The Role of mRNA Alternative Splicing in Macrophages Infected with Mycobacterium tuberculosis: A Field Needing to Be Discovered
Abstract
1. Introduction
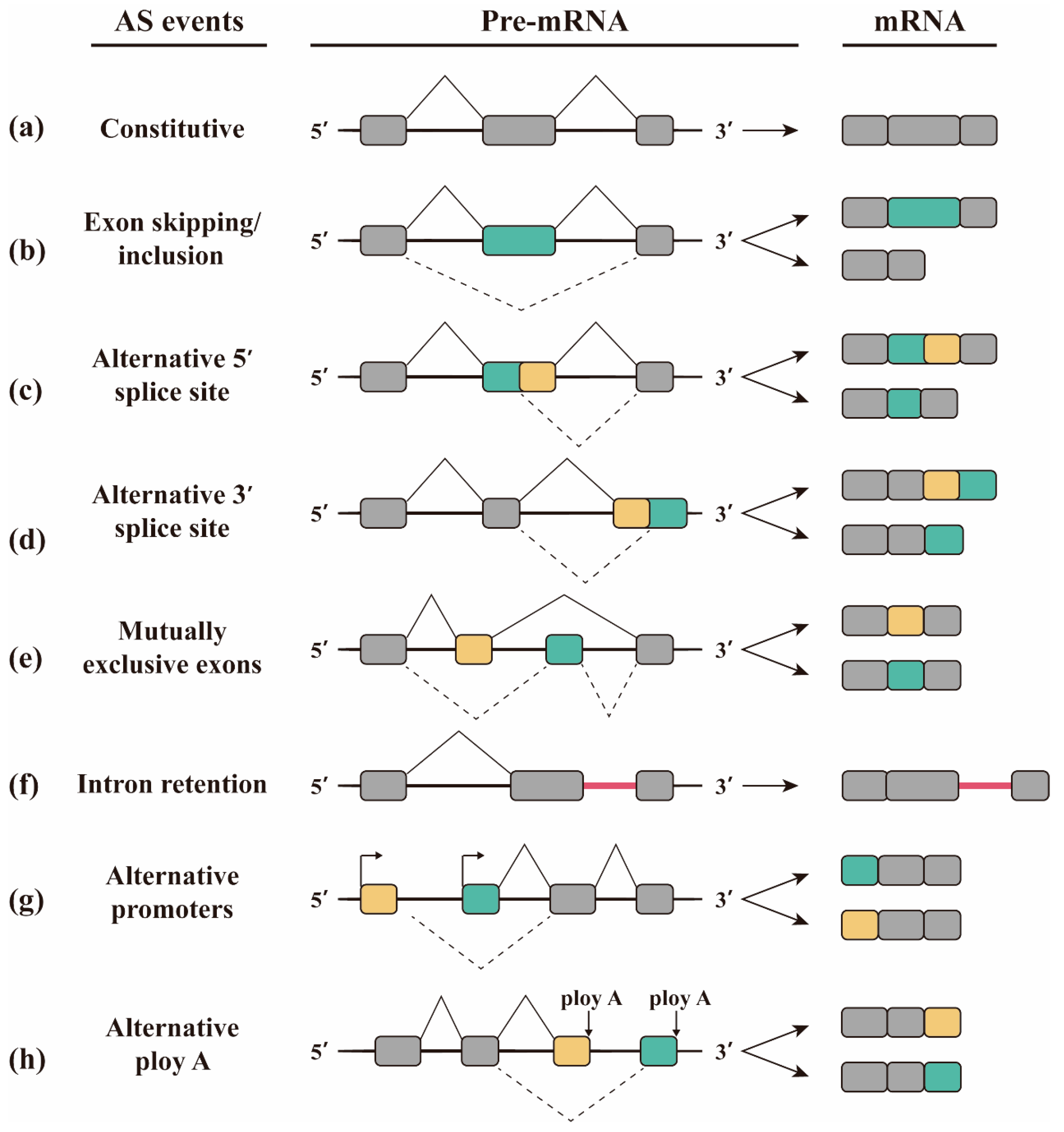
2. Alternative Splicing
3. Global Changes in Gene Expression upon Infection with Mtb
3.1. Alternative Splicing Causes Changes in Protein Activity
3.2. TB-Specific Alternatively Spliced Isoforms
3.3. The Expression of Splicing Factors Changes in Cells Infected with Mtb
