Acid-Polyethyleneimine Carbon Dots Carrying the PINK1 Gene Regulate ATP-Related Metabolic Dysfunction in APP/PS1-N2a Cells
Abstract
1. Introduction
2. Results
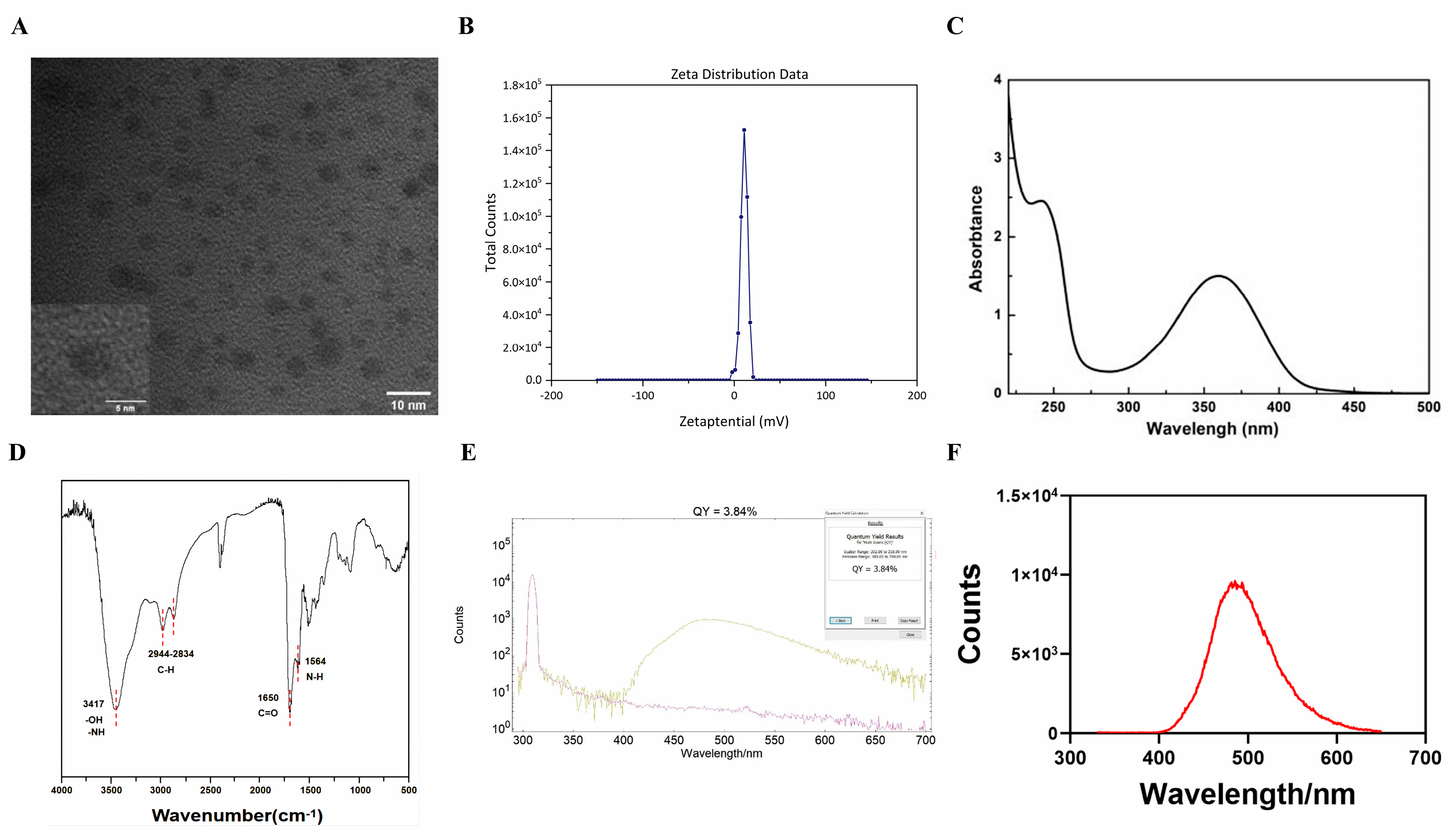
2.1. Characterization of the CA-PEI CDs
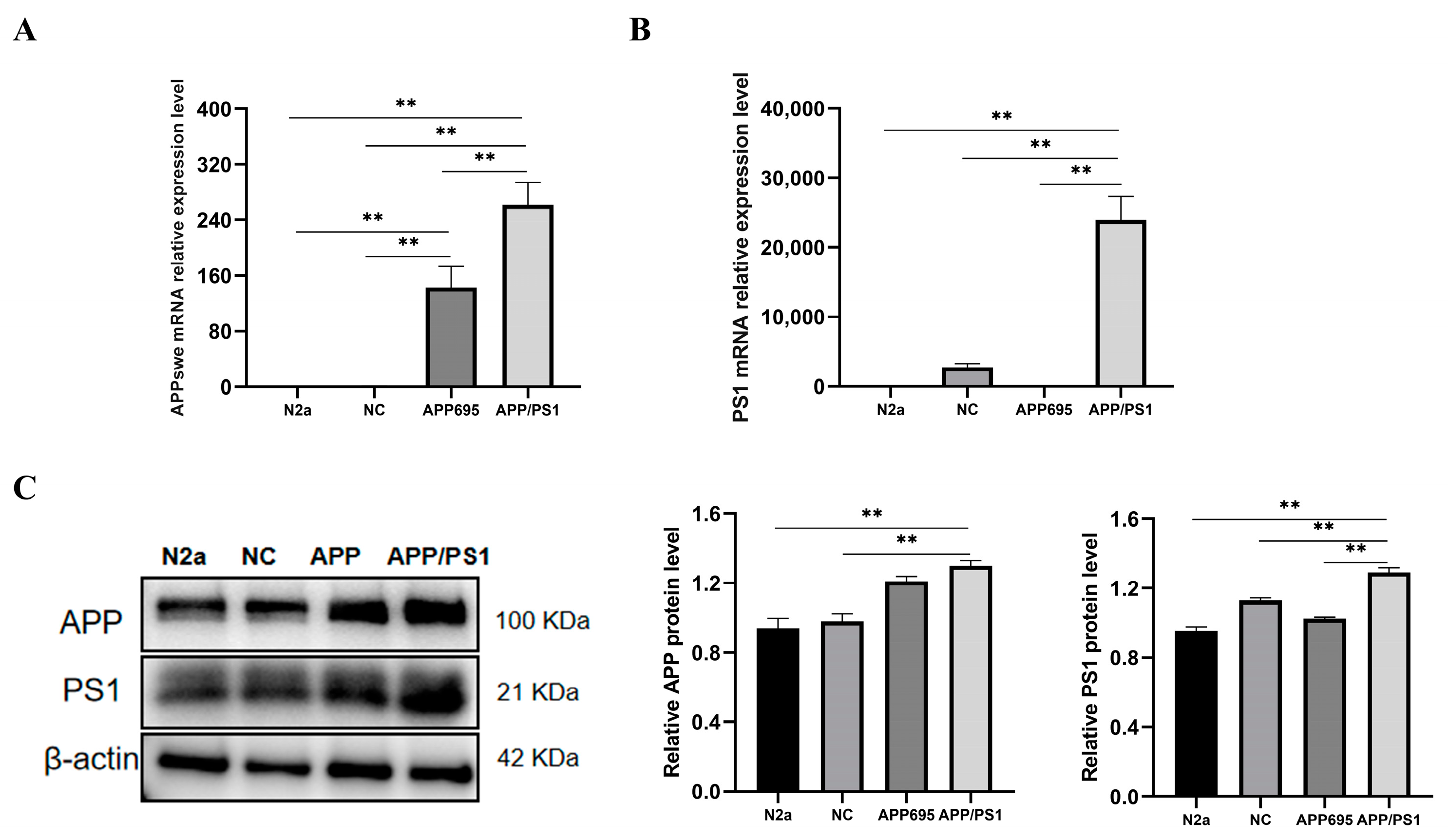
2.2. Establishment of the APP/PS1-N2a Cell Line
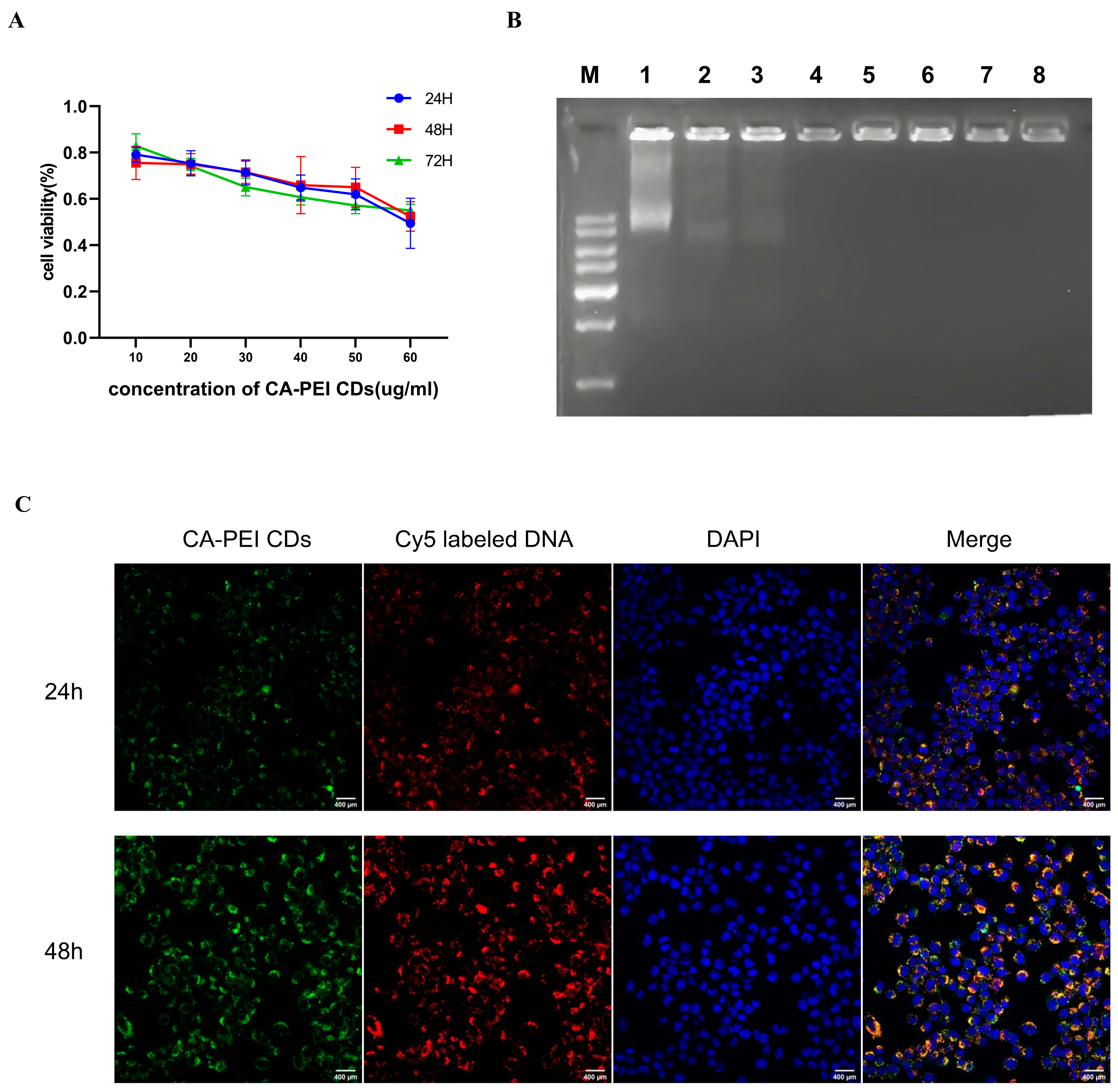
2.3. Cytotoxic Effect Assessment of the CA-PEI CDs
2.4. CA-PEI CD/pDNA Complex Formation
2.5. Cell Imaging Study
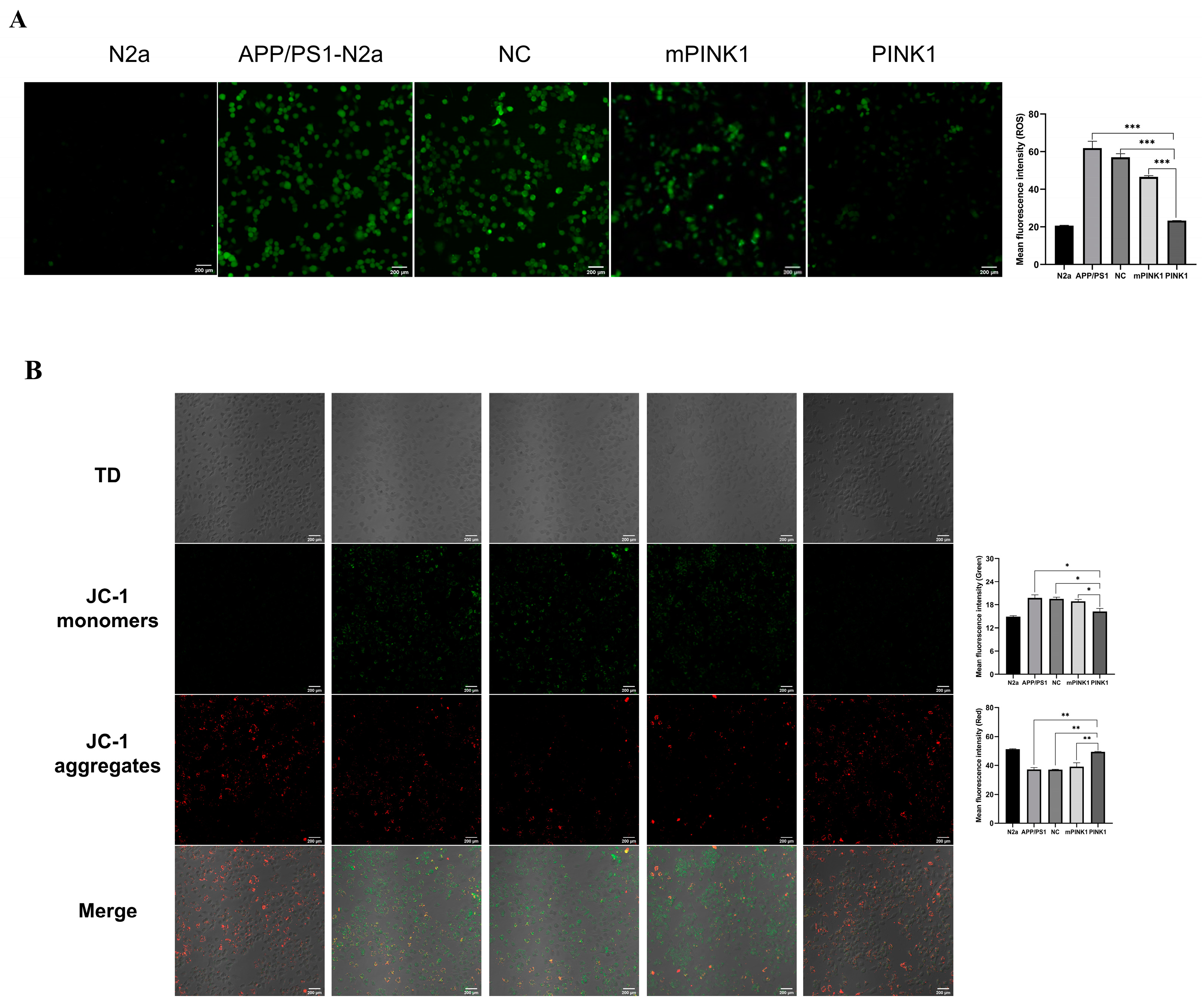
2.6. PINK1 Reduced ROS Levels in APP/PS1-N2a Cells
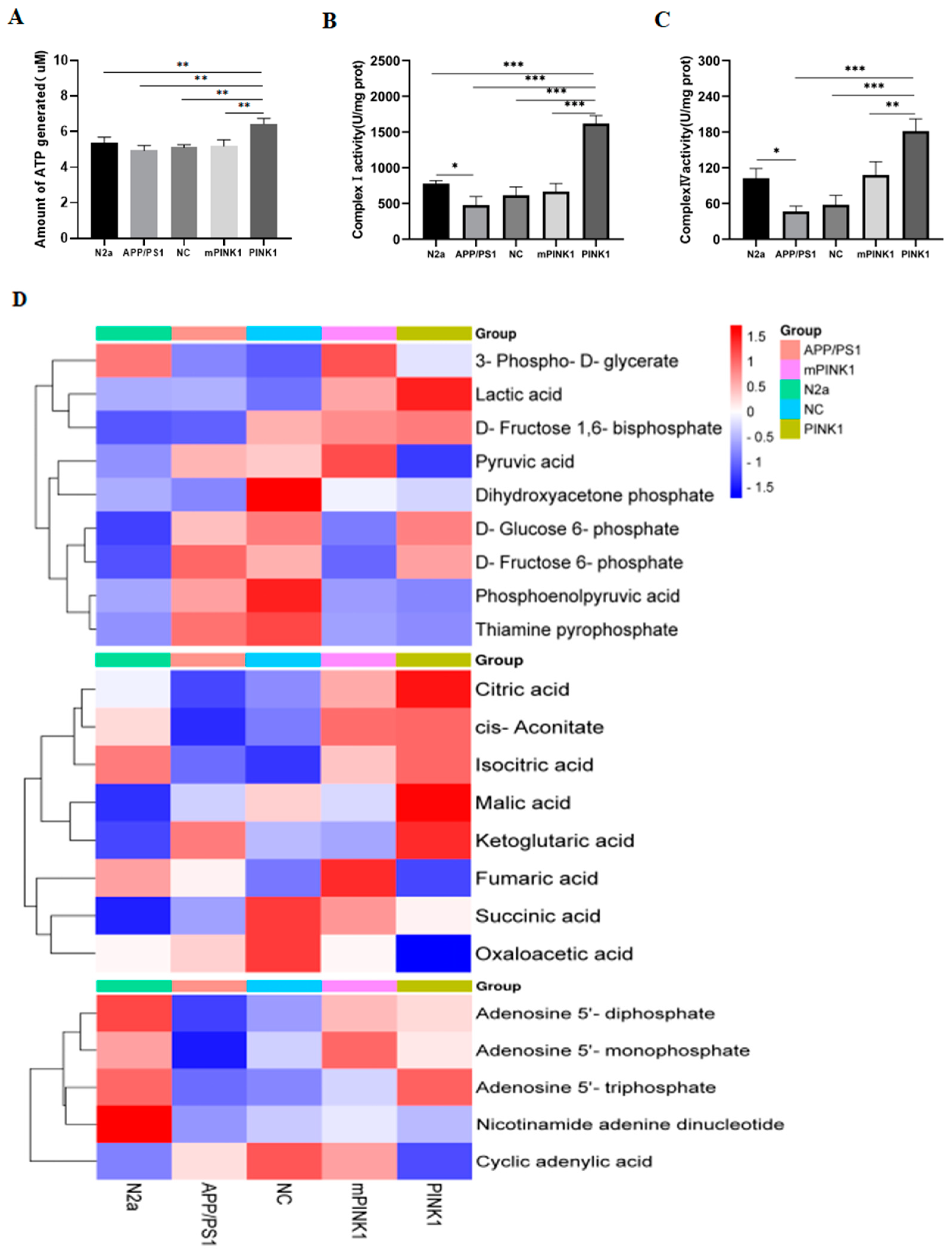
2.7. PINK1 Improves Mitochondrial Function by Increasing the MMP and Intracellular ATP Levels
2.8. PINK1 Induces ATP-Related Metabolic Changes in APP/PS1-N2a Cells
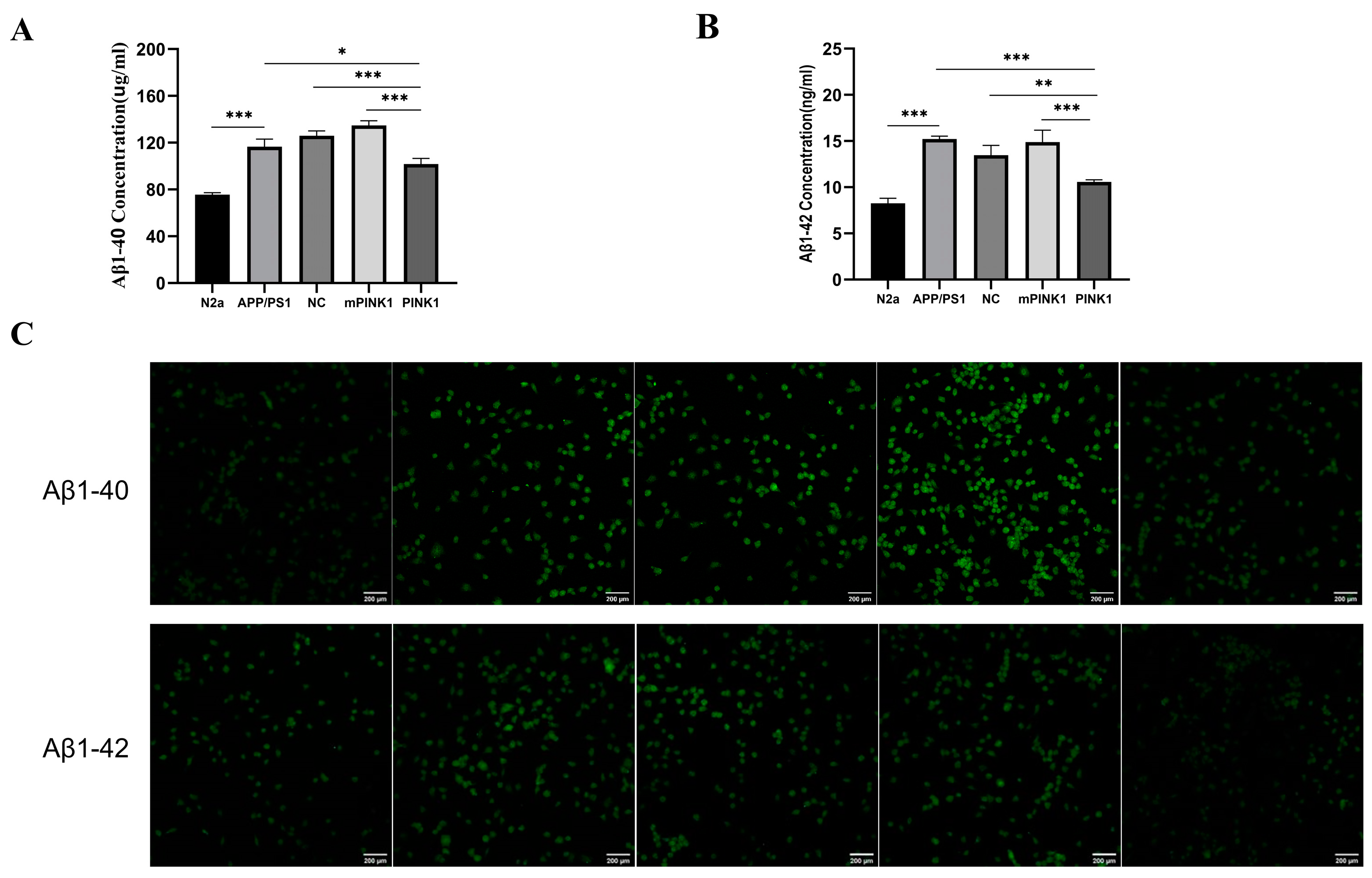
2.9. PINK1 Decreased Aβ1-40 and Aβ1-42 Levels in APP/PS1-N2a Cells
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation of CA–PEI CDs
4.3. Instrumentation and Characterization
4.4. Measurement of Fluorescence QY
4.5. Agarose Gel Retardation Assay
4.6. Cell Culture
4.7. PS1 Gene-Transfected N2a/APP695 Cells
4.8. mRNA Quantification Analysis (qRT-PCR)
4.9. Western Blot Analysis
4.10. Cell Viability Assay
4.11. In Vitro Fluorescence Imaging
4.12. Intracellular ROS Measurement
4.13. MMP
4.14. ATP Assay
4.15. Determination of Mitochondrial CI-IV Activity
4.16. LC-MS/MS Analysis
4.17. Immunofluorescence Assay and Confocal Microscopy
4.18. Quantification of Aβ1-40 and Aβ1-42 by ELISA
4.19. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, B.L.; Li, W.W.; Zhu, C.; Jin, W.S.; Zeng, F.; Liu, Y.H.; Bu, X.L.; Zhu, J.; Yao, X.Q.; Wang, Y.J. Clinical research on Alzheimer’s disease: Progress and perspectives. Neurosci. Bull. 2018, 34, 1111–1118. [Google Scholar] [CrossRef]
- Li, Y.; Xia, X.; Wang, Y.; Zheng, J.C. Mitochondrial dysfunction in microglia: A novel perspective for pathogenesis of Alzheimer’s disease. J. Neuroinflammation 2022, 19, 248. [Google Scholar] [CrossRef]
