Multi-Omics Study on Molecular Mechanisms of Single-Atom Fe-Doped Two-Dimensional Conjugated Phthalocyanine Framework for Photocatalytic Antibacterial Performance
Abstract
1. Introduction
2. Results and Discussion
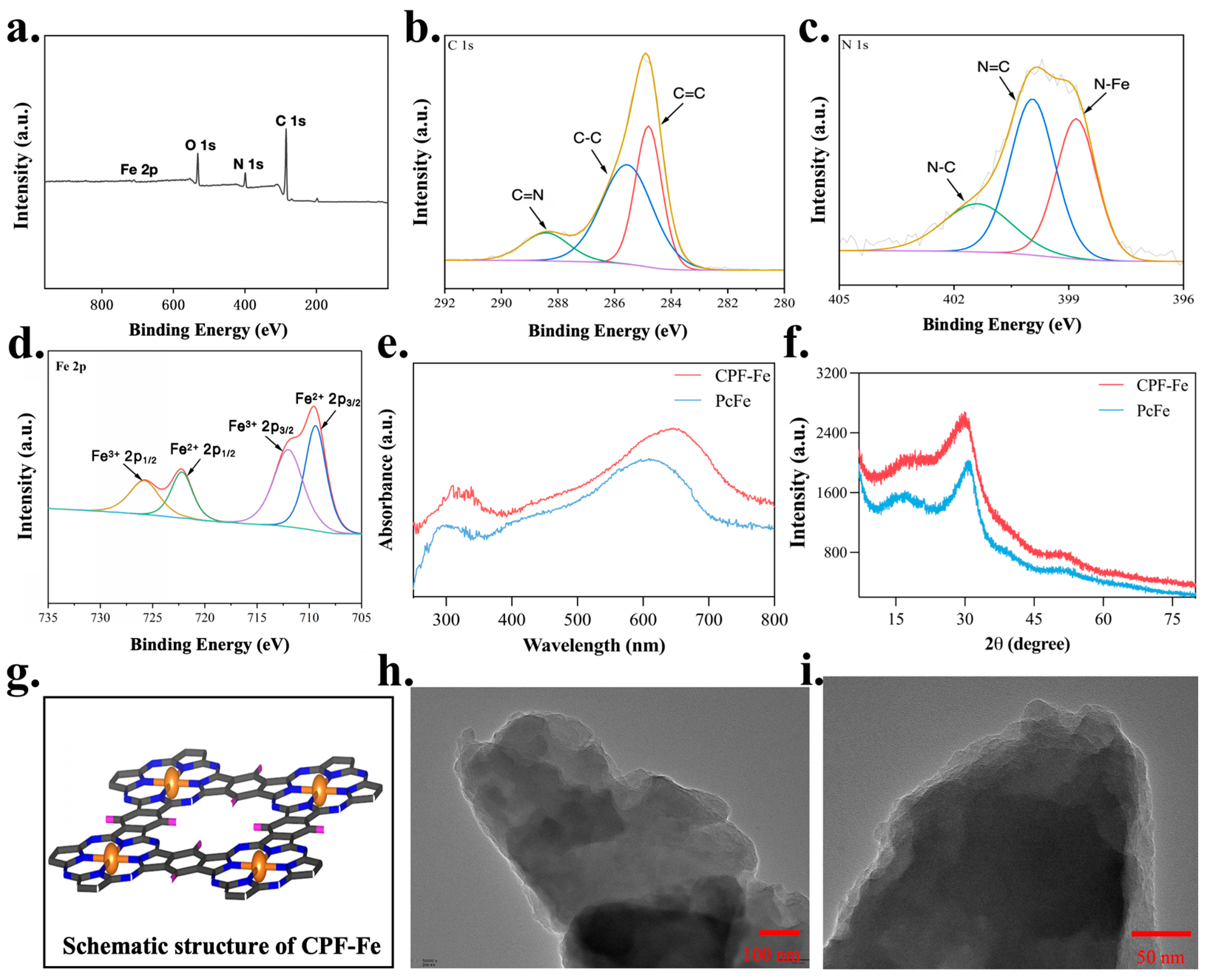
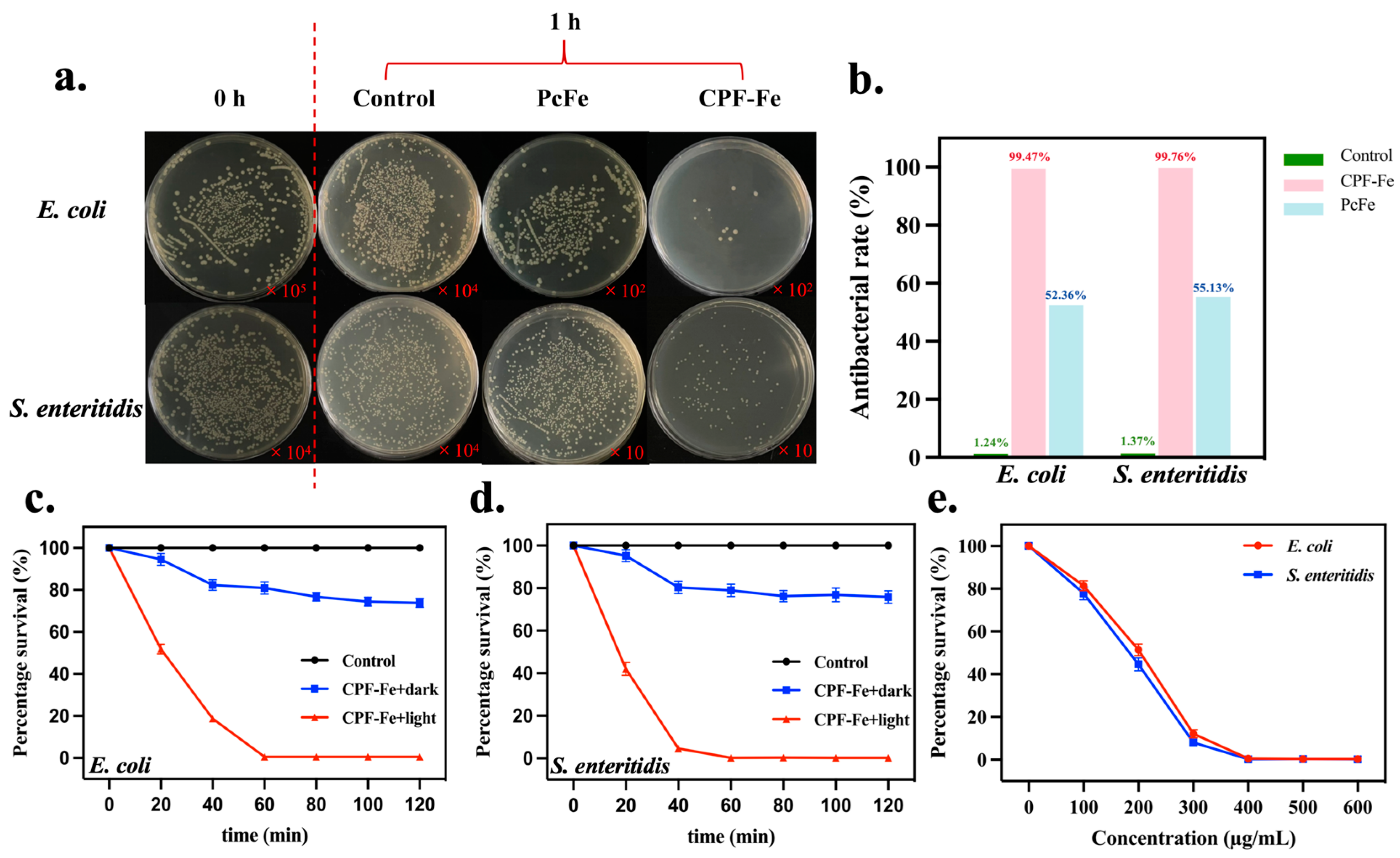
2.1. Characterization of CPF-Fe and Antibacterial Activity Tests
2.2. CPF-Fe Treatment of Farm Sewage
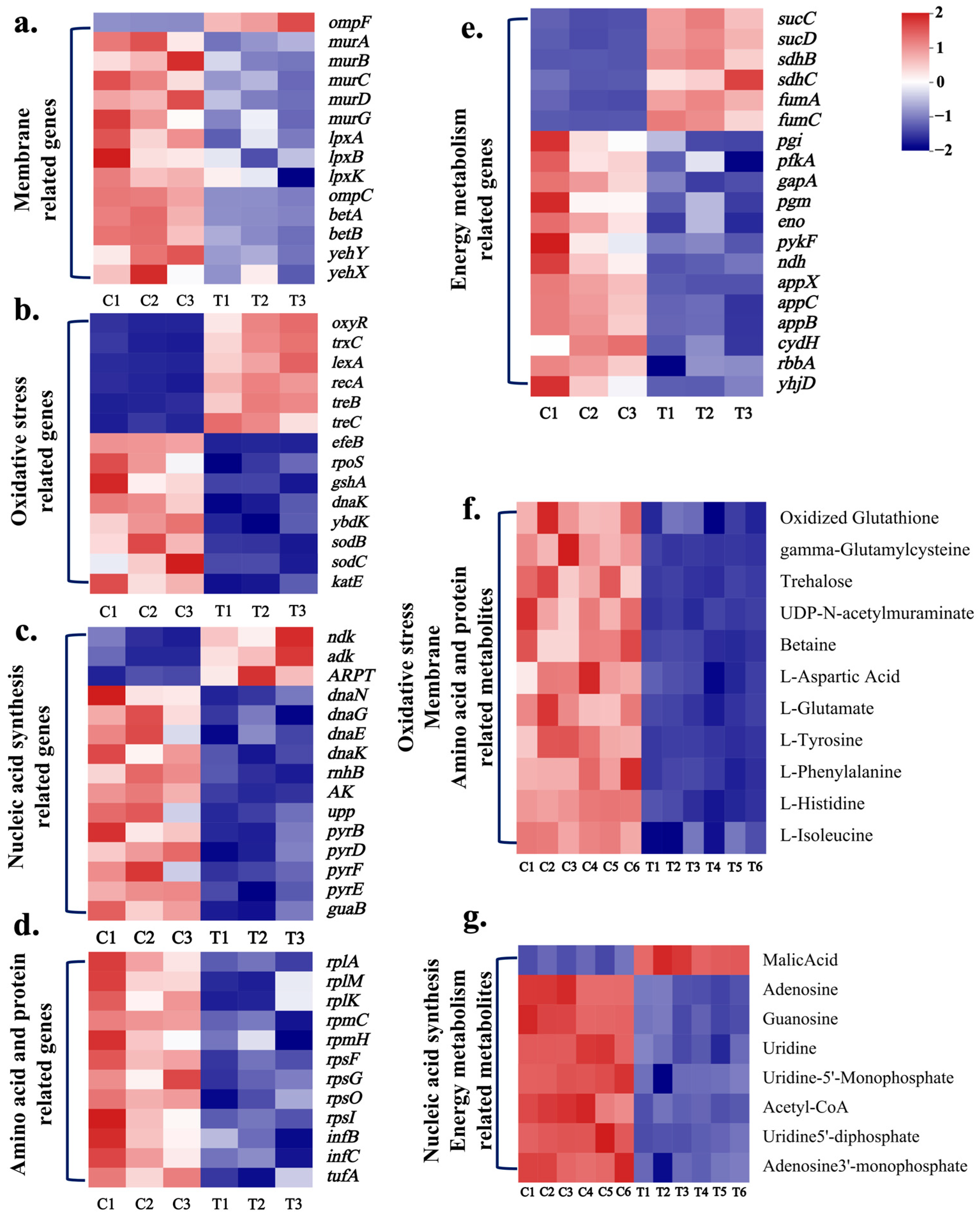
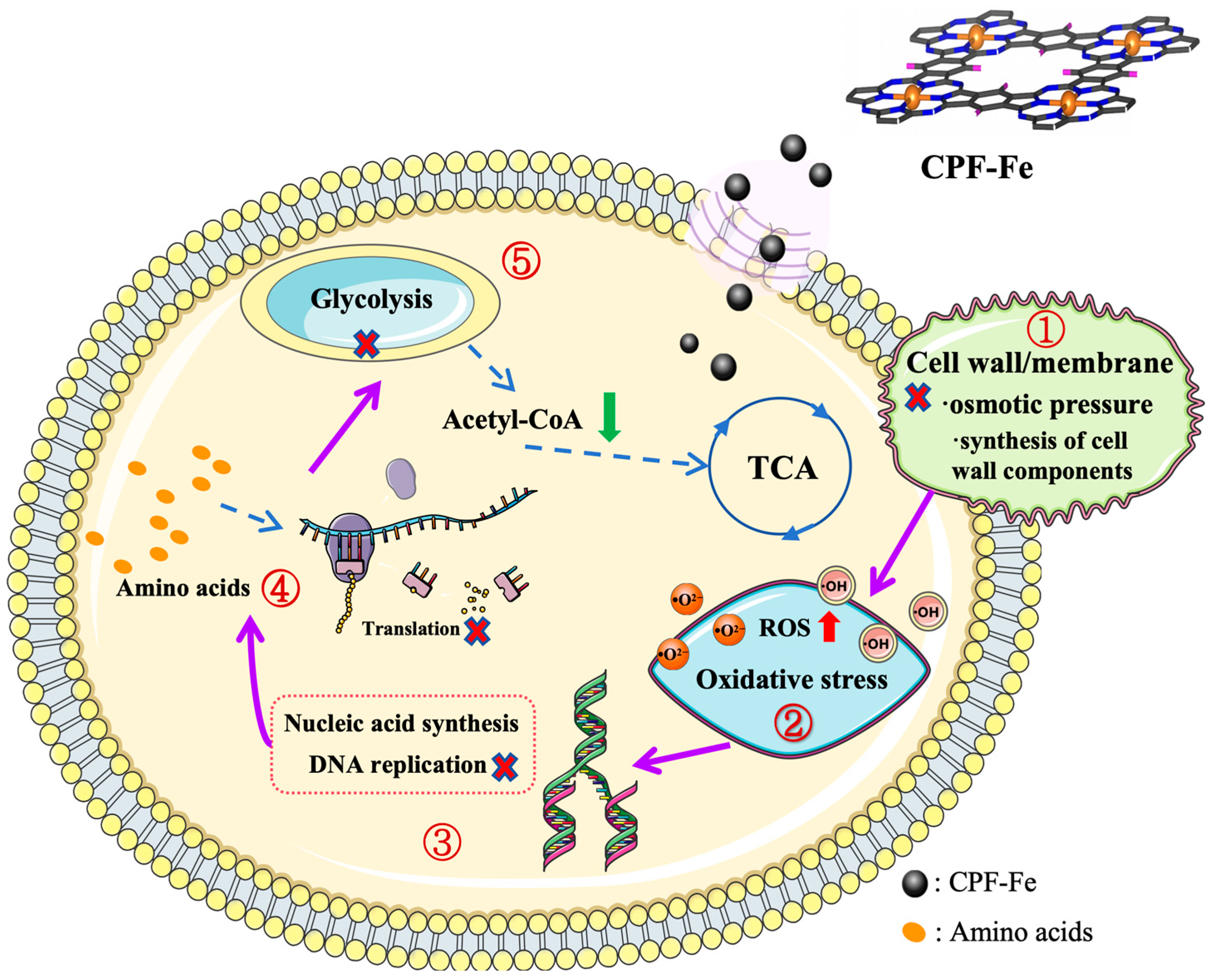
2.3. Antibacterial Molecular Mechanisms of CPF-Fe
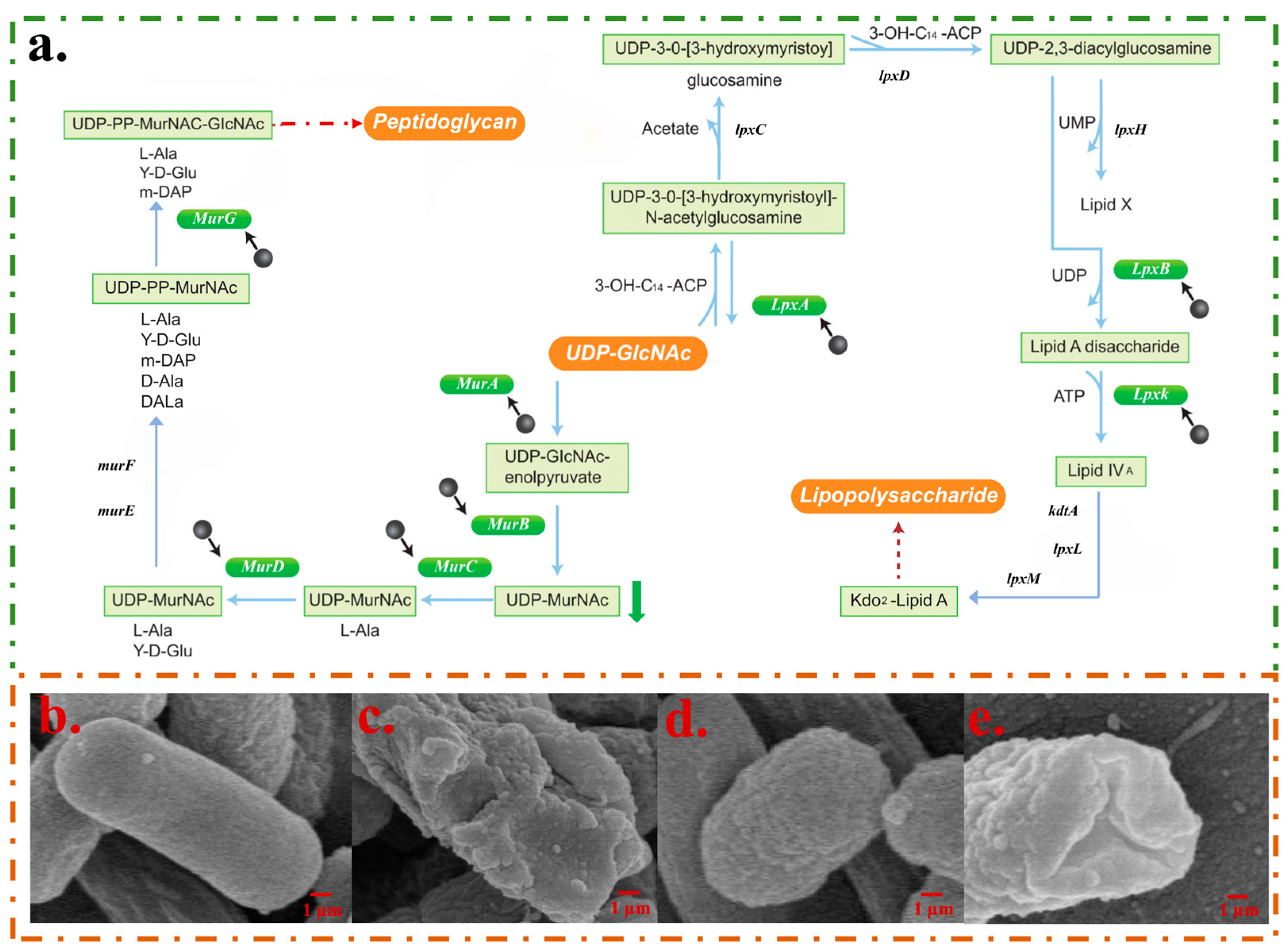
2.3.1. CPF-Fe Destroyed the Cell Membrane/Wall
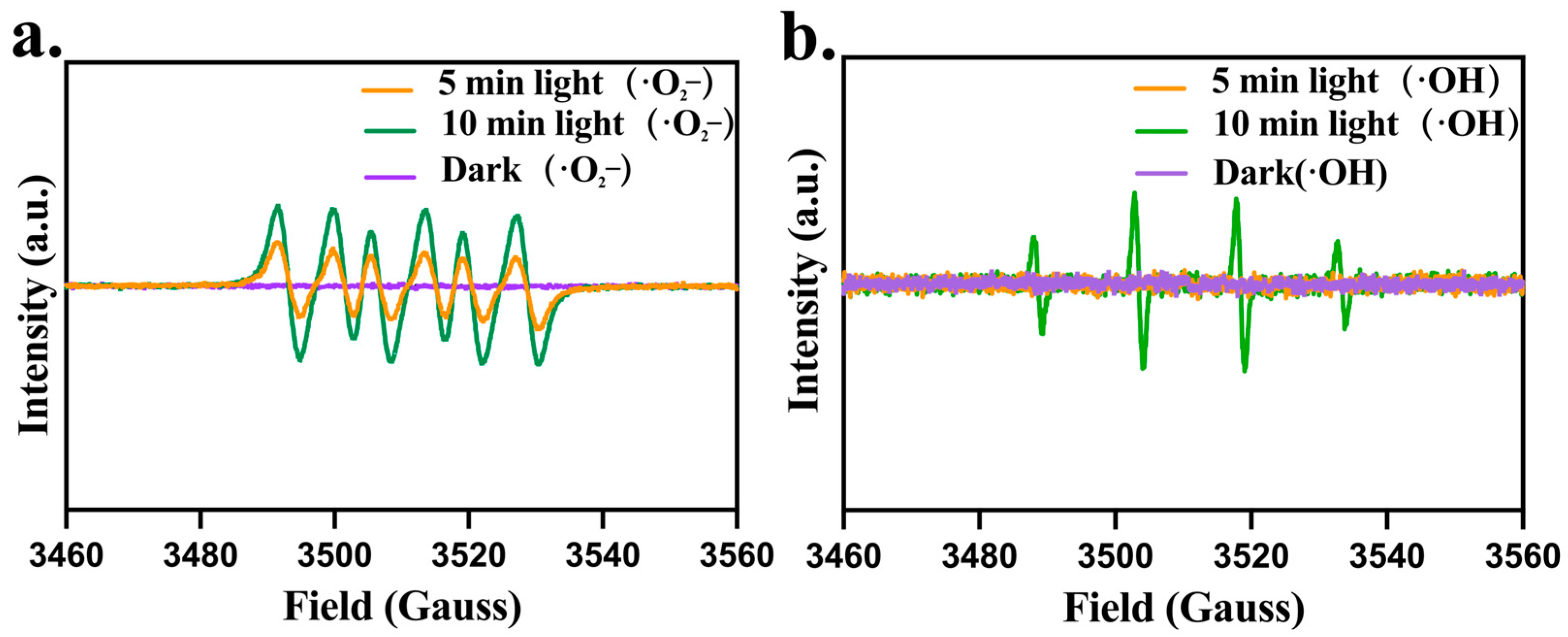
2.3.2. CPF-Fe Resulted in the Oxidative Stress
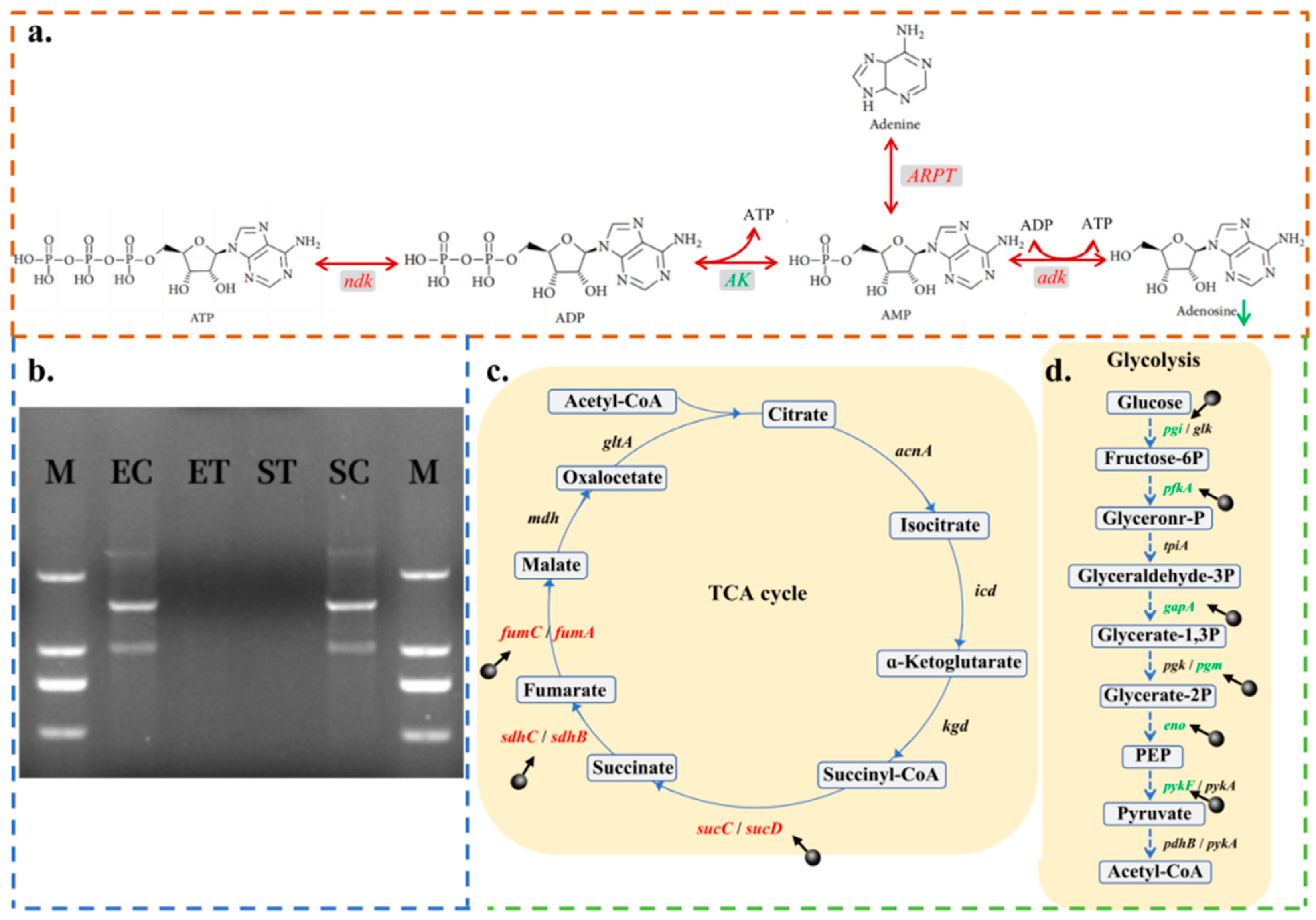
2.3.3. CPF-Fe Disrupted Synthesis of Nucleic Acid
2.3.4. CPF-Fe Inhibited Amino Acid Metabolism and Protein Synthesis
2.3.5. Energy Metabolism Disorder
3. Materials and Methods
3.1. Materials
3.2. Sample Preparation
3.3. Bacterial Culture
3.4. Photocatalysis of Antibacterial Activity Assessments
3.5. Sewage Treatment
3.6. Features of Bacterial Morphology
3.7. Measurement of ROS
3.8. PCR Amplification and Gel Electrophoresis of 16s rDNA
3.9. Transcriptomics Research
3.10. Non-Targeted Metabolomics Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xin, X.; Qi, C.; Xu, L.; Gao, Q.; Liu, X. Green synthesis of silver nanoparticles and their antibacterial effects. Front. Chem. Eng. 2022, 4, 941240. [Google Scholar] [CrossRef]
- Antoniadou, A.; Kontopidou, F.; Poulakou, G.; Koratzanis, E.; Galani, I. Colistin-resistant isolates of Klebsiella pneumoniae emerging in intensive care unit patients: First report of a multiclonal cluster. J. Antimicrob. Chemother. 2007, 59, 786–790. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Robinson, S.M.; Gupta, A. Functional Gold Nanoparticles as Potent Antimicrobial Agents against Multi-Drug-Resistant Bacteria. ACS Nano 2023, 8, 10682–10686. [Google Scholar] [CrossRef] [PubMed]