4. Alternative Splicing in Host Response to Mtb Infection
4.1. The Role of IL-4/IL-4δ2 Ratio in Mtb Infection
4.2. IL-7 and IL-7 R Isoforms Play Different Roles in Control of Mtb Infection
4.3. IL12-Rβ1 Is Essential for Host Defence against Mtb Infection
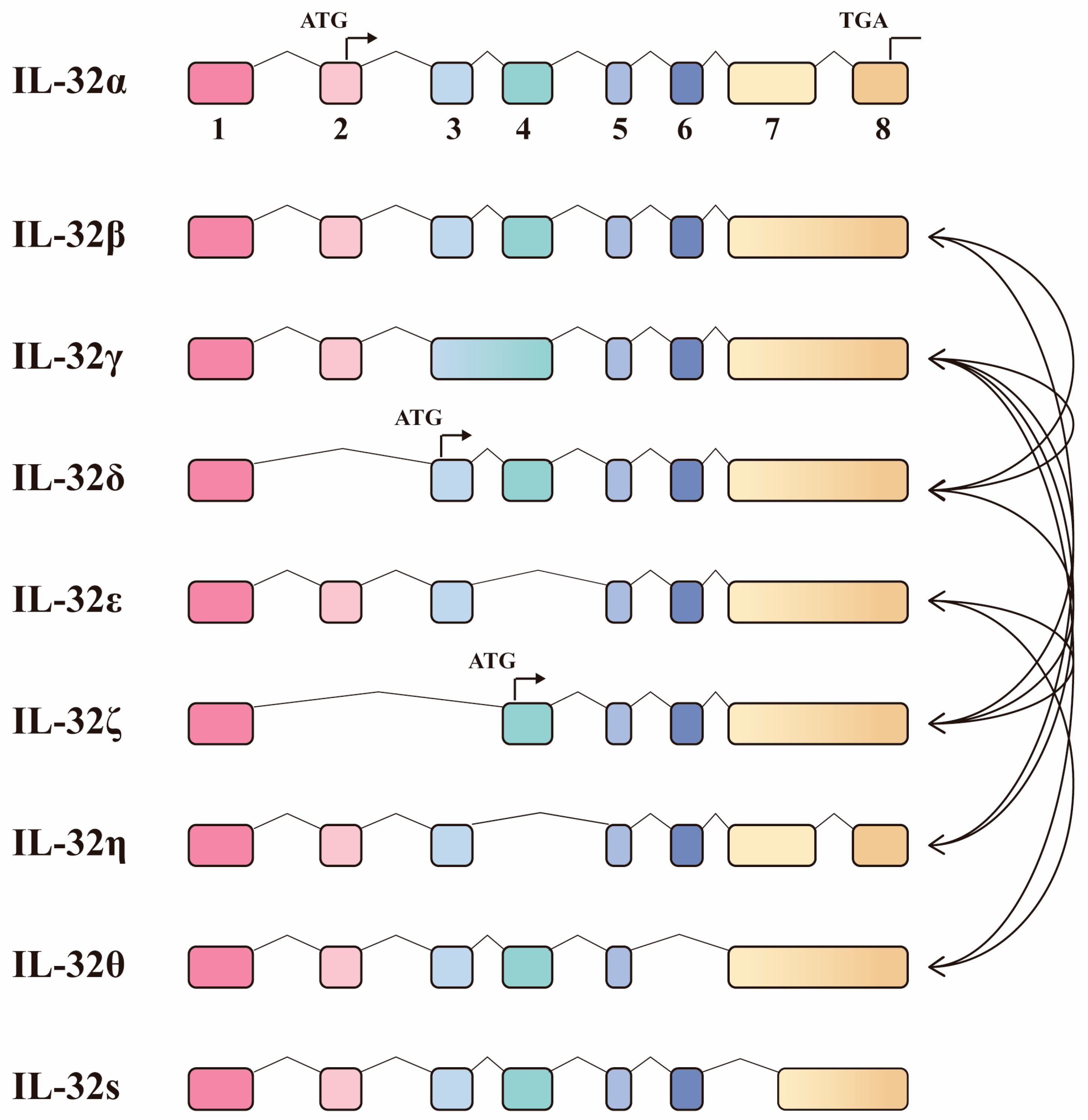
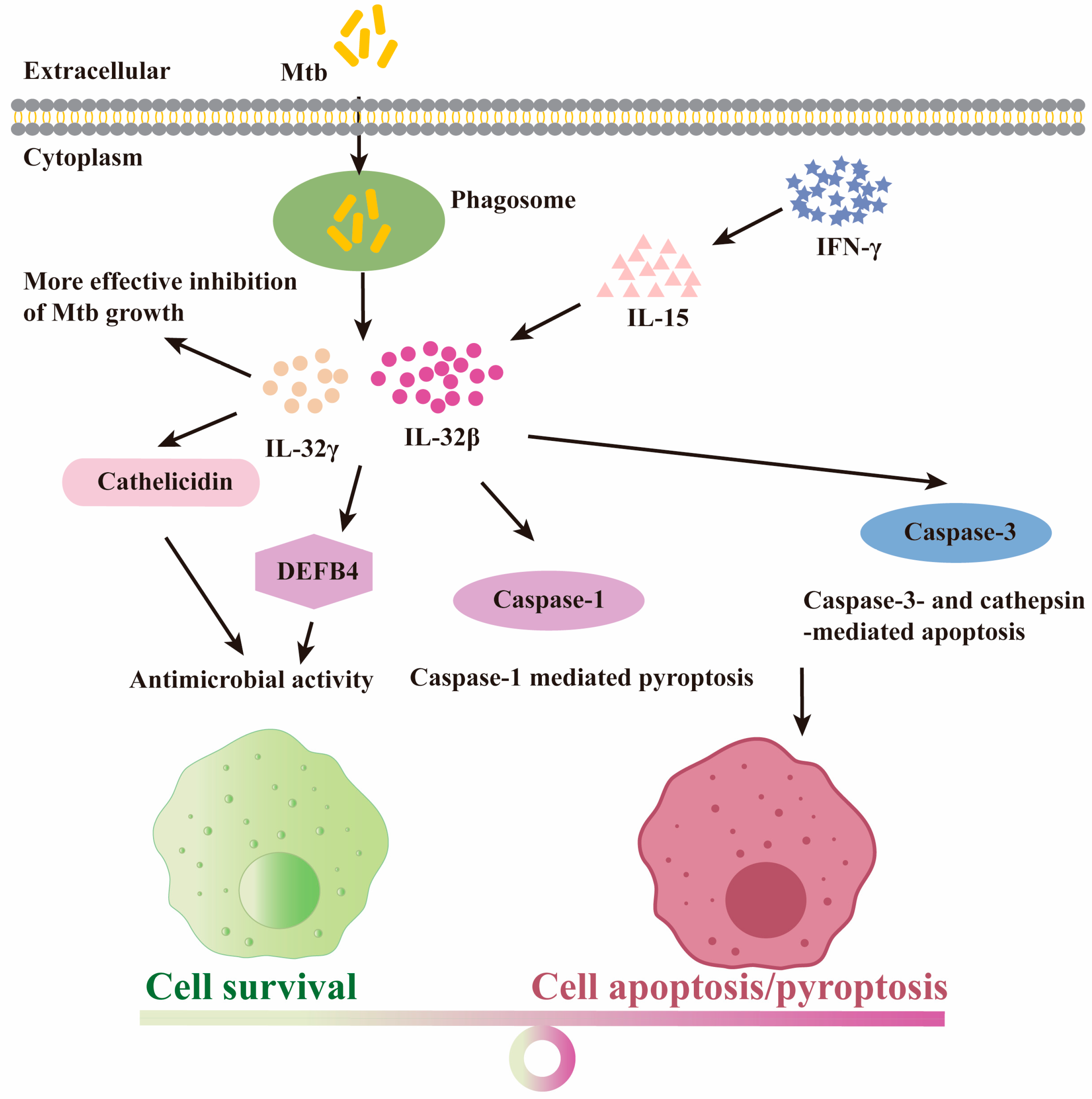
4.4. The Protective Role for IL-32 Isoforms upon Mtb Infection
5. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bagcchi, S. WHO’s Global Tuberculosis Report 2022. Lancet Microbe 2023, 4, e20. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.G. Mycobacterium tuberculosis and the intimate discourse of a chronic infection. Immunol. Rev. 2015, 240, 252–268. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Tuberculosis Report 2023; World Health Organization: Geneva, Switzerland, 2023.