- Nunnari, J.; Suomalainen, A. Mitochondria: In sickness and in health. Cell 2012, 148, 1145–1159. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Huo, S.; Mizuhara, T.; Das, R.; Lee, Y.W.; Hou, S.; Moyano, D.F.; Duncan, B.; Liang, X.J.; Rotello, V.M. The interplay of size and surface functionality on the cellular uptake of sub-10 nm gold nanoparticles. ACS Nano 2015, 9, 9986–9993. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S. Aducanumab: First approval. Drugs 2021, 81, 1437–1443. [Google Scholar] [CrossRef]
- Hoy, S.M. Lecanemab: First approval. Drugs 2023, 83, 359–365. [Google Scholar] [CrossRef]
- Shi, M.; Chu, F.; Zhu, F.; Zhu, J. Impact of anti-amyloid-beta monoclonal antibodies on the pathology and clinical profile of alz-heimer’s disease: A focus on aducanumab and lecanemab. Front. Aging Neurosci. 2022, 14, 870517. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.T.; Beal, M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006, 443, 787–795. [Google Scholar] [CrossRef]
- Shoshan-Barmatz, V.; Nahon-Crystal, E.; Shteinfer-Kuzmine, A.; Gupta, R. VDAC1, mitochondrial dysfunction, and Alzheimer’s disease. Pharmacol. Res. 2018, 131, 87–101. [Google Scholar] [CrossRef]
- Dagda, R.K.; Pien, I.; Wang, R.; Zhu, J.; Wang, K.Z.; Callio, J.; Banerjee, T.D.; Dagda, R.Y.; Chu, C.T. Beyond the mitochondrion: Cytosolic PINK1 remodels dendrites through protein kinase A. J. Neurochem. 2014, 128, 864–877. [Google Scholar] [CrossRef]
- Jiang, X.; Jin, T.; Zhang, H.; Miao, J.; Zhao, X.; Su, Y.; Zhang, Y. Current progress of mitochondrial quality control pathways underlying the pathogenesis of parkinson’s disease. Oxidative Med. Cell. Longev. 2019, 2019, 4578462. [Google Scholar] [CrossRef]