- Vimbela, G.V.; Ngo, S.M.; Fraze, C.; Yang, L.; Stout, D.A. Antibacterial properties and toxicity from metallic nanomaterials. Int. J. Nanomed. 2017, 12, 3941–3965. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Zhang, L. Nanomaterials arising amid antibiotic resistance. Nat. Rev. Microbiol. 2021, 19, 5–6. [Google Scholar] [CrossRef]
- Panja, S.; Adams, D.J. Pathway Dependence in Redox-Driven Metal-Organic Gels. Chemistry 2020, 26, 6130–6135. [Google Scholar] [CrossRef]
- Zhou, L.; Zhu, X.; Yang, J.; Cai, L.; Zhang, L. Deciphering the photoactive species-directed antibacterial mechanism of bismuth oxychloride with modulated nanoscale thickness. J. Environ. Manag. 2023, 333, 117411. [Google Scholar] [CrossRef]
- Gao, M.; Zhang, D.; Pu, X.; Li, H.; Li, W. Combustion synthesis of Fe-doped BiOCl with high visible-light photocatalytic activities. Sep. Purif. Technol. 2016, 162, 114–119. [Google Scholar] [CrossRef]
- Wang, X.; Yu, R.; Wang, P.; Chen, F.; Yu, H. Co-modification of F− and Fe(III) ions as a facile strategy towards effective separation of photogenerated electrons and holes. Appl. Surf. Sci. 2015, 351, 66–73. [Google Scholar] [CrossRef]
- Tang, N.; Li, Y.P.; Kurosu, S.; Koizumi, Y.; Matsumoto, H. Interfacial reactions of solid Co and solid Fe with liquid Al. Corros. Sci. 2012, 60, 32–37. [Google Scholar] [CrossRef]
- Cao, L.; Zhang, Q.; Miao, R.; Lin, J.; Feng, R. Application of omics technology in the research on edible fungi. Curr. Res. Food Sci. 2023, 6, 100430. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhao, X.; Li, R.; Yang, H. Integrated metabolomics and transcriptomics reveal the adaptive responses of Salmonella enterica serovar Typhimurium to thyme and cinnamon oils. Food Res. Int. 2022, 157, 111241. [Google Scholar] [CrossRef] [PubMed]
- Ashokan, M.; Rana, E.; Sneha, K.; Namith, C.; Naveen Kumar, G.S. Metabolomics-a powerful tool in livestock research. Anim. Biotechnol. 2022, 34, 3237–3249. [Google Scholar] [CrossRef]