- Ghebreyesus, T.A.; Kasaeva, T. Global Tuberculosis Report 2022; World Health Organization: Geneva, Switzerland, 2022.
- van Leth, F.; Van der Werf, M.J.; Borgdorff, M.W. Prevalence of tuberculous infection and incidence of tuberculosis: A re-assessment of the Styblo rule. Bull. World Health Organ. 2008, 86, 20–26. [Google Scholar] [CrossRef]
- Eddo, K.; Alon, M.; Gil, A. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2006, 35, 125–131. [Google Scholar]
- Kalam, H.; Singh, K.; Chauhan, K.; Fontana, M.F.; Kumar, D. Alternate splicing of transcripts upon Mycobacterium tuberculosis infection impacts the expression of functional protein domains. IUBMB Life 2018, 70, 845–854. [Google Scholar] [CrossRef]
- Kim, E.; Goren, A.; Ast, G. Alternative splicing: Current perspectives. Bioessays 2008, 30, 38–47. [Google Scholar] [CrossRef]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef] [PubMed]
- Merkhofer, E.C.; Hu, P.; Johnson, T.L. Introduction to cotranscriptional RNA splicing. Methods Mol. Biol. 2014, 1126, 83–96. [Google Scholar]
- Yan, C.; Wan, R.; Shi, Y. Molecular Mechanisms of pre-mRNA Splicing through Structural Biology of the Spliceosome. Cold Spring Harb. Perspect. Biol. 2019, 11, a032409. [Google Scholar] [CrossRef]
- Gilbert, W. Why genes in pieces? Nature 1978, 271, 501. [Google Scholar] [CrossRef]
- Early, P.; Rogers, J.; Davis, M.; Calame, K.; Bond, M.; Wall, R.; Hood, L. Two mRNAs can be produced from a single immunoglobulin μ gene by alternative RNA processing pathways. Cell 1980, 20, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Werneke, J.M.; Chatfield, J.M.; Ogren, W.L. Alternative mRNA splicing generates the two ribulosebisphosphate carboxylase/oxygenase activase polypeptides in spinach and Arabidopsis. Plant Cell 1989, 1, 815–825. [Google Scholar] [PubMed]
- Watanabe, Y.; Yokobori, S.; Inaba, T.; Yamagishi, A.; Oshima, T.; Kawarabayasi, Y.; Kikuchi, H.; Kita, K. Introns in protein-coding genes in Archaea. FEBS Lett. 2002, 510, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Yokobori, S.I.; Itoh, T.; Yoshinari, S.; Nomura, N.; Sako, Y.; Yamagishi, A.; Oshima, T.; Kita, K.; Watanabe, Y.I. Gain and loss of an intron in a protein-coding gene in Archaea: The case of an archaeal RNA pseudouridine synthase gene. BMC Evol. Biol. 2009, 9, 198. [Google Scholar] [CrossRef] [PubMed]
- Artamonova, I.I.; Gelfand, M.S. Comparative Genomics and Evolution of Alternative Splicing: The Pessimists’Science. Chem. Rev. 2007, 107, 3407–3430. [Google Scholar] [CrossRef] [PubMed]
- Alekseyenko, A.V.; Namshin, K.; Lee, C.J. Global analysis of exon creation versus loss and the role of alternative splicing in 17 vertebrate genomes. RNA 2007, 13, 661–670. [Google Scholar] [CrossRef]
- Sharp, P.A. Split genes and RNA splicing. Cell 1994, 77, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Huang, B.O.; Xu, Y.M.; Li, J.; Huang, L.F.; Lin, J.; Zhang, J.; Min, Q.H.; Yang, W.M.; et al. Mechanism of alternative splicing and its regulation. Biomed. Rep. 2015, 3, 152–158. [Google Scholar] [CrossRef]