- Han, Y.; Liu, D.; Cheng, Y.; Ji, Q.; Liu, M.; Zhang, B.; Zhou, S. Maintenance of mitochondrial homeostasis for Alzheimer’s disease: Strategies and challenges. Redox Biol. 2023, 63, 102734. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Yu, Q.; Yan, S.; Hu, G.; Lue, L.F.; Walker, D.G.; Wu, L.; Yan, S.F.; Tieu, K.; Yan, S.S. PINK1 signalling rescues amyloid pathology and mitochondrial dysfunction in Alzheimer’s disease. Brain 2017, 140, 3233–3251. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xue, Y.; Yao, Y.; Li, Y.; Ji, X.; Chi, T.; Liu, P.; Zou, L. PINK1 regulates mitochondrial fission/fusion and neuroinflammation in β-amyloid-induced Alzheimer’s disease models. Neurochem. Int. 2022, 154, 105298. [Google Scholar] [CrossRef]
- Patro, S.; Ratna, S.; Yamamoto, H.A.; Ebenezer, A.T.; Ferguson, D.S.; Kaur, A.; McIntyre, B.C.; Snow, R.; Solesio, M.E. Atp synthase and mitochondrial bioenergetics dysfunction in Alzheimer’s disease. Int. J. Mol. Sci. 2021, 22, 11185. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Halliwell, B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat. Rev. Neurosci. 2019, 20, 148–160. [Google Scholar] [CrossRef]
- Ardanaz, C.G.; Ramírez, M.J.; Solas, M. Brain metabolic alterations in Alzheimer’s disease. Int. J. Mol. Sci. 2022, 23, 3785. [Google Scholar] [CrossRef]
- Cunnane, S.C.; Trushina, E.; Morland, C.; Prigione, A.; Casadesus, G.; Andrews, Z.B.; Beal, M.F.; Bergersen, L.H.; Brinton, R.D.; de la Monte, S.; et al. Brain energy rescue: An emerging therapeutic concept for neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 2020, 19, 609–633. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Sigdel, G.; Mintz, K.J.; Seven, E.S.; Zhou, Y.; Wang, C.; Leblanc, R.M. Carbon dots: A future blood-brain barrier penetrating nanomedicine and drug nanocarrier. Int. J. Nanomed. 2021, 16, 5003–5016. [Google Scholar] [CrossRef]
- Lamptey, R.N.L.; Chaulagain, B.; Trivedi, R.; Gothwal, A.; Layek, B.; Singh, J. A review of the common neurodegenerative disorders: Current therapeutic approaches and the potential role of nanotherapeutics. Int. J. Mol. Sci. 2022, 23, 1851. [Google Scholar] [CrossRef]