- Miao, H.; Gao, W.; Xu, L.; Zhu, Y. Supramolecular iron phthalocyanine organic polymer with robust built-in electric field and shorter migration distance for photocatalytic pollutant degradation and antibacterial. Sep. Purif. Technol. 2022, 301, 122026. [Google Scholar] [CrossRef]
- Zang, Y.; Lu, D.Q.; Wang, K. A pyrolysis-free Ni/Fe bimetallic electrocatalyst for overall water splitting. Nat. Commun. 2023, 14, 1792. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Guo, C.; Liu, J.; Hu, B.; Dai, J.; Wang, M.; Jin, R.; Luo, Z.; Li, H.; Chen, C. Accelerating the oxygen adsorption kinetics to regulate the oxygen reduction catalysis via Fe3C nanoparticles coupled with single Fe-N4 sites. Energy Storage Mater. 2022, 51, 149–158. [Google Scholar] [CrossRef]
- Pomposiello, P.J.; Koutsolioutsou, A.; Carrasco, D.; Demple, B. SoxRS-regulated expression and genetic analysis of the yggX gene of Escherichia coli. J. Bacteriol. 2003, 185, 6624–6632. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ji, F.; Qiao, J.; Dong, X.; Wu, Y. Overexpression of the key genes in the biosynthetic pathways of lipid A and peptidoglycan in Escherichia coli. Biotechnol. Appl. Biochem. 2023, 70, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.S.; Bulawa, C.E.; Raetz, C.R. The biosynthesis of gram-negative endotoxin. Formation of lipid A precursors from UDP-GlcNAc in extracts of Escherichia coli. J. Biol. Chem. 1985, 260, 15536–15541. [Google Scholar] [CrossRef]
- Egger, L.A.; Park, H.; Inouye, M. Signal transduction via the histidyl-aspartyl phosphorelay. Genes Cells 1997, 2, 167–184. [Google Scholar] [CrossRef]
- Miller, E.N.; Ingram, L.O. Combined effect of betaine and trehalose on osmotic tolerance of Escherichia coli in mineral salts medium. Biotechnol. Lett. 2007, 29, 213–217. [Google Scholar] [CrossRef]