- Xing, Y.; Lee, C. Alternative splicing and RNA selection pressure—Evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006, 7, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Dai, X.; Wu, J. Alternative splicing: An important mechanism in stem cell biology. World J. Stem Cells 2015, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Dong, H.; Shi, Y.; Bian, L. Mutually exclusive alternative splicing of pre-mRNAs. Wiley Interdiscip. Rev. RNA 2018, 9, e1468. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Pham, M.H.C.; Ko, K.S.; Rhee, B.D.; Han, J. Alternative splicing isoforms in health and disease. Pflügers Arch. 2018, 470, 995–1016. [Google Scholar] [CrossRef]
- Hatje, K.; Kollmar, M. Expansion of the mutually exclusive spliced exome in Drosophila. Nat. Commun. 2013, 4, 2460. [Google Scholar] [CrossRef]
- Sugnet, C.W.; Kent, W.J.; Ares, M., Jr.; Haussler, D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput. 2004, 9, 66–77. [Google Scholar]
- Hong, W.; Zhang, J.; Dong, H.; Shi, Y.; Ma, H.; Zhou, F.; Xu, B.; Fu, Y.; Zhang, S.; Hou, S.; et al. Intron-targeted mutagenesis reveals roles for Dscam1 RNA pairing architecture-driven splicing bias in neuronal wiring. Cell Rep. 2021, 36, 109373. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.D.; Berglund, J.A. Alternative Pre-mRNA Splicing. Acta Physiol. 2013, 1126, 45–54. [Google Scholar]
- Sakabe, N.J.; Souza, S.J.D. Sequence features responsible for intron retention in human. Bmc Genom. 2007, 8, 59. [Google Scholar] [CrossRef]
- Kalam, H.; Fontana, M.F.; Kumar, D. Alternate splicing of transcripts shape macrophage response to Mycobacterium tuberculosis infection. PLoS Pathog. 2017, 13, e1006236. [Google Scholar] [CrossRef]
- Mvubu, N.E.; Pillay, B.; Pillay, M. Infection of pulmonary epithelial cells by clinical strains of M. tuberculosis induces alternate splicing events. Gene 2020, 750, 144755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Niu, C.; Fu, R.Y.; Peng, Z.Y. Mycobacterium tuberculosis H37Rv infection regulates alternative splicing in Macrophages. Bioengineered 2018, 9, 203–208. [Google Scholar] [CrossRef]
- Lyu, M.; Zhou, J.; Zhou, Y.; Chong, W.; Xu, W.; Lai, H.; Niu, L.; Hai, Y.; Yao, X.; Gong, S.; et al. From tuberculosis bedside to bench: UBE2B splicing as a potential biomarker and its regulatory mechanism. Signal Transduct. Target. Ther. 2023, 8, 82. [Google Scholar] [CrossRef]
- Danelishvili, L.; Yamazaki, Y.; Selker, J.; Bermudez, L.E. Secreted Mycobacterium tuberculosis Rv3654c and Rv3655c proteins participate in the suppression of macrophage apoptosis. PLoS ONE 2010, 5, e10474. [Google Scholar] [CrossRef]
- Long, J.C.; Caceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009, 417, 15–27. [Google Scholar] [CrossRef]
- Lai, M.C.; Peng, T.Y.; Tarn, W.Y. Functional interplay between viral and cellular SR proteins in control of post-transcriptional gene regulation. Febs J. 2009, 276, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Arinobu, Y.; Atamas, S.P.; Otsuka, T.; Niiro, H.; Yamaoka, K.; Mitsuyasu, H.; Niho, Y.; Hamasaki, N.; White, B.; Izuhara, K. Antagonistic effects of an alternative splice variant of human IL-4, IL-4delta2, on IL-4 activities in human monocytes and B cells. Cell. Immunol. 1999, 191, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Seah, G.T.; Rook, G.A. High levels of mRNA encoding IL-4 in unstimulated peripheral blood mononuclear cells from tuberculosis patients revealed by quantitative nested reverse transcriptase-polymerase chain reaction; correlations with serum IgE levels. Scand. J. Infect. Dis. 2001, 33, 106–109. [Google Scholar]