- Seven, E.S.; Seven, Y.B.; Zhou, Y.; Poudel-Sharma, S.; Diaz-Rucco, J.J.; Kirbas Cilingir, E.; Mitchell, G.S.; Van Dyken, J.D.; Leblanc, R.M. Crossing the blood-brain barrier with carbon dots: Uptake mechanism and in vivo cargo delivery. Nanoscale Adv. 2021, 3, 3942–3953. [Google Scholar] [CrossRef]
- Zhang, W.; Kandel, N.; Zhou, Y.; Smith, N.; Ferreira, B.C.; Perez, M.; Claure, M.L.; Mintz, K.J.; Wang, C.; Leblanc, R.M. Drug delivery of memantine with carbon dots for Alzheimer’s disease: Blood-brain barrier penetration and inhibition of tau aggregation. J. Colloid Interface Sci. 2022, 617, 20–31. [Google Scholar] [CrossRef]
- Wang, N.; Zhu, P.; Huang, R.; Wang, C.; Sun, L.; Lan, B.; He, Y.; Zhao, H.; Gao, Y. PINK1: The guard of mitochondria. Life Sci. 2020, 259, 118247. [Google Scholar] [CrossRef]
- Boakye-Yiadom, K.O.; Kesse, S.; Opoku-Damoah, Y.; Filli, M.S.; Aquib, M.; Joelle, M.M.B.; Farooq, M.A.; Mavlyanova, R.; Raza, F.; Bavi, R.; et al. Carbon dots: Applications in bioimaging and theranostics. Int. J. Pharm. 2019, 564, 308–317. [Google Scholar] [CrossRef]
- Song, T.; Song, X.; Zhu, C.; Patrick, R.; Skurla, M.; Santangelo, I.; Green, M.; Harper, D.; Ren, B.; Forester, B.P.; et al. Mitochondrial dysfunction, oxidative stress, neuroinflammation, and metabolic alterations in the progression of Alzheimer’s disease: A meta-analysis of in vivo magnetic resonance spectroscopy studies. Ageing Res. Rev. 2021, 72, 101503. [Google Scholar] [CrossRef] [PubMed]
- Breijyeh, Z.; Karaman, R. Comprehensive review on Alzheimer’s disease: Causes and treatment. Molecules 2020, 25, 5789. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.; Long, H.; Zhou, T.; Liu, Y.; Zhao, J.; Han, J.; Yang, X.; Yu, Y.; Chen, F.; Shi, S. Blood-brain barrier Permeable nanoparticles for Alzheimer’s disease treatment by selective mitophagy of microglia. Biomaterials 2022, 288, 121690. [Google Scholar] [CrossRef]