- Holmstrom, K.O.; Welin, B.; Mandal, A.; Kristiansdottir, I.; Teeri, T.H. Production of the Escherichia coli betaine-aldehyde dehydrogenase, an enzyme required for the synthesis of the osmoprotectant glycine betaine, in transgenic plants. Plant J. 1994, 6, 749–758. [Google Scholar] [CrossRef]
- Checroun, C.; Gutierrez, C. Sigma(s)-dependent regulation of yehZYXW, which encodes a putative osmoprotectant ABC transporter of Escherichia coli. FEMS Microbiol. Lett. 2004, 236, 221–226. [Google Scholar] [PubMed]
- Wang, Y.; Wu, J.E.; Yang, H. Comparison of the metabolic responses of eight Escherichia coli strains including the “big six” in pea sprouts to low concentration electrolysed water by NMR spectroscopy. Food Control 2022, 131, 108458. [Google Scholar] [CrossRef]
- Dalrymple, O.K.; Stefanakos, E.; Trotz, M.A. A review of the mechanisms and modeling of photocatalytic disinfection. Appl. Catal. B-Environ. 2010, 98, 27–38. [Google Scholar] [CrossRef]
- Wu, W.; Qin, Y.; Fang, Y.; Zhang, Y.; Shao, S. Based on multi-omics technology study the antibacterial mechanisms of pH-dependent N-GQDs beyond ROS. J. Hazard Mater. 2023, 441, 129954. [Google Scholar] [CrossRef] [PubMed]
- Ju, X.; Fang, X.; Xiao, Y.; Li, B.; Shi, R. Small RNA GcvB Regulates Oxidative Stress Response of Escherichia coli. Antioxidants 2021, 10, 1774. [Google Scholar] [CrossRef] [PubMed]
- Ritz, D.; Patel, H.; Doan, B.; Zheng, M.; Aslund, F. Thioredoxin 2 is involved in the oxidative stress response in Escherichia coli. J. Biol. Chem. 2000, 275, 2505–2512. [Google Scholar] [CrossRef]
- Patil, S.; Valdramidis, V.P.; Karatzas, K.A.; Cullen, P.J.; Bourke, P. Assessing the microbial oxidative stress mechanism of ozone treatment through the responses of Escherichia coli mutants. J. Appl. Microbiol. 2011, 111, 136–144. [Google Scholar] [CrossRef]
- Imlay, J.A.; Linn, S.T. Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J. Bacteriol. 1987, 169, 2967–2976. [Google Scholar] [CrossRef]
