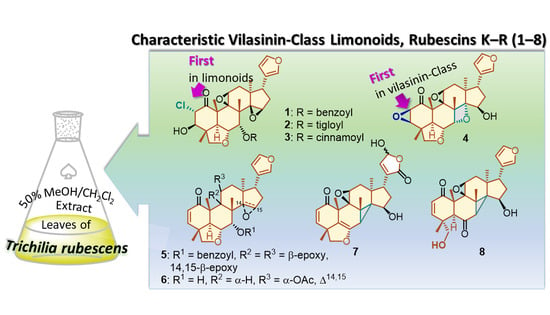
Unusual Vilasinin-Class Limonoids from Trichilia rubescens
Abstract
1. Introduction
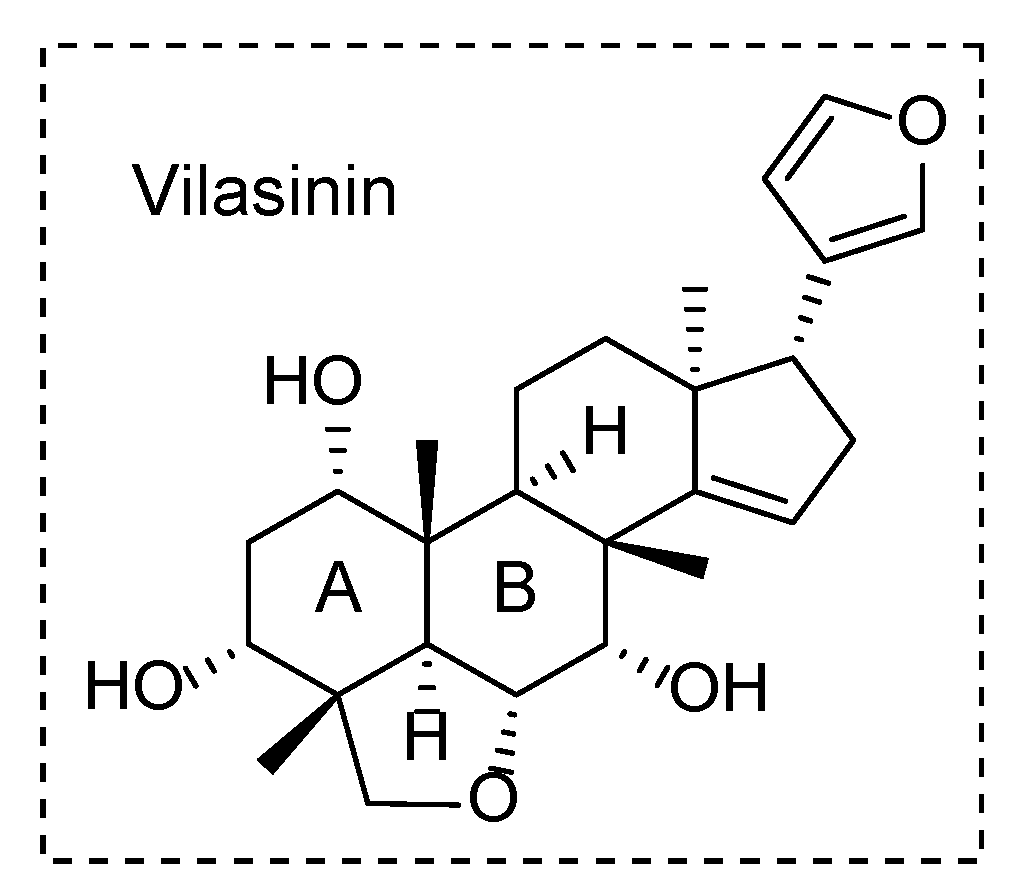
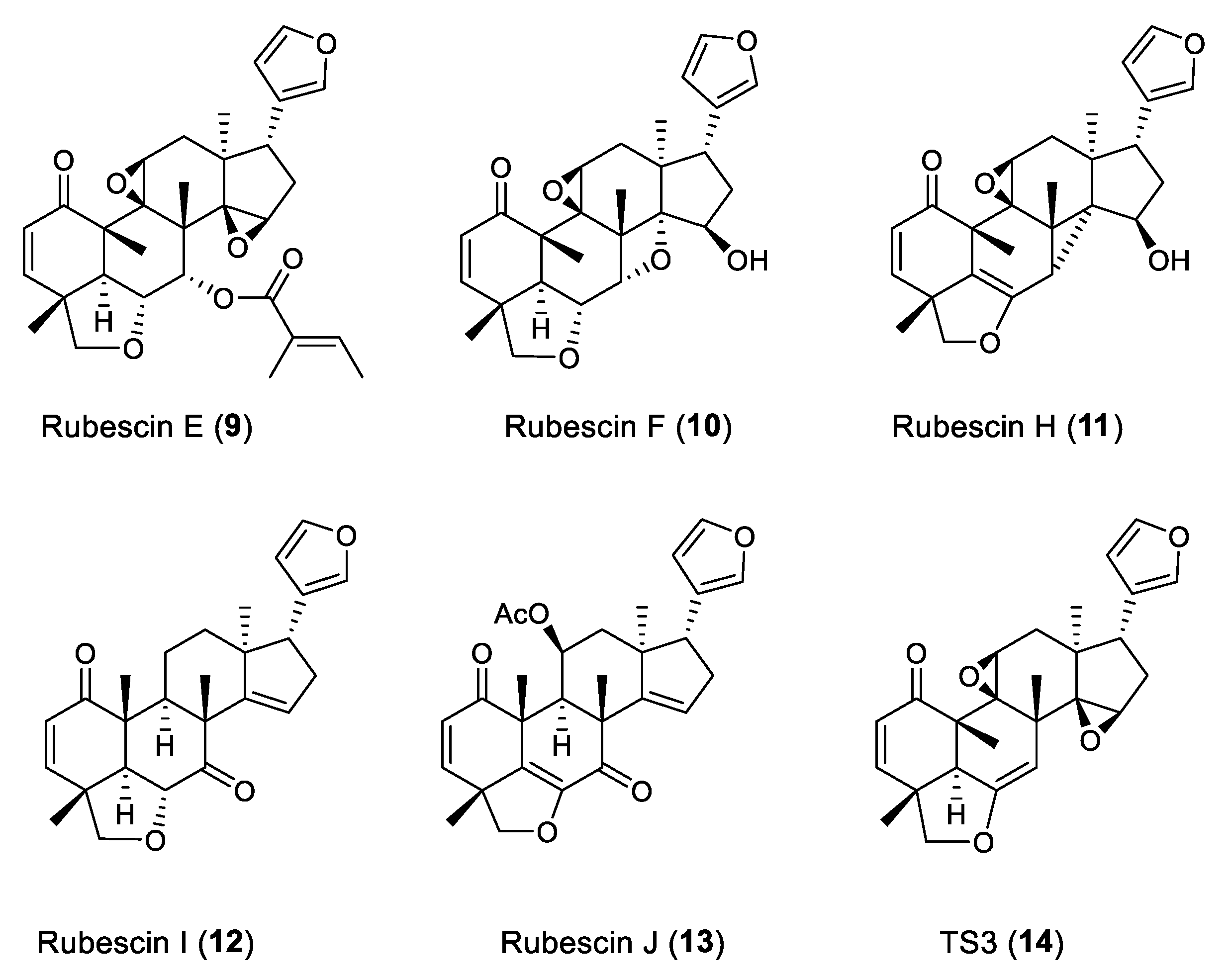
