Neuroprotection of Truncated Peptide IIAVE from Isochrysis zhanjiangensis: Quantum Chemical, Molecular Docking, and Bioactivity Studies
Abstract
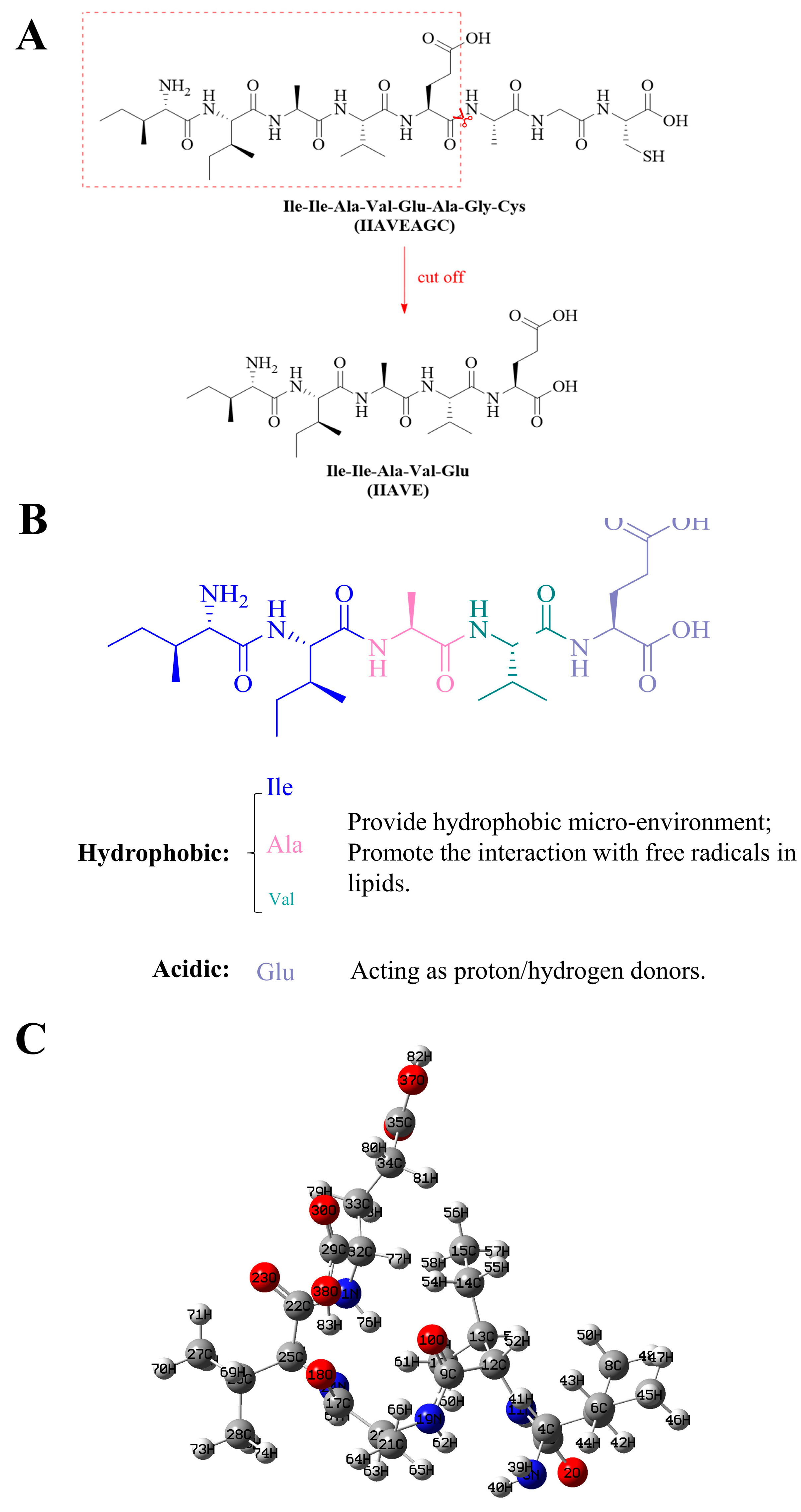
1. Introduction
2. Results
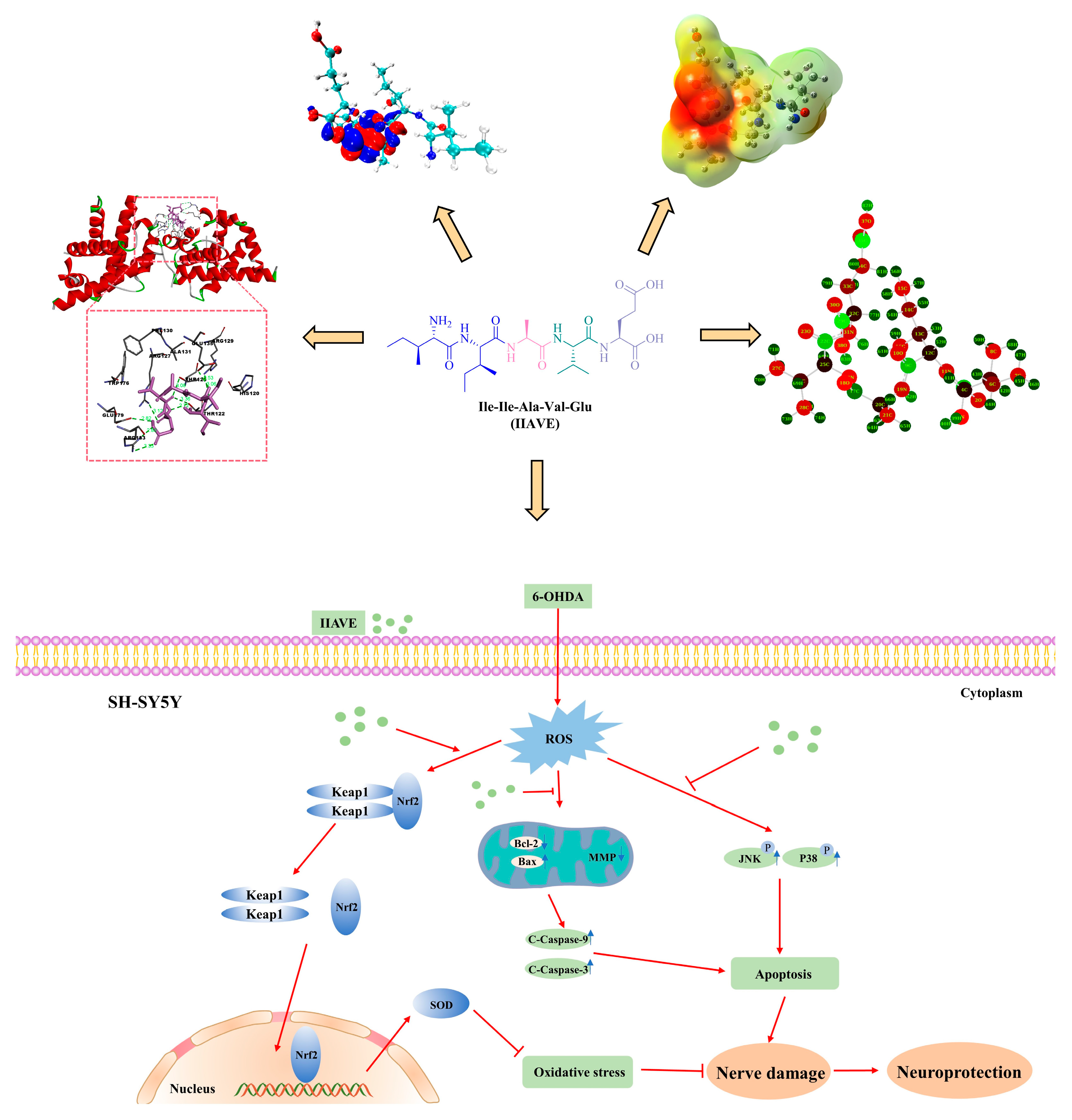
2.1. Molecular Docking Analysis
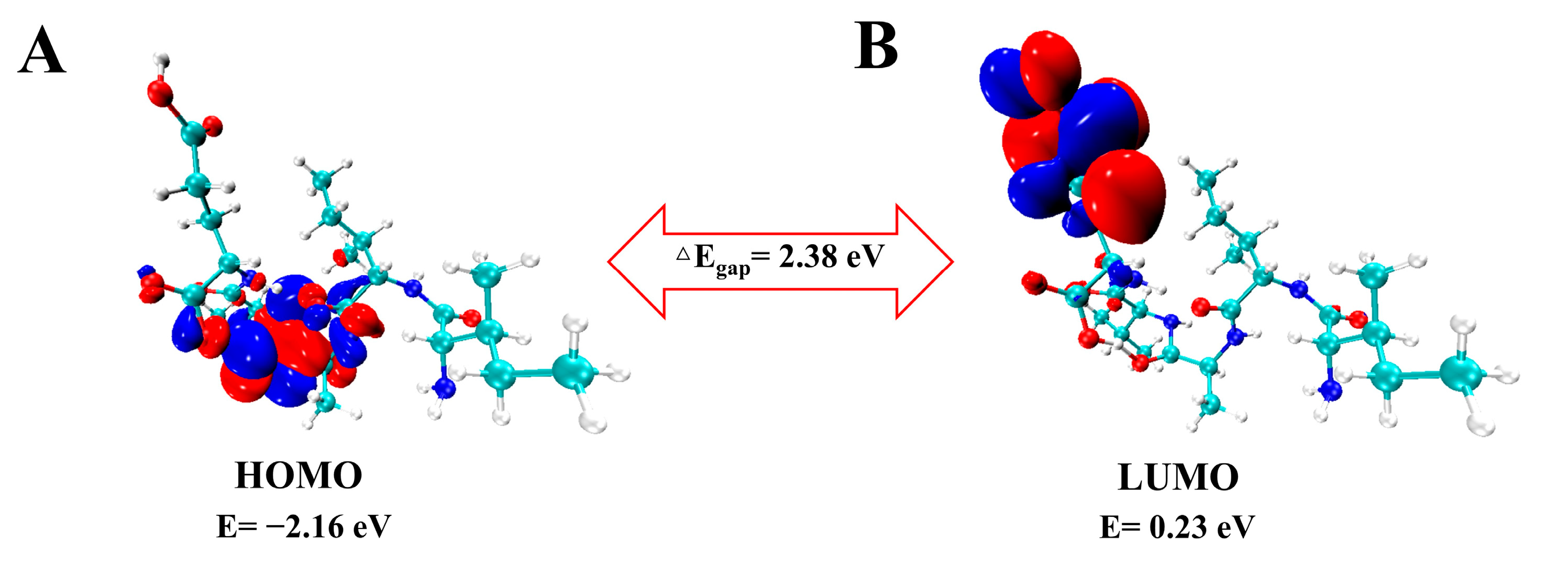
2.2. Frontier Molecular Orbital (FMO) and Chemical Reactivity
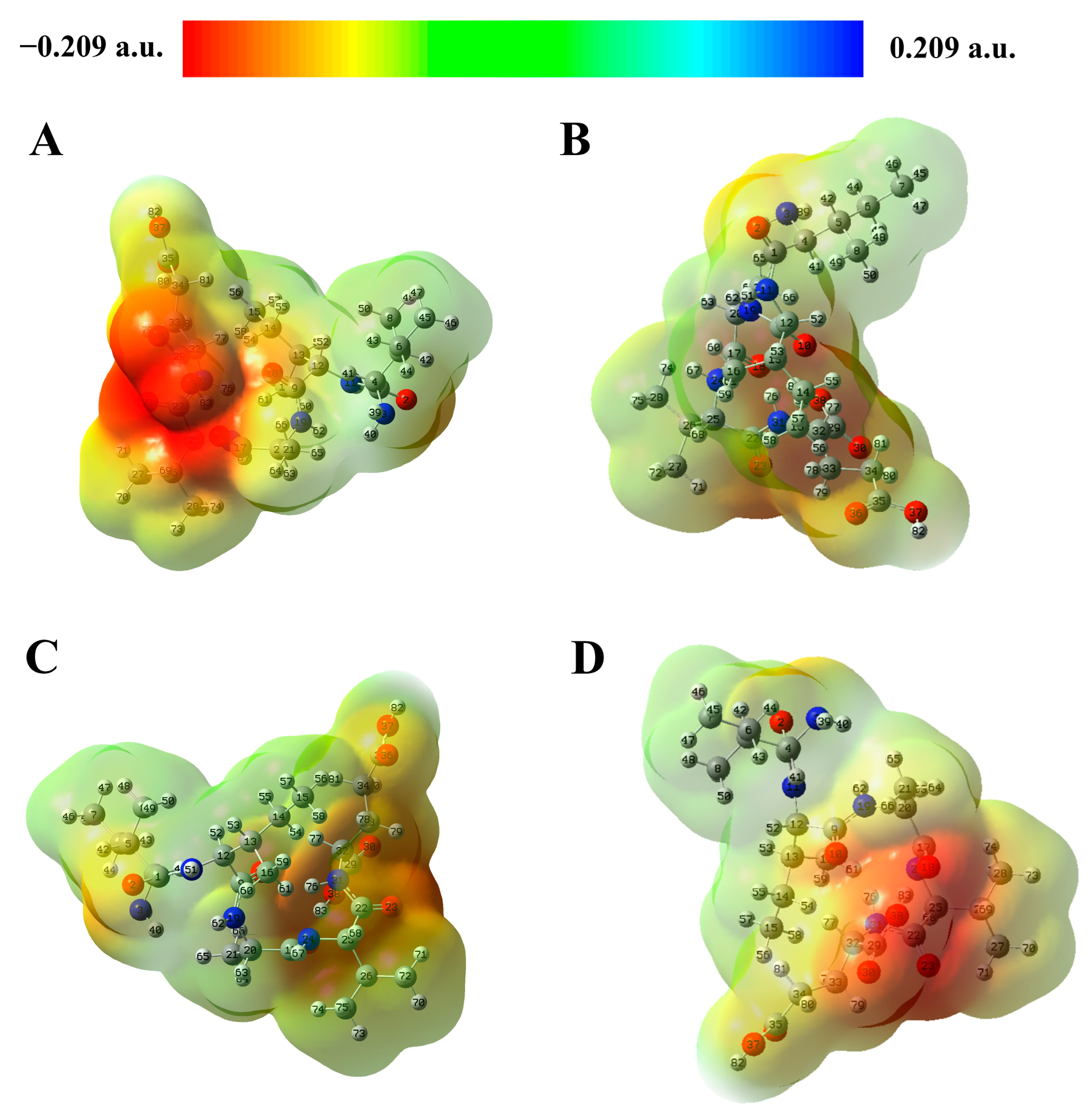
2.3. Molecular Electrostatic Potential (MEP) Analysis
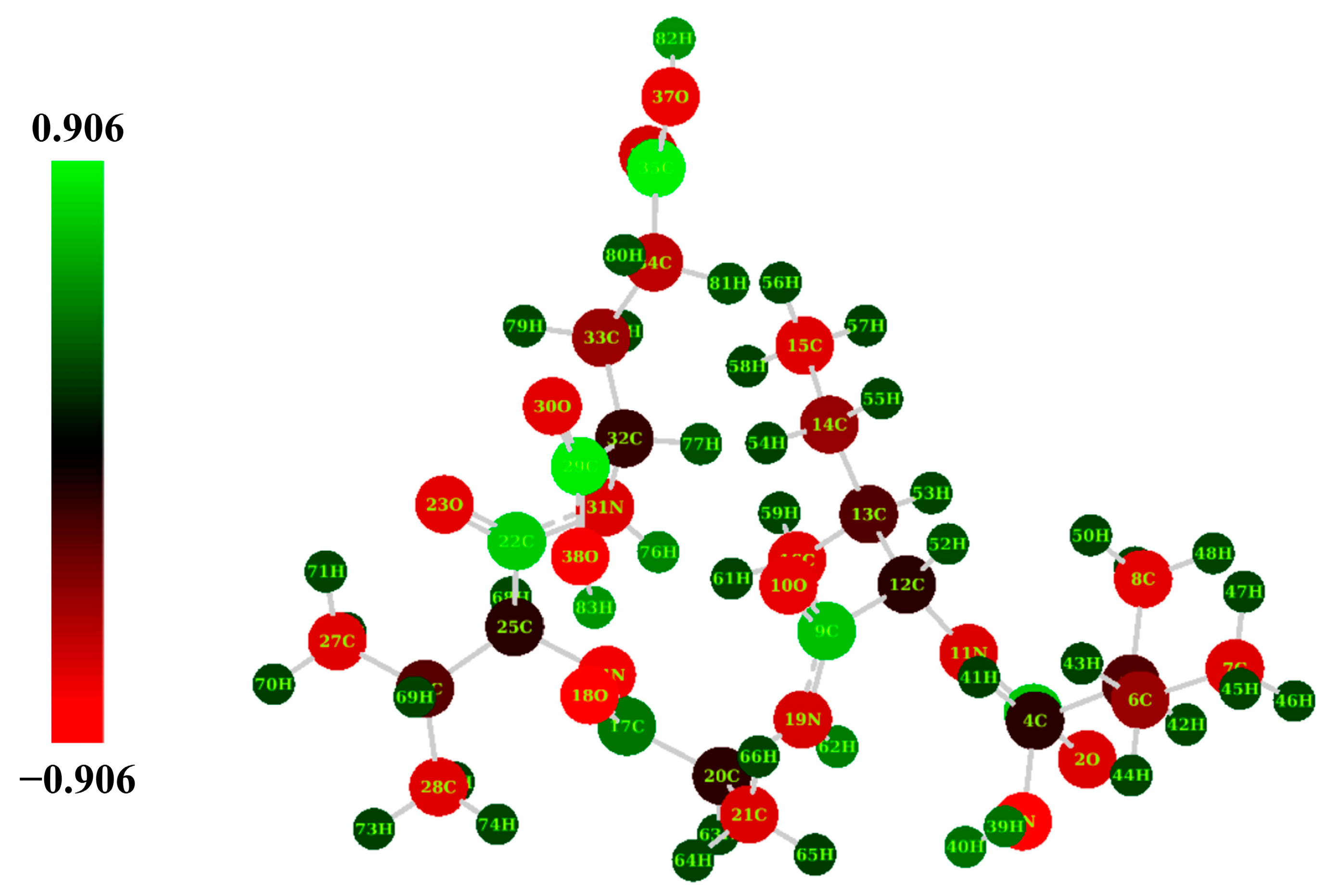
2.4. Natural Population Analysis (NPA)
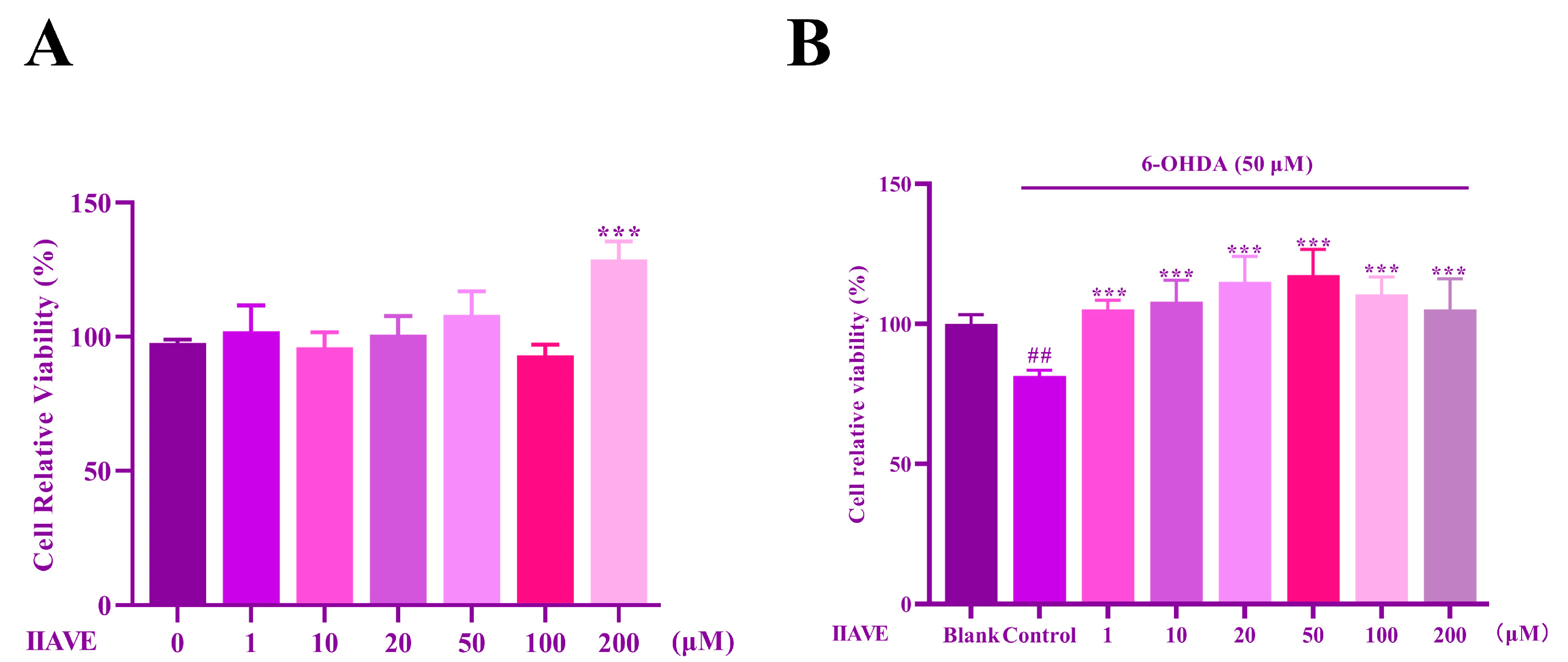
2.5. Effects of IIAVE and 6-OHDA on the Viability of SH-SY5Y Cells
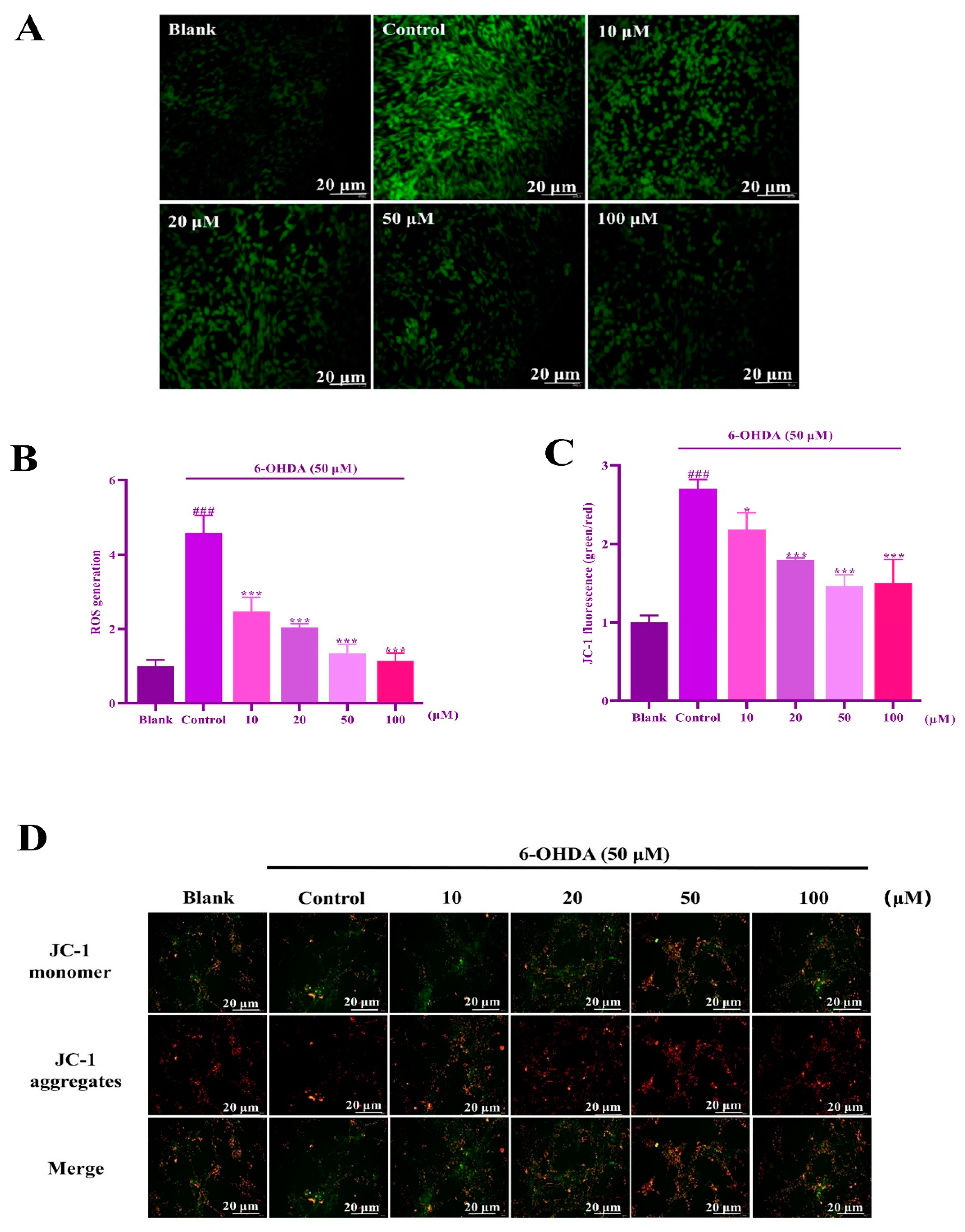
2.6. IIAVE Alleviates Mitochondrial Dysfunction
2.7. IIAVE Alleviates Oxidative Stress via the Keap1/Nrf2/SOD-1 Pathway
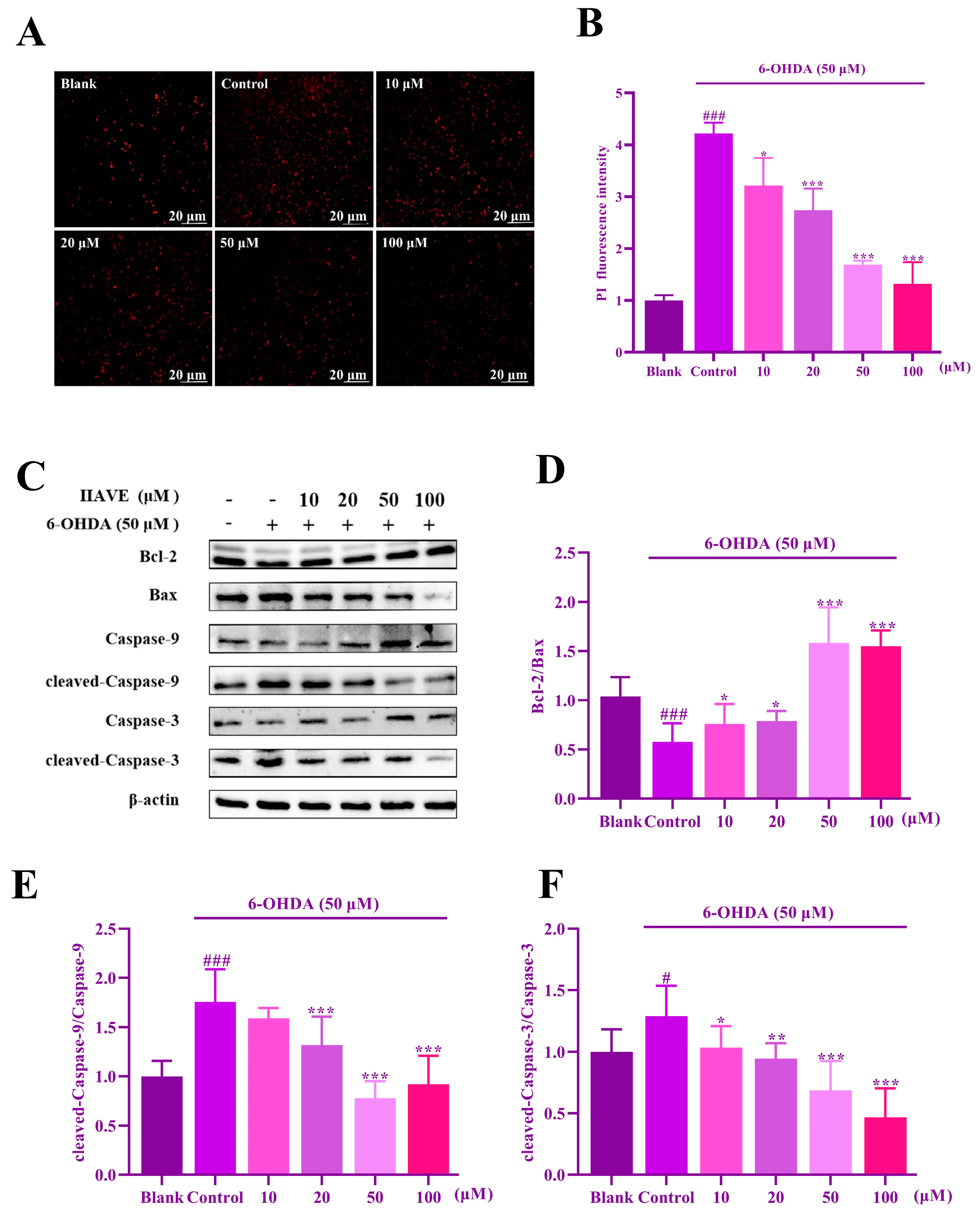
2.8. IIAVE Inhibits Apoptosis in SH-SY5Y Cells
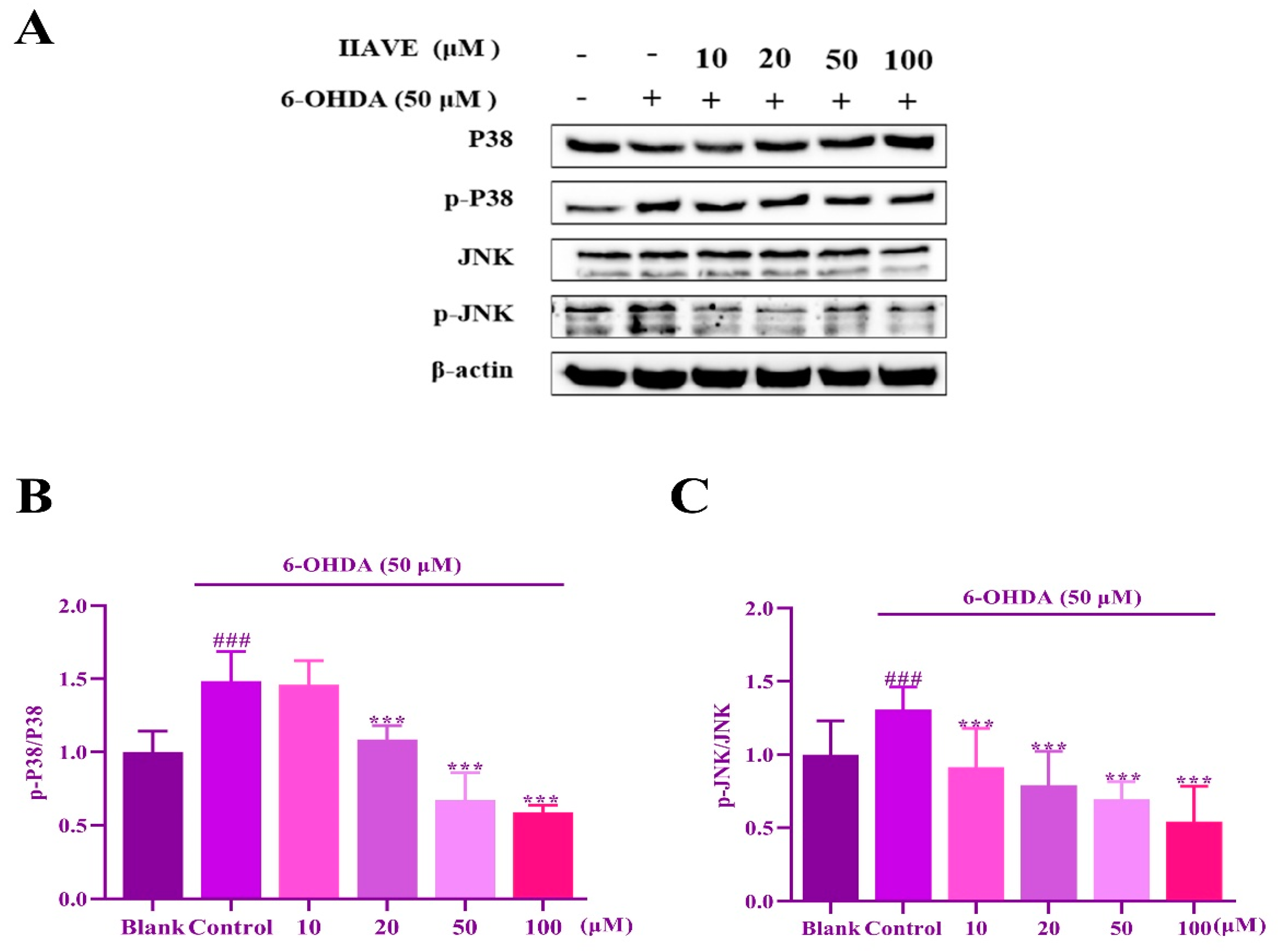
2.9. IIAVE Inhibit JNK and P38 Phosphorylation Elicited by 6-OHDA
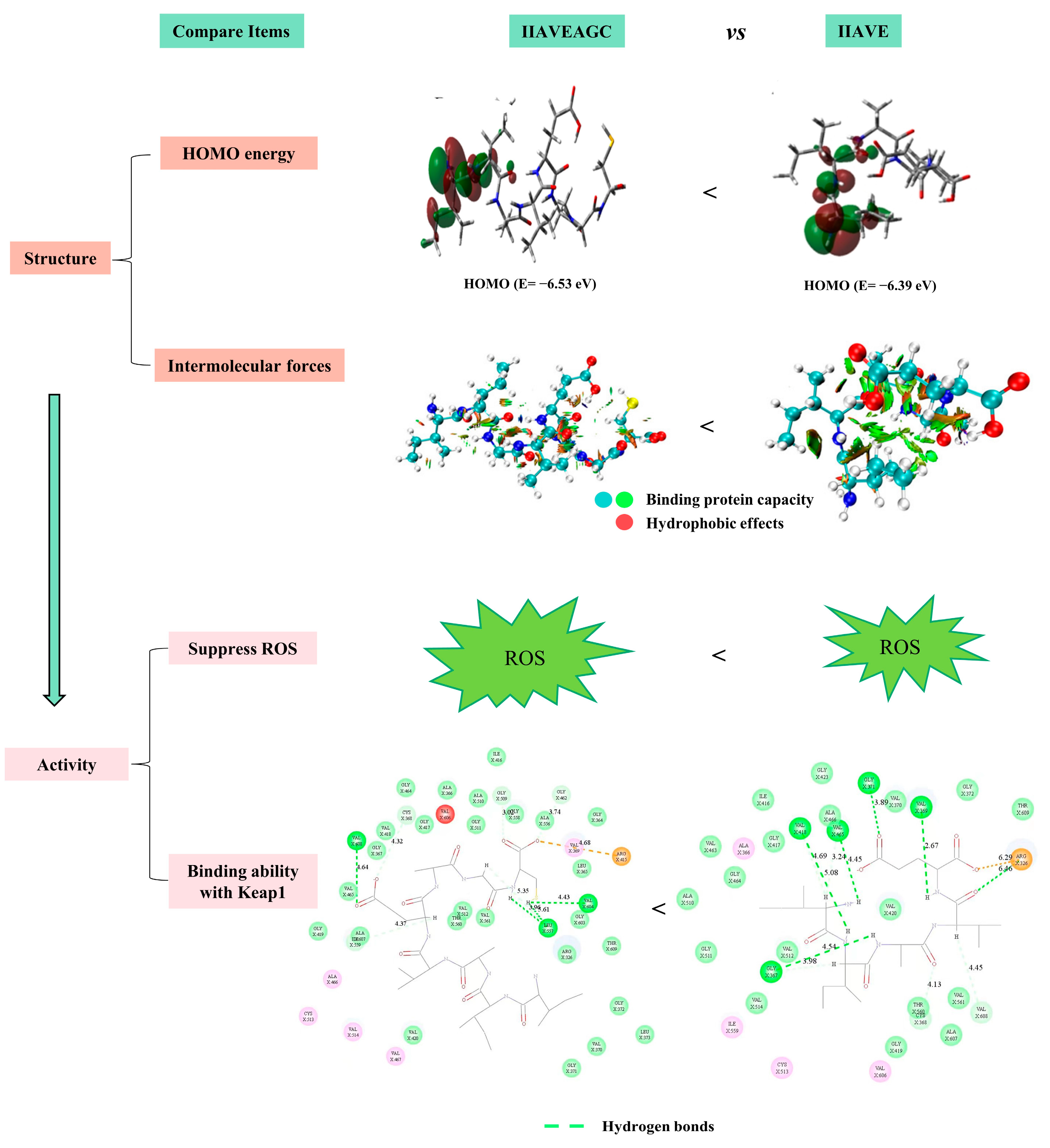
2.10. Comparison of Antioxidant Properties of Two Peptides
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Chemicals
4.2. Research Methods
4.2.1. Cell Viability and Toxicity Assay (CCK-8)
4.2.2. ROS Generation Detection
4.2.3. PI Apoptosis Analysis
4.2.4. Mitochondrial Membrane Potential Detection
4.2.5. Immunofluorescence
4.2.6. Western Blotting
4.2.7. Molecular Docking
4.2.8. Quantum Chemistry Research
4.2.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jankovic, J. Parkinson’s disease: Clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 2008, 79, 368. [Google Scholar] [CrossRef]
- Jenner, P. Oxidative stress in Parkinson’s disease. Ann. Neurol. 2003, 53 (Suppl. S3), S26–S36; discussion S36–S38. [Google Scholar] [CrossRef] [PubMed]
- Cadenas, E.; Davies, K.J.A. Mitochondrial free radical generation, oxidative stress, and aging11This article is dedicated to the memory of our dear friend, colleague, and mentor Lars Ernster (1920–1998), in gratitude for all he gave to us. Free Radic. Biol. Med. 2000, 29, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, S.M.; Pereira, C.; Oliveira, C.R. Mitochondrial function is differentially affected upon oxidative stress. Free Radic. Biol. Med. 1999, 26, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Cecarini, V.; Gee, J.; Fioretti, E.; Amici, M.; Angeletti, M.; Eleuteri, A.M.; Keller, J.N. Protein oxidation and cellular homeostasis: Emphasis on metabolism. Biochim. Et Biophys. Acta (BBA) Mol. Cell Res. 2007, 1773, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.D.; Zhao, X.; Li, Y.; Li, G.R.; Liu, X.L. Damage to dopaminergic neurons by oxidative stress in Parkinson’s disease (Review). Int. J. Mol. Med. 2018, 41, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Mythri, R.B.; Venkateshappa, C.; Harish, G.; Mahadevan, A.; Muthane, U.B.; Yasha, T.C.; Srinivas Bharath, M.M.; Shankar, S.K. Evaluation of Markers of Oxidative Stress, Antioxidant Function and Astrocytic Proliferation in the Striatum and Frontal Cortex of Parkinson’s Disease Brains. Neurochem. Res. 2011, 36, 1452–1463. [Google Scholar] [CrossRef] [PubMed]
- Hauser, D.N.; Hastings, T.G. Mitochondrial dysfunction and oxidative stress in Parkinson’s disease and monogenic parkinsonism. Neurobiol. Dis. 2013, 51, 35–42. [Google Scholar] [CrossRef]
- Perfeito, R.; Cunha-Oliveira, T.; Rego, A.C. Revisiting oxidative stress and mitochondrial dysfunction in the pathogenesis of Parkinson disease-resemblance to the effect of amphetamine drugs of abuse. Free Radic. Biol. Med. 2012, 53, 1791–1806. [Google Scholar] [CrossRef]
- Simola, N.; Morelli, M.; Carta, A.R. The 6-Hydroxydopamine model of parkinson’s disease. Neurotoxic. Res. 2007, 11, 151–167. [Google Scholar] [CrossRef]
- Garver, D.L.; Cedarbaum, J.; Maas, J.W. Blood-brain barrier to 6-hydroxydopamine: Uptake by heart and brain. Life Sci. 1975, 17, 1081–1084. [Google Scholar] [CrossRef]
- Perier, C.; Vila, M. Mitochondrial biology and Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012, 2, a009332. [Google Scholar] [CrossRef]
- Cohen, G. Oxidative stress, mitochondrial respiration, and Parkinson’s disease. Ann. N. Y. Acad. Sci. 2000, 899, 112–120. [Google Scholar] [CrossRef]
- Blandini, F.; Armentero, M.-T.; Martignoni, E. The 6-hydroxydopamine model: News from the past. Park. Relat. Disord. 2008, 14, S124–S129. [Google Scholar] [CrossRef]
- Choi, D.; Hwang, S.; Lee, E.; Yoon, S.; Yoon, B.K.; Bae, D. Expression of mitochondria-dependent apoptosis genes (p53, Bax, and Bcl-2) in rat granulosa cells during follicular development. J. Soc. Gynecol. Investig. 2004, 11, 311–317. [Google Scholar] [CrossRef]