- Oktyabrskii, O.N.; Muzyka, N.G.; Ushakov, V.Y.; Smirnova, G.V. The role of thiol redox systems in the peroxide stress response of Escherichia coli. Microbiology 2007, 76, 669–675. [Google Scholar] [CrossRef]
- Miethke, M.; Monteferrante, C.G.; Marahiel, M.A.; van Dijl, J.M. The Bacillus subtilis EfeUOB transporter is essential for high-affinity acquisition of ferrous and ferric iron. Biochim. Biophys. Acta 2013, 1833, 2267–2278. [Google Scholar] [CrossRef] [PubMed]
- Otero, V.; Vilarigues, M.; Carlyle, L.; Cotte, M.; De Nolf, W. A little key to oxalate formation in oil paints: Protective patina or chemical reactor? Photochem. Photobiol. Sci. 2018, 17, 266–270. [Google Scholar] [CrossRef]
- Kanai, T.; Takahashi, K.; Inoue, H. Three distinct-type glutathione S-transferases from Escherichia coli important for defense against oxidative stress. J. Biochem. 2006, 140, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Strom, A.R.; Kaasen, I. Trehalose metabolism in Escherichia coli: Stress protection and stress regulation of gene expression. Mol. Microbiol. 1993, 8, 205–210. [Google Scholar] [CrossRef]
- Jin, K.; Peng, G.; Liu, Y.; Xia, Y. The acid trehalase, ATM1, contributes to the in vivo growth and virulence of the entomopathogenic fungus, Metarhizium acridum. Fungal Genet. Biol. 2015, 77, 61–67. [Google Scholar] [CrossRef]
- Laursen, B.S.; de A Steffensen, S.A.; Hedegaard, J.; Moreno, J.M.; Mortensen, K.K. Structural requirements of the mRNA for intracistronic translation initiation of the enterobacterial infB gene. Genes Cells 2002, 7, 901–910. [Google Scholar] [CrossRef]
- Sims, J.; Benz, E.W. Initiation of DNA replication by the Escherichia coli dnaG protein: Evidence that tertiary structure is involved. Proc. Natl. Acad. Sci. USA 1980, 77, 900–904. [Google Scholar] [CrossRef]
- Uchida, K.H. Functional cooperation of the dnaE and dnaN gene products in Escherichia coli. Proc. Natl. Acad. Sci. USA 1981, 78, 5764–5767. [Google Scholar]