- Sun, J.; Roy, S. Gene-based therapies for neurodegenerative diseases. Nat. Neurosci. 2021, 24, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Butt, M.H.; Zaman, M.; Ahmad, A.; Khan, R.; Mallhi, T.H.; Hasan, M.M.; Khan, Y.H.; Hafeez, S.; Massoud, E.E.S.; Rahman, M.H.; et al. Appraisal for the potential of viral and nonviral vectors in gene therapy: A review. Genes 2022, 13, 1370. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, J.; Gu, J.; Bartoli, M.; Domena, J.B.; Zhou, Y.; Ferreira, B.C.; Kirbas Cilingir, E.; McGee, C.M.; Sampson, R.; et al. Nano-carrier for gene delivery and bioimaging based on pentaetheylenehexamine modified carbon dots. J. Colloid Interface Sci. 2023, 639, 180–192. [Google Scholar] [CrossRef]
- Brase, L.; You, S.F.; D’Oliveira Albanus, R.; Del-Aguila, J.L.; Dai, Y.; Novotny, B.C.; Soriano-Tarraga, C.; Dykstra, T.; Fernandez, M.V.; Budde, J.P.; et al. Single-nucleus RNA-sequencing of autosomal dominant Alzheimer disease and risk variant carriers. Nat. Commun. 2023, 14, 2314. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Bezprozvanny, I. Conformational models of APP processing by gamma secretase based on analysis of pathogenic mutations. Int. J. Mol. Sci. 2021, 22, 13600. [Google Scholar] [CrossRef]
- Kwart, D.; Gregg, A.; Scheckel, C.; Murphy, E.A.; Paquet, D.; Duffield, M.; Fak, J.; Olsen, O.; Darnell, R.B.; Tessier-Lavigne, M. A large panel of isogenic APP and PSEN1 mutant human iPSC neurons reveals shared endosomal abnormalities mediated by APP β-CTFs, not Aβ. Neuron 2019, 104, 256–270. [Google Scholar] [CrossRef]
- Guo, F.; Li, Q.; Zhang, X.; Liu, Y.; Jiang, J.; Cheng, S.; Yu, S.; Zhang, X.; Liu, F.; Li, Y.; et al. Applications of carbon dots for the treatment of Alzheimer’s disease. Int. J. Nanomed. 2022, 17, 6621–6638. [Google Scholar] [CrossRef] [PubMed]