- Choi, H.J.; Lee, S.Y.; Cho, Y.; Hwang, O. JNK activation by tetrahydrobiopterin: Implication for Parkinson’s disease. J. Neurosci. Res. 2004, 75, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Kahle, P.J.; Waak, J.; Gasser, T. DJ-1 and prevention of oxidative stress in Parkinson’s disease and other age-related disorders. Free Radic. Biol. Med. 2009, 47, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Orth, M.; Schapira, A. Mitochondrial involvement in Parkinson’s disease. Neurochem. Int. 2002, 40, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Giaime, E.; Yamaguchi, H.; Gautier, C.A.; Kitada, T.; Shen, J. Loss of DJ-1 does not affect mitochondrial respiration but increases ROS production and mitochondrial permeability transition pore opening. PLoS ONE 2012, 7, e40501. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, J.; Chen, S.; Wang, Y.; Cao, L.; Zhang, Y.; Kang, W.; Li, H.; Gui, Y.; Chen, S. DJ-1 modulates the expression of Cu/Zn-superoxide dismutase-1 through the Erk1/2–Elk1 pathway in neuroprotection. Ann. Neurol. 2011, 70, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Klintworth, H.; Newhouse, K.; Li, T.; Choi, W.S.; Faigle, R.; Xia, Z. Activation of c-Jun N-Terminal Protein Kinase Is a Common Mechanism Underlying Paraquat- and Rotenone-Induced Dopaminergic Cell Apoptosis. Toxicol. Sci. 2007, 97, 149–162. [Google Scholar] [CrossRef]
- Korsmeyer, S.J.; Gross, A.; Harada, H.; Zha, J.; Zinkel, S. Death and Survival Signals Determine Active/Inactive Conformations of Pro-apoptotic BAX, BAD, and BID Molecules. Cold Spring Harb. Symp. Quant. Biol. 1999, 64, 343–350. [Google Scholar] [CrossRef]
- Deepika, N.P.; Rahman, H.M.; Chipurupalli, S.; Shilpa, T.N.; Duraiswamy, B. The Emerging Role of Marine Natural Products for the Treatment of Parkinson’s Disease. CNS Neurol. Disord. Drug Targets 2023, 22, 801–816. [Google Scholar] [CrossRef]
- Miyashita, K. Marine antioxidants: Polyphenols and carotenoids from algae. In Antioxidant and Funcional. Components in Aquatic. Food; Wiley Online Library: West Sussex, UK, 2014; pp. 219–235. [Google Scholar] [CrossRef]