- Lin, S.; Zou, Z.; Zhou, C.; Zhang, H.; Cai, Z. Transcriptome Analysis Reveals the Molecular Mechanisms Underlying Adenosine Biosynthesis in Anamorph Strain of Caterpillar Fungus. BioMed Res. Int. 2019, 2019, 1864168. [Google Scholar] [CrossRef]
- Martinussen, H. Cloning and characterization of upp, a gene encoding uracil phosphoribosyltransferase from Lactococcus lactis. J. Bacteriol. 1994, 176, 6457–6463. [Google Scholar] [CrossRef] [PubMed]
- Husnain, S.I.; Busby, S.J.; Thomas, M.S. Downregulation of the Escherichia coli guaB promoter by upstream-bound cyclic AMP receptor protein. J. Bacteriol. 2009, 191, 6094–6104. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Weiss, B. YjjG, a dUMP phosphatase, is critical for thymine utilization by Escherichia coli K-12. J. Bacteriol. 2007, 189, 2186–2189. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gokey, T.; Halavaty, A.S.; Minasov, G.; Anderson, W.F.; Kuhn, M.L. Structure of the Bacillus anthracis dTDP-l-rhamnose biosynthetic pathway enzyme: dTDP-alpha-d-glucose 4,6-dehydratase, RfbB. J. Struct. Biol. 2018, 202, 175–181. [Google Scholar] [CrossRef]
- O’Connell, T.C. ‘Trophic’ and ‘source’ amino acids in trophic estimation: A likely metabolic explanation. Oecologia 2017, 184, 317–326. [Google Scholar] [CrossRef]
- Lamolle, G.; Simon, D.; Iriarte, A.; Musto, H. Main Factors Shaping Amino Acid Usage Across Evolution. J. Mol. Evol. 2023, 91, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.E. Sialic Acid Catabolism in Staphylococcus aureus. J. Bacteriol. 2013, 195, 1779–1788. [Google Scholar] [CrossRef]
- Virgil, H. A new access to alkyl-α-ketoglutaric acids, precursors of glutamic acid analogues by enzymatic transamination. Application to the synthesis of (2S,4R)-4-propyl-glutamic acid. Tetrahedron Lett. 1999, 40, 6577–6580. [Google Scholar]
- Fernandes, P.A.; Eriksson, L.A.; Ramos, M.J. Pyruvate formate lyase: A new perspective. J. Phys. Chem. B 2003, 107, 5751–5757. [Google Scholar]