- Massaro, M.; Barone, G.; Biddeci, G.; Cavallaro, G.; Di Blasi, F.; Lazzara, G.; Nicotra, G.; Spinella, C.; Spinelli, G.; Riela, S. Halloysite nanotubes-carbon dots hybrids multifunctional nanocarrier with positive cell target ability as a potential non-viral vector for oral gene therapy. J. Colloid Interface Sci. 2019, 552, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Mohammadinejad, R.; Dadashzadeh, A.; Moghassemi, S.; Ashrafizadeh, M.; Dehshahri, A.; Pardakhty, A.; Sassan, H.; Sohrevardi, S.M.; Mandegary, A. Shedding light on gene therapy: Carbon dots for the minimally invasive image-guided delivery of plasmids and noncoding RNAs—A review. J. Adv. Res. 2019, 18, 81–93. [Google Scholar] [CrossRef]
- Lee, S.Y.; An, H.J.; Kim, J.M.; Sung, M.J.; Kim, D.K.; Kim, H.K.; Oh, J.; Jeong, H.Y.; Lee, Y.H.; Yang, T.; et al. PINK1 deficiency impairs osteoblast differentiation through aberrant mitochondrial homeostasis. Stem Cell Res. Ther. 2021, 12, 589. [Google Scholar] [CrossRef]
- Soman, S.K.; Dagda, R.K. Role of cleaved PINK1 in neuronal development, synaptogenesis, and plasticity: Implications for Parkinson’s disease. Front. Neurosci. 2021, 15, 769331. [Google Scholar] [CrossRef]
- Li, J.; Yang, D.; Li, Z.; Zhao, M.; Wang, D.; Sun, Z.; Wen, P.; Dai, Y.; Gou, F.; Ji, Y.; et al. PINK1/Parkin-mediated mitophagy in neurodegenerative diseases. Ageing Res. Rev. 2023, 84, 101817. [Google Scholar] [CrossRef]
- Wang, X.J.; Qi, L.; Cheng, Y.F.; Ji, X.F.; Chi, T.Y.; Liu, P.; Zou, L.B. PINK1 overexpression prevents forskolin-induced tau hyperphosphorylation and oxidative stress in a rat model of Alzheimer’s disease. Acta Pharmacol. Sin. 2022, 43, 1916–1927. [Google Scholar] [CrossRef]
- Lin, Q.; Li, S.; Jiang, N.; Shao, X.; Zhang, M.; Jin, H.; Zhang, Z.; Shen, J.; Zhou, Y.; Zhou, W.; et al. PINK1-parkin pathway of mitophagy protects against contrast-induced acute kidney injury via decreasing mitochondrial ROS and NLRP3 inflammasome activation. Redox Biol. 2019, 26, 101254. [Google Scholar] [CrossRef] [PubMed]