- Li, Y.; Qian, Z.J.; Ryu, B.; Lee, S.H.; Kim, M.M.; Kim, S.K. Chemical components and its antioxidant properties in vitro: An edible marine brown alga, Ecklonia cava. Bioorg. Med. Chem. 2009, 17, 1963–1973. [Google Scholar] [CrossRef] [PubMed]
- Matos, J.; Cardoso, C.; Bandarra, N.M.; Afonso, C. Microalgae as healthy ingredients for functional food: A review. Food Funct. 2017, 8, 2672–2685. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tan, L.; Li, C.; Zhou, C.; Hong, P.; Sun, S.; Qian, Z.J. Mechanism analysis of a novel angiotensin-I-converting enzyme inhibitory peptide from Isochrysis zhanjiangensis microalgae for suppressing vascular injury in human umbilical vein endothelial cells. J. Agric. Food Chem. 2020, 68, 4411–4423. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; He, Y.-L.; Tang, Y.; Hong, P.; Zhou, C.; Sun, S.; Qian, Z.J. Mechanism analysis of octapeptide from microalgae, Isochrysis zhanjiangensis for suppressing vascular injury and angiogenesis in human umbilical vein endothelial cell. Int. Immunopharmacol. 2022, 111, 109149. [Google Scholar] [CrossRef]
- Pei, Y.; Lui, Y.; Cai, S.; Zhou, C.; Hong, P.; Qian, Z.J. A novel peptide isolated from microalgae Isochrysis zhanjiangensis exhibits anti-apoptosis and anti-inflammation in ox-LDL induced HUVEC to improve atherosclerosis. Plant Foods Hum. Nutr. 2022, 77, 181–189. [Google Scholar] [CrossRef]
- He, Y.L.; Lin, L.; Zheng, H.; Mo, Y.; Zhou, C.; Sun, S.; Hong, P.; Qian, Z.J. Potential anti-skin aging effect of a peptide AYAPE isolated from Isochrysis zhanjiangensis on UVB-induced HaCaT cells and H2O2-induced BJ cells. J. Photochem. Photobiol. B Biol. 2022, 233, 112481. [Google Scholar] [CrossRef]
- Chen, M.F.; Zhang, Y.Y.; Di He, M.; Li, C.Y.; Zhou, C.X.; Hong, P.Z.; Qian, Z.J. Antioxidant Peptide Purified from Enzymatic Hydrolysates of Isochrysis Zhanjiangensis and Its Protective Effect against Ethanol Induced Oxidative Stress of HepG2 Cells. Biotechnol. Bioprocess Eng. 2019, 24, 308–317. [Google Scholar] [CrossRef]
- Sun, J.; He, H.; Xie, B.J. Novel antioxidant peptides from fermented mushroom Ganoderma lucidum. J. Agric. Food Chem. 2004, 52, 6646–6652. [Google Scholar] [CrossRef]
- Meisel, H.; FitzGerald, R.J. Biofunctional peptides from milk proteins: Mineral binding and cytomodulatory effects. Curr. Pharm. Des. 2003, 9, 1289–1296. [Google Scholar] [PubMed]