- Fletcher, H.A.; Owiafe, P.; Jeffries, D.; Hill, P.; Rook, G.A.; Zumla, A.; Doherty, T.M.; Brookes, R.H.; Vacsel Study, G. Increased expression of mRNA encoding interleukin (IL)-4 and its splice variant IL-4delta2 in cells from contacts of Mycobacterium tuberculosis, in the absence of in vitro stimulation. Immunology 2004, 112, 669–673. [Google Scholar] [CrossRef]
- Demissie, A.; Abebe, M.; Aseffa, A.; Rook, G.; Fletcher, H.; Zumla, A.; Weldingh, K.; Brock, I.; Andersen, P.; Doherty, T.M.; et al. Healthy individuals that control a latent infection with Mycobacterium tuberculosis express high levels of Th1 cytokines and the IL-4 antagonist IL-4delta2. J. Immunol. 2004, 172, 6938–6943. [Google Scholar] [CrossRef]
- Dheda, K.; Chang, J.S.; Huggett, J.F.; Kim, L.U.; Johnson, M.A.; Zumla, A.; Rook, G.A. The stability of mRNA encoding IL-4 is increased in pulmonary tuberculosis, while stability of mRNA encoding the antagonistic splice variant, IL-4delta2, is not. Tuberculosis 2007, 87, 237–241. [Google Scholar] [CrossRef]
- Krawczenko, A.; Kieda, C.; Dus, D. The biological role and potential therapeutic application of interleukin 7. Arch. Immunol. Ther. Exp. 2005, 53, 518–525. [Google Scholar]
- Adankwah, E.; Harelimana, J.D.; Minadzi, D.; Aniagyei, W.; Abass, M.K.; Batsa Debrah, L.; Owusu, D.O.; Mayatepek, E.; Phillips, R.O.; Jacobsen, M. Lower IL-7 Receptor Expression of Monocytes Impairs Antimycobacterial Effector Functions in Patients with Tuberculosis. J. Immunol. 2021, 206, 2430–2440. [Google Scholar] [CrossRef] [PubMed]
- Rane, L.; Rahman, S.; Magalhaes, I.; Ahmed, R.; Spangberg, M.; Kondova, I.; Verreck, F.; Andersson, J.; Brighenti, S.; Maeurer, M.J. Increased (6 exon) interleukin-7 production after M. tuberculosis infection and soluble interleukin-7 receptor expression in lung tissue. Genes Immun. 2011, 12, 513–522. [Google Scholar] [CrossRef]
- Maeurer, M.J.; Trinder, P.; Hommel, G.; Walter, W.; Freitag, K.; Atkins, D.; Storkel, S. Interleukin-7 or interleukin-15 enhances survival of Mycobacterium tuberculosis-infected mice. Infect. Immun. 2000, 68, 2962–2970. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vudattu, N.K.; Magalhaes, I.; Hoehn, H.; Pan, D.; Maeurer, M.J. Expression analysis and functional activity of interleukin-7 splice variants. Genes Immun. 2009, 10, 132–140. [Google Scholar] [CrossRef]
- Crawley, A.M.; Faucher, S.; Angel, J.B. Soluble IL-7R alpha (sCD127) inhibits IL-7 activity and is increased in HIV infection. J. Immunol. 2010, 184, 4679–4687. [Google Scholar] [CrossRef]
- Chua, A.O.; Wilkinson, V.L.; Presky, D.H.; Gubler, U. Cloning and characterization of a mouse IL-12 receptor-beta component. J. Immunol. 1995, 155, 4286–4294. [Google Scholar] [CrossRef]