- Fang, E.F.; Hou, Y.; Palikaras, K.; Adriaanse, B.A.; Kerr, J.S.; Yang, B.; Lautrup, S.; Hasan-Olive, M.M.; Caponio, D.; Dan, X.; et al. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci. 2019, 22, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Jin, J.; Xu, Y.; Zhu, X. Aberrant energy metabolism in Alzheimer’s disease. J. Transl. Intern. Med. 2022, 10, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhao, F.; Ma, X.; Perry, G.; Zhu, X. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: Recent advances. Mol. Neurodegener. 2020, 15, 30. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.L. Glucose, glycolysis, and neurodegenerative diseases. J. Cell. Physiol. 2020, 235, 7653–7662. [Google Scholar] [CrossRef] [PubMed]
- Nolfi-Donegan, D.; Braganza, A.; Shiva, S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol. 2020, 37, 101674. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Zhang, M.; Jeong, Y.Y.; Margolis, D.J.; Cai, Q. The role of mitophagy in the regulation of mitochondrial energetic status in neurons. Autophagy 2021, 17, 4182–4201. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Lee, J.; Liu, Z.; Kim, H.; Martin, D.R.; Wu, D.; Liu, M.; Xue, X. Mitophagy protein PINK1 suppresses colon tumor growth by metabolic reprogramming via p53 activation and reducing acetyl-CoA production. Cell Death Differ. 2021, 28, 2421–2435. [Google Scholar] [CrossRef] [PubMed]
- Hansen, G.E.; Gibson, G.E. The alpha-ketoglutarate dehydrogenase complex as a hub of plasticity in neurodegeneration and regeneration. Int. J. Mol. Sci. 2022, 23, 12403. [Google Scholar] [CrossRef]
- Arnold, P.K.; Finley, L.W.S. Regulation and function of the mammalian tricarboxylic acid cycle. J. Biol. Chem. 2023, 299, 102838. [Google Scholar] [CrossRef]
- Chhimpa, N.; Singh, N.; Puri, N.; Kayath, H.P. The novel role of mitochondrial citrate synthase and citrate in the pathophysiology of Alzheimer’s disease. J. Alzheimers Dis. 2023, 94, S453–S472. [Google Scholar] [CrossRef] [PubMed]