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Peptides 2010, 31, 1949–1956. [Google Scholar] [CrossRef] [PubMed]
- Elias, R.J.; Kellerby, S.S.; Decker, E.A. Antioxidant Activity of Proteins and Peptides. Crit. Rev. Food Sci. Nutr. 2008, 48, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Power, O.; Jakeman, P.; FitzGerald, R.J. Antioxidative peptides: Enzymatic production, in vitro and in vivo antioxidant activity and potential applications of milk-derived antioxidative peptides. Amino Acids 2013, 44, 797–820. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; He, Y.L.; Liu, Y.; Hong, P.; Zhou, C.; Sun, S.; Qian, Z.J. Comparative in silico and in vitro study of the stability and biological activity of an octapeptide from microalgae Isochrysis zhanjiangensis and its truncated short peptide. Food Funct. 2023, 14, 3659–3672. [Google Scholar] [CrossRef] [PubMed]
- Emamian, S.; Lu, T.; Kruse, H.; Emamian, H. Exploring nature and predicting strength of hydrogen bonds: A correlation analysis between atoms-in-molecules descriptors, binding energies, and energy components of symmetry-adapted perturbation theory. J. Comput. Chem. 2019, 40, 2868–2881. [Google Scholar] [CrossRef] [PubMed]
- Benayahoum, A.; Amira-Guebailia, H.; Houache, O. A DFT method for the study of the antioxidant action mechanism of resveratrol derivatives. J. Mol. Model. 2013, 19, 2285–2298. [Google Scholar] [CrossRef] [PubMed]
- Barbouchi, M.; Benzidia, B.; Aouidate, A.; Ghaleb, A.; El Idrissi, M.; Choukrad, M. Theoretical modeling and experimental studies of Terebinth extracts as green corrosion inhibitor for iron in 3% NaCl medium. J. King Saud. Univ. Sci. 2020, 32, 2995–3004. [Google Scholar] [CrossRef]
- Koopmans, T. Ordering of wave functions and eigenenergies to the individual electrons of an atom. Physica 1933, 1, 104–113. [Google Scholar] [CrossRef]
- Jordaan, M.A.; Ebenezer, O.; Mthiyane, K.; Damoyi, N.; Shapi, M. Amide imidic prototropic tautomerization of efavirenz, NBO analysis, hyperpolarizability, polarizability and HOMO–LUMO calculations using density functional theory. Comput. Theor. Chem. 2021, 1201, 113273. [Google Scholar] [CrossRef]
- Alyar, S.; Şen, T.; Özmen, Ü.Ö.; Alyar, H.; Adem, Ş.; Şen, C. Synthesis, spectroscopic characterizations, enzyme inhibition, molecular docking study and DFT calculations of new Schiff bases of sulfa drugs. J. Mol. Struct. 2019, 1185, 416–424. [Google Scholar] [CrossRef]