- de Jong, R.; Altare, F.; Haagen, I.A.; Elferink, D.G.; Boer, T.; van Breda Vriesman, P.J.; Kabel, P.J.; Draaisma, J.M.; van Dissel, J.T.; Kroon, F.P.; et al. Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science 1998, 280, 1435–1438. [Google Scholar] [CrossRef]
- Ford, N.R.; Miller, H.E.; Reeme, A.E.; Waukau, J.; Bengtson, C.; Routes, J.M.; Robinson, R.T. Inflammatory signals direct expression of human IL12RB1 into multiple distinct isoforms. J. Immunol. 2012, 189, 4684–4694. [Google Scholar] [CrossRef]
- Robinson, R.T.; Khader, S.A.; Martino, C.A.; Fountain, J.J.; Teixeira-Coelho, M.; Pearl, J.E.; Smiley, S.T.; Winslow, G.M.; Woodland, D.L.; Walter, M.J.; et al. Mycobacterium tuberculosis infection induces il12rb1 splicing to generate a novel IL-12Rbeta1 isoform that enhances DC migration. J. Exp. Med. 2010, 207, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.A.; Fountain, J.J.; Miller, H.E.; Cooper, A.M.; Robinson, R.T.; Flynn, J.L. IL12Rβ1ΔTM is a secreted product of il12rb1 that promotes control of extrapulmonary tuberculosis. Infect. Immun. 2015, 83, 560–571. [Google Scholar] [CrossRef] [PubMed]
- Aass, K.R.; Kastnes, M.H.; Standal, T. Molecular interactions and functions of IL-32. J. Leukoc. Biol. 2021, 109, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Dahl, C.A.; Schall, R.P.; He, H.L.; Cairns, J.S. Identification of a novel gene expressed in activated natural killer cells and T cells. J. Immunol. 1992, 148, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Han, S.; Azam, T.; Yoon, D.; Dinarello, C. Interleukin-32: A cytokine and inducer of TNFalpha. Immunity 2005, 22, 131–142. [Google Scholar] [PubMed]
- Kang, J.W.; Park, Y.S.; Lee, D.H.; Kim, M.S.; Bak, Y.; Ham, S.Y.; Park, S.H.; Kim, H.; Ahn, J.H.; Hong, J.T.J.B. Interaction network mapping among IL-32 isoforms. Biochimie 2014, 101, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Azam, T.; Lewis, E.C.; Joosten, L.; Wang, M.; Langenberg, D.; Meng, X.; Chan, E.D.; Yoon, D.Y.; Ottenhoff, T. Mycobacterium tuberculosis Induces Interleukin-32 Production through a Caspase-1/IL-18/Interferon-γ-Dependent Mechanism. PLoS Med. 2006, 3, e277. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Kim, S.H.; Azam, T.; McGibney, M.T.; Huang, H.; Dinarello, C.A.; Chan, E.D. IL-32 is a host protective cytokine against Mycobacterium tuberculosis in differentiated THP-1 human macrophages. J. Immunol. 2010, 184, 3830–3840. [Google Scholar] [CrossRef]
- Bai, X.; Shang, S.; Henao-Tamayo, M.; Basaraba, R.J.; Ovrutsky, A.R.; Matsuda, J.L.; Takeda, K.; Chan, M.M.; Dakhama, A.; Kinney, W.H.; et al. Human IL-32 expression protects mice against a hypervirulent strain of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2015, 112, 5111–5116. [Google Scholar] [CrossRef]
- Koeken, V.; Verrall, A.J.; Ardiansyah, E.; Apriani, L.; Dos Santos, J.C.; Kumar, V.; Alisjahbana, B.; Hill, P.C.; Joosten, L.A.B.; van Crevel, R.; et al. IL-32 and its splice variants are associated with protection against Mycobacterium tuberculosis infection and skewing of Th1/Th17 cytokines. J. Leukoc. Biol. 2020, 107, 113–118. [Google Scholar] [CrossRef]