- Chandel, N.S. Glycolysis. Cold Spring Harb. Perspect. Biol. 2021, 13, a040535. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Acín-Peréz, R.; Geghman, K.D.; Manfredi, G.; Lu, B.; Li, C. Pink1 regulates the oxidative phosphorylation machinery via mitochondrial fission. Proc. Natl. Acad. Sci. USA 2011, 108, 12920–12924. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, H.; Li, R.; Sterling, K.; Song, W. Amyloid β-based therapy for Alzheimer’s disease: Challenges, successes and future. Signal Transduct. Target. Ther. 2023, 8, 248. [Google Scholar] [CrossRef]
- Guo, T.; Zhang, D.; Zeng, Y.; Huang, T.Y.; Xu, H.; Zhao, Y. Molecular and cellular mechanisms underlying the pathogenesis of Alzheimer’s disease. Mol. Neurodegener. 2020, 15, 40. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Guo, F.; Luo, Y.; Zhang, X.; Wang, C.; Liu, Y.; Zhang, H. Electropositive Citric Acid-Polyethyleneimine Carbon Dots Carrying the PINK1 Gene Regulate ATP-Related Metabolic Dysfunction in APP/PS1-N2a Cells. Molecules 2024, 29, 1907. https://doi.org/10.3390/molecules29091907
Yu S, Guo F, Luo Y, Zhang X, Wang C, Liu Y, Zhang H. Electropositive Citric Acid-Polyethyleneimine Carbon Dots Carrying the PINK1 Gene Regulate ATP-Related Metabolic Dysfunction in APP/PS1-N2a Cells. Molecules. 2024; 29(9):1907. https://doi.org/10.3390/molecules29091907
Chicago/Turabian StyleYu, Si, Feng Guo, Yuzhen Luo, Xingfang Zhang, Chenyu Wang, Yiheng Liu, and Haiying Zhang. 2024. "Electropositive Citric Acid-Polyethyleneimine Carbon Dots Carrying the PINK1 Gene Regulate ATP-Related Metabolic Dysfunction in APP/PS1-N2a Cells" Molecules 29, no. 9: 1907. https://doi.org/10.3390/molecules29091907
APA StyleYu, S., Guo, F., Luo, Y., Zhang, X., Wang, C., Liu, Y., & Zhang, H. (2024). Electropositive Citric Acid-Polyethyleneimine Carbon Dots Carrying the PINK1 Gene Regulate ATP-Related Metabolic Dysfunction in APP/PS1-N2a Cells. Molecules, 29(9), 1907. https://doi.org/10.3390/molecules29091907