- Shahab, S.; Sheikhi, M.; Filippovich, L.; Alnajjar, R.; Ihnatovich, Z.; Laznev, K.; Strogova, A.; Atroshko, M.; Drachilovskaya, M. Quantum-chemical modeling, spectroscopic (FT-IR, excited states, UV/Vis, polarization, and Dichroism) studies of two new benzo[d]oxazole derivatives. J. Mol. Struct. 2020, 1202, 127352. [Google Scholar] [CrossRef]
- Li, L.; Wu, C.; Wang, Z.; Zhao, L.; Li, Z.; Sun, C.; Sun, T. Density functional theory (DFT) and natural bond orbital (NBO) study of vibrational spectra and intramolecular hydrogen bond interaction of l-ornithine–l-aspartate. Spectrochim. Acta Part A 2015, 136, 338–346. [Google Scholar] [CrossRef]
- Weinhold, F.; Landis, C.R. Natural bond orbitals and extensions of localized bonding concepts. Chem. Educ. Res. Pract. 2001, 2, 91–104. [Google Scholar] [CrossRef]
- Arunan, E.; Desiraju, G.R.; Klein, R.A.; Sadlej, J.; Scheiner, S.; Alkorta, I.; Clary, D.C.; Crabtree, R.H.; Dannenberg, J.J.; Hobza, P. Definition of the hydrogen bond (IUPAC Recommendations 2011). Pure Appl. Chem. 2011, 83, 1637–1641. [Google Scholar] [CrossRef]
- Blum, D.; Torch, S.; Lambeng, N.; Nissou, M.F.; Benabid, A.L.; Sadoul, R.; Verna, J.M. Molecular pathways involved in the neurotoxicity of 6-OHDA, dopamine and MPTP: Contribution to the apoptotic theory in Parkinson’s disease. Prog. Neurobiol. 2001, 65, 135–172. [Google Scholar] [CrossRef]
- Huang, L.; Li, Y.; Lin, Q.; Lou, B.; Chen, Y. Enantioselective permeations of amino acids through l-proline-modified gold nanochannel membrane: An experimental and theoretical study. Amino Acids 2018, 50, 1549–1556. [Google Scholar] [CrossRef]
- Kim, S.K.; Mendis, E. Bioactive compounds from marine processing byproducts—A review. Food Res. Int. 2006, 39, 383–393. [Google Scholar] [CrossRef]
- Sun, Y.X.; Hao, Q.L.; Wei, W.X.; Yu, Z.X.; Lu, L.D.; Wang, X.; Wang, Y.S. Experimental and density functional studies on 4-(3,4-dihydroxybenzylideneamino)antipyrine, and 4-(2,3,4-trihydroxybenzylideneamino)antipyrine. J. Mol. Struct. THEOCHEM 2009, 904, 74–82. [Google Scholar] [CrossRef]