- Bai, X.; Ovrutsky, A.R.; Kartalija, M.; Chmura, K.; Kamali, A.; Honda, J.R.; Oberley-Deegan, R.E.; Dinarello, C.A.; Crapo, J.D.; Chang, L.Y.; et al. IL-32 expression in the airway epithelial cells of patients with Mycobacterium avium complex lung disease. Int. Immunol. 2011, 23, 679–691. [Google Scholar] [CrossRef] [PubMed]
- Schenk, M.; Mahapatra, S.; Le, P.; Kim, H.J.; Choi, A.W.; Brennan, P.J.; Belisle, J.T.; Modlin, R.L.J.I. Immunity, Human NOD2 Recognizes Structurally Unique Muramyl Dipeptides from Mycobacterium leprae. Infect. Immun. 2016, 84, 2429–2438. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Yang, W.; Jie, Z.; Elgaili, A.A.; Xie, J.J.O. Mycobacterium tuberculosis PPE32 promotes cytokines production and host cell apoptosis through caspase cascade accompanying with enhanced ER stress response. Oncotarget 2016, 7, 67347–67359. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Kinney, W.H.; Su, W.L.; Bai, A.; Ovrutsky, A.R.; Honda, J.R.; Netea, M.G.; Henao-Tamayo, M.; Ordway, D.J.; Dinarello, C.A.; et al. Caspase-3-independent apoptotic pathways contribute to interleukin-32gamma-mediated control of Mycobacterium tuberculosis infection in THP-1 cells. BMC Microbiol. 2015, 15, 39. [Google Scholar] [CrossRef] [PubMed]
- Montoya, D.; Inkeles, M.S.; Liu, P.T.; Realegeno, S.; Teles, R.M.; Vaidya, P.; Munoz, M.A.; Schenk, M.; Swindell, W.R.; Chun, R.; et al. IL-32 is a molecular marker of a host defense network in human tuberculosis. Sci. Transl. Med. 2014, 6, 250ra114. [Google Scholar] [CrossRef] [PubMed]
- Goda, C.; Kanaji, T.; Kanaji, S.; Tanaka, G.; Arima, K.; Ohno, S.; Izuhara, K. Involvement of IL-32 in activation-induced cell death in T cells. Int. Immunol. 2006, 18, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Deng, W.; Xie, J. The Biology and Role of Interleukin-32 in Tuberculosis. J. Immunol. Res. 2018, 2018, 1535194. [Google Scholar] [CrossRef]
- Her, E.; Kim, S.J.; Kim, B.K.; Choi, W.S.; Yoon, D.Y.; Kim, S.H.; Bae, S.Y.; Choi, J.D.; Dinarello, C.A.; Lee, C.K. Identification of the most active interleukin-32 isoform. Immunology 2017, 126, 535–542. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, W.; Yang, H.; Wang, X.; Shi, J.; Zhang, J.; Xie, J. The Role of mRNA Alternative Splicing in Macrophages Infected with Mycobacterium tuberculosis: A Field Needing to Be Discovered. Molecules 2024, 29, 1798. https://doi.org/10.3390/molecules29081798
Hong W, Yang H, Wang X, Shi J, Zhang J, Xie J. The Role of mRNA Alternative Splicing in Macrophages Infected with Mycobacterium tuberculosis: A Field Needing to Be Discovered. Molecules. 2024; 29(8):1798. https://doi.org/10.3390/molecules29081798
Chicago/Turabian StyleHong, Weiling, Hongxing Yang, Xiao Wang, Jingyi Shi, Jian Zhang, and Jianping Xie. 2024. "The Role of mRNA Alternative Splicing in Macrophages Infected with Mycobacterium tuberculosis: A Field Needing to Be Discovered" Molecules 29, no. 8: 1798. https://doi.org/10.3390/molecules29081798
APA StyleHong, W., Yang, H., Wang, X., Shi, J., Zhang, J., & Xie, J. (2024). The Role of mRNA Alternative Splicing in Macrophages Infected with Mycobacterium tuberculosis: A Field Needing to Be Discovered. Molecules, 29(8), 1798. https://doi.org/10.3390/molecules29081798