- Stanley, W.M.; Salas, M.; Wahba, A.J. Translation of the genetic message: Factors involved in the initiation of protein synthesis. Proc. Natl. Acad. Sci. USA 1966, 56, 290–295. [Google Scholar] [CrossRef]
- Kaltschmidt, E.; Dzionara, M.; Wittmann, H.G. Ribosomal proteins. XV. Amino acid compositions of isolated ribosomal proteins from 30S and 50S subunits of Escherichia coli. Mol. Gen. Genet. 1970, 109, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Küster, C.; Piepersberg, W.; Distler, J. Cloning and transcriptional analysis of the rplKA-or f31-rplJL gene cluster of Streptomyces griseus. Mol. Gen. Genet. MGG 1998, 257, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Kondo, N.; Yoshida, M.; Nishiyama, M.; Kosono, S. Dynamic changes in lysine acetylation and succinylation of the elongation factor Tu in Bacillus subtilis. Microbiology 2019, 165, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Xu, J.; Jiao, L.; Liu, M.; Zhang, T. Acid adaptive response of Alicyclobacillus acidoterrestris: A strategy to survive lethal heat and acid stresses. Food Res. Int. 2022, 157, 111364. [Google Scholar] [CrossRef] [PubMed]
- Seebacher, F. Is Endothermy an Evolutionary By-Product? Trends Ecol. Evol. 2020, 35, 503–511. [Google Scholar] [CrossRef]
- Bulusu, V.; Aulehla, A. Metabolic Control of Cellular Differentiation. Dev. Cell 2016, 39, 286–287. [Google Scholar] [CrossRef]
- Chen, X.; Dong, X.; Liu, J.; Luo, Q.; Liu, L. Pathway engineering of Escherichia coli for alpha-ketoglutaric acid production. Biotechnol. Bioeng. 2020, 117, 2791–2801. [Google Scholar] [CrossRef] [PubMed]
- Usui, Y.; Hirasawa, T.; Furusawa, C. Investigating the effects of perturbations topgi and eno gene expression on central carbon metabolism in Escherichia coli using 13C metabolic flux analysis. Microb. Cell Factories 2012, 11, 87. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, Y.; Shen, Y.; Zhang, X.; Xu, S. Efficient production of androstenedione by repeated batch fermentation in waste cooking oil media through regulating NAD(+)/NADH ratio and strengthening cell vitality of Mycobacterium neoaurum. Bioresour. Technol. 2019, 279, 209–217. [Google Scholar] [CrossRef]