- Freeman, S.; Grinstein, S.; Orlowski, J. Determinants, Maintenance and Function of Organellar pH. Physiol. Rev. 2022, 103, 515–606. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Yuan, W.; Liu, F.; Feng, J.; Guo, Y. Acylated ghrelin is protective against 6-OHDA-induced neurotoxicity by regulating autophagic flux. Front. Pharmacol. 2021, 11, 586302. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.C.; Liu, W.Y.; Liou, S.S.; Liu, I.M. p-Hydroxybenzyl Alcohol Antagonized the ROS-Dependent JNK/Jun/Caspase-3 Pathway to Produce Neuroprotection in a Cellular Model of Parkinson’s Disease. Nutrients 2022, 14, 5002. [Google Scholar] [PubMed]
- Chen, Q.X.; Zhou, L.; Long, T.; Qin, D.L.; Wang, Y.L.; Ye, Y.; Zhou, X.G.; Wu, J.M.; Wu, A.G. Galangin Exhibits Neuroprotective Effects in 6-OHDA-Induced Models of Parkinson’s Disease via the Nrf2/Keap1 Pathway. Pharmaceuticals 2022, 15, 1014. [Google Scholar] [PubMed]
- Sun, Y.; He, L.; Wang, T.; Hua, W.; Qin, H.; Wang, J.; Wang, L.; Gu, W.; Li, T.; Li, N.; et al. Activation of p62-Keap1-Nrf2 Pathway Protects 6-Hydroxydopamine-Induced Ferroptosis in Dopaminergic Cells. Mol. Neurobiol. 2020, 57, 4628–4641. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cui, W.; Li, G.; Yuan, S.; Xu, D.; Hoi, M.P.M.; Lin, Z.; Dou, J.; Han, Y.; Lee, S.M.Y. Baicalein Protects against 6-OHDA-Induced Neurotoxicity through Activation of Keap1/Nrf2/HO-1 and Involving PKCα and PI3K/AKT Signaling Pathways. J. Agric. Food Chem. 2012, 60, 8171–8182. [Google Scholar] [CrossRef] [PubMed]
- Ha Kim, K.; Sadikot, R.T.; Yeon Lee, J.; Jeong, H.S.; Oh, Y.K.; Blackwell, T.S.; Joo, M. Suppressed ubiquitination of Nrf2 by p47phox contributes to Nrf2 activation. Free Radic. Biol. Med. 2017, 113, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Di Muzio, E.; Toti, D.; Polticelli, F. DockingApp: A user friendly interface for facilitated docking simulations with AutoDock Vina. J. Comput.-Aided Mol. Des. 2017, 31, 213–218. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Akhtari, K.; Hassanzadeh, K.; Fakhraei, B.; Fakhraei, N.; Hassanzadeh, H.; Zarei, S.A. A density functional theory study of the reactivity descriptors and antioxidant behavior of Crocin. Comput. Theor. Chem. 2013, 1013, 123–129. [Google Scholar] [CrossRef]
- Ouaket, A.; Chraka, A.; Raissouni, I.; Amrani, M.A.E.; Berrada, M.; Knouzi, N. Synthesis, spectroscopic (13C/1H-NMR, FT-IR) investigations, quantum chemical modelling (FMO, MEP, NBO analysis), and antioxidant activity of the bis-benzimidazole molecule. J. Mol. Struct. 2022, 1259, 132729. [Google Scholar] [CrossRef]
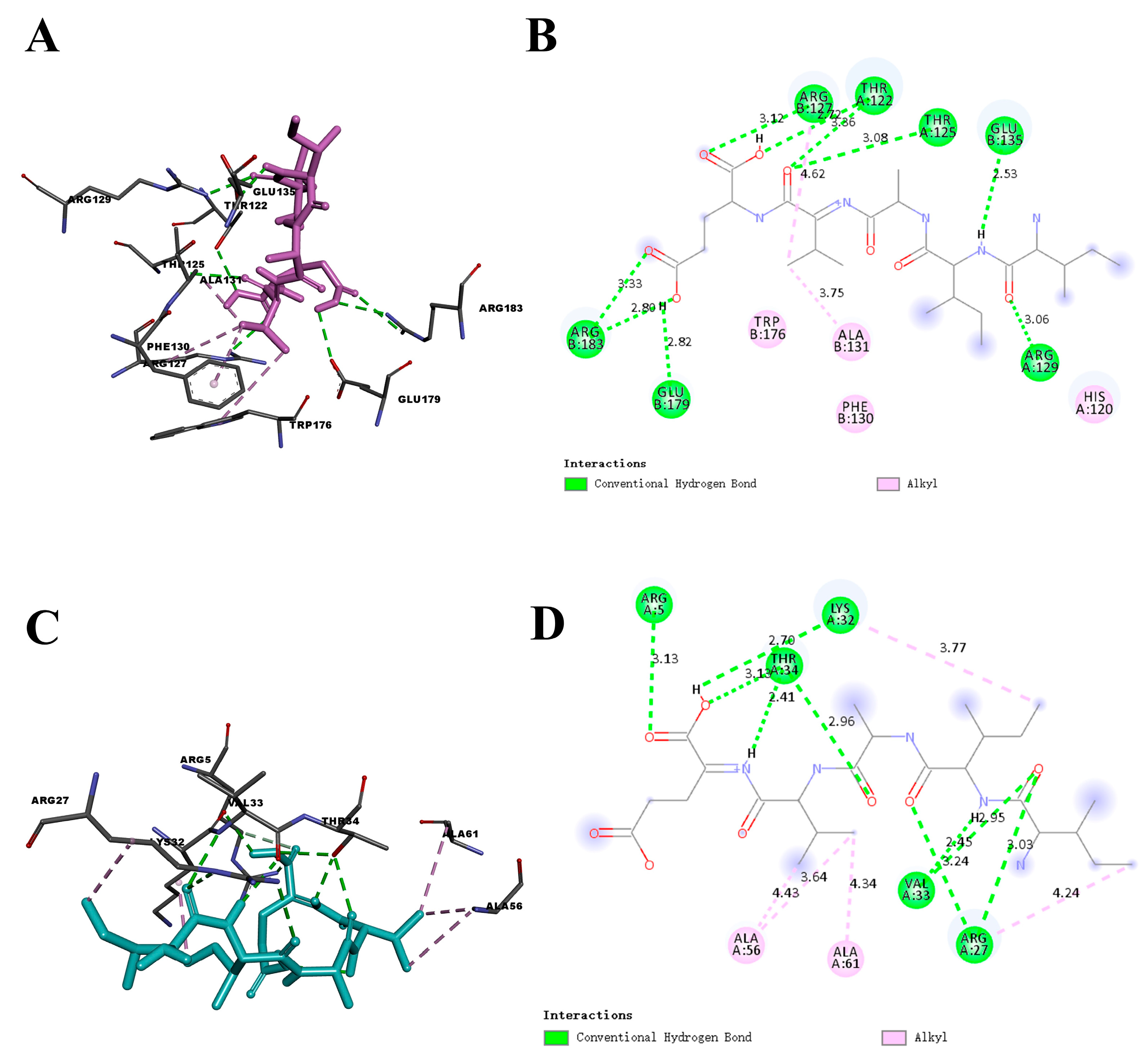

| Protein | Residue | Interaction Type | Distance (Å) | Protein | Residue | Interaction Type | Distance (Å) |
|---|---|---|---|---|---|---|---|
| Bcl-2 | Arg-183 | H-Bond | 2.80 | DJ-1 | Arg-27 | H-Bond | 3.03 |
| Arg-183 | H-Bond | 3.33 | Val-33 | H-Bond | 2.95 | ||
| Glu-179 | H-Bond | 2.82 | Arg-27 | H-Bond | 3.24 | ||
| Arg-127 | H-Bond | 3.12 | Val-33 | H-Bond | 2.45 | ||
| Thr-125 | H-Bond | 3.08 | Lys-32 | H-Bond | 2.70 | ||
| Thr-122 | H-Bond | 2.72 | Arg-5 | H-Bond | 3.13 | ||
| Thr-122 | H-Bond | 3.36 | Thr-34 | H-Bond | 2.41 | ||
| Arg-129 | H-Bond | 3.06 | Thr-34 | H-Bond | 2.96 | ||
| Glu-135 | H-Bond | 2.53 | Ala-61 | Hydrophobic | 4.34 | ||
| Arg-127 | Hydrophobic | 4.62 | Ala-56 | Hydrophobic | 4.43 | ||
| Ala-131 | Hydrophobic | 3.75 | Ala-56 | Hydrophobic | 3.64 | ||
| Lys-32 | Hydrophobic | 3.77 | |||||
| Arg-27 | Hydrophobic | 4.24 |
| Parameter (eV) | Calculated Result |
|---|---|
| EHOMO | −2.16 |
| ELUMO | 0.23 |
| ΔEgap | 2.38 |
| I | 2.16 |
| A | −0.23 |
| χ | 0.97 |
| µ | −0.97 |
| η | 1.19 |
| ω | 0.40 |
| S | 0.40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Lin, L.; Li, H.; Qian, Z.-J. Neuroprotection of Truncated Peptide IIAVE from Isochrysis zhanjiangensis: Quantum Chemical, Molecular Docking, and Bioactivity Studies. Molecules 2024, 29, 692. https://doi.org/10.3390/molecules29030692
Liu Q, Lin L, Li H, Qian Z-J. Neuroprotection of Truncated Peptide IIAVE from Isochrysis zhanjiangensis: Quantum Chemical, Molecular Docking, and Bioactivity Studies. Molecules. 2024; 29(3):692. https://doi.org/10.3390/molecules29030692
Chicago/Turabian StyleLiu, Qiuqi, Liyuan Lin, Huijuan Li, and Zhong-Ji Qian. 2024. "Neuroprotection of Truncated Peptide IIAVE from Isochrysis zhanjiangensis: Quantum Chemical, Molecular Docking, and Bioactivity Studies" Molecules 29, no. 3: 692. https://doi.org/10.3390/molecules29030692
APA StyleLiu, Q., Lin, L., Li, H., & Qian, Z.-J. (2024). Neuroprotection of Truncated Peptide IIAVE from Isochrysis zhanjiangensis: Quantum Chemical, Molecular Docking, and Bioactivity Studies. Molecules, 29(3), 692. https://doi.org/10.3390/molecules29030692