- Sousa, P.M.; Silva, S.T.; Hood, B.L.; Charro, N.; Carita, J.N. Supramolecular organizations in the aerobic respiratory chain of Escherichia coli. Biochimie 2011, 93, 418–425. [Google Scholar] [CrossRef][Green Version]
| CODcr (mg/L) | Total Bacteria (CFU/100 mL) | Escherichia coli (CFU/100 mL) | |
|---|---|---|---|
| Control | 1387.7 ± 6.11 A | 1501.3 ± 18.82 A | 160.3 ± 4.16 A |
| CPF-Fe | 244.3 ± 2.50 B | 302.7 ± 9.87 B | 2.3 ± 0.58 B |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diao, S.; Duan, Y.; Wang, M.; Feng, Y.; Miao, H.; Zhao, Y. Multi-Omics Study on Molecular Mechanisms of Single-Atom Fe-Doped Two-Dimensional Conjugated Phthalocyanine Framework for Photocatalytic Antibacterial Performance. Molecules 2024, 29, 1601. https://doi.org/10.3390/molecules29071601
Diao S, Duan Y, Wang M, Feng Y, Miao H, Zhao Y. Multi-Omics Study on Molecular Mechanisms of Single-Atom Fe-Doped Two-Dimensional Conjugated Phthalocyanine Framework for Photocatalytic Antibacterial Performance. Molecules. 2024; 29(7):1601. https://doi.org/10.3390/molecules29071601
Chicago/Turabian StyleDiao, Shihong, Yixin Duan, Mengying Wang, Yuanjiao Feng, Hong Miao, and Yongju Zhao. 2024. "Multi-Omics Study on Molecular Mechanisms of Single-Atom Fe-Doped Two-Dimensional Conjugated Phthalocyanine Framework for Photocatalytic Antibacterial Performance" Molecules 29, no. 7: 1601. https://doi.org/10.3390/molecules29071601
APA StyleDiao, S., Duan, Y., Wang, M., Feng, Y., Miao, H., & Zhao, Y. (2024). Multi-Omics Study on Molecular Mechanisms of Single-Atom Fe-Doped Two-Dimensional Conjugated Phthalocyanine Framework for Photocatalytic Antibacterial Performance. Molecules, 29(7), 1601. https://doi.org/10.3390/molecules29071601