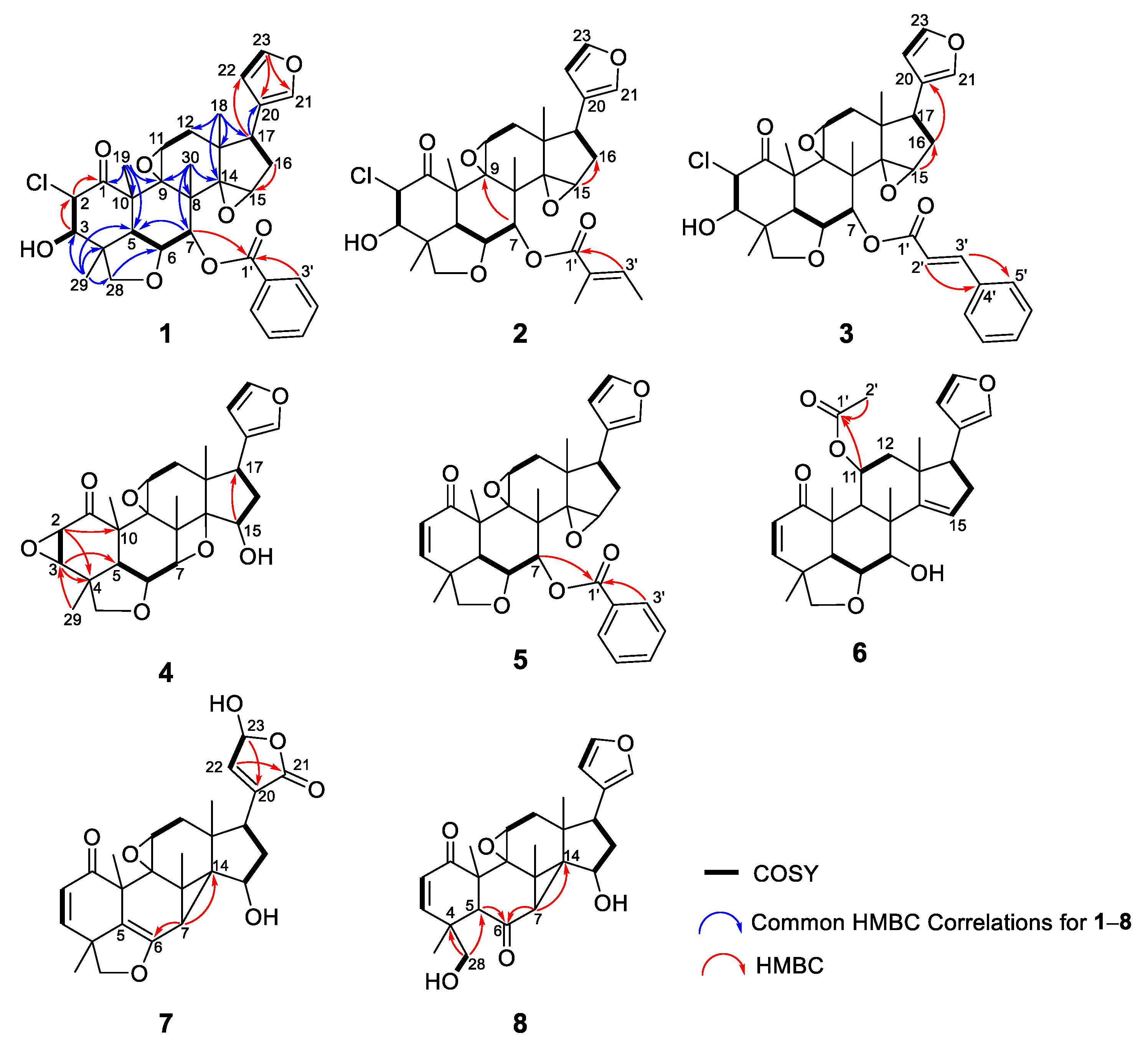
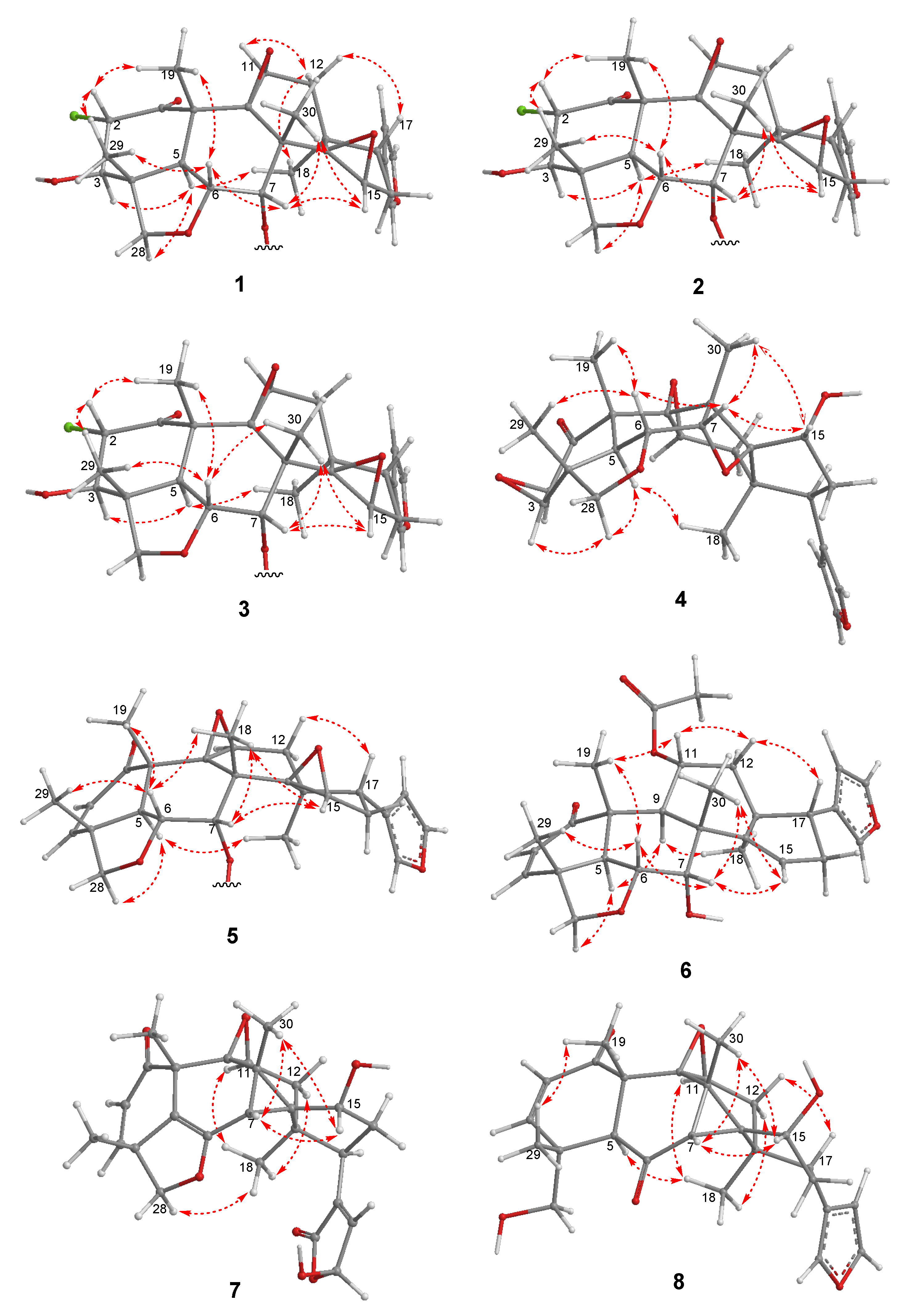
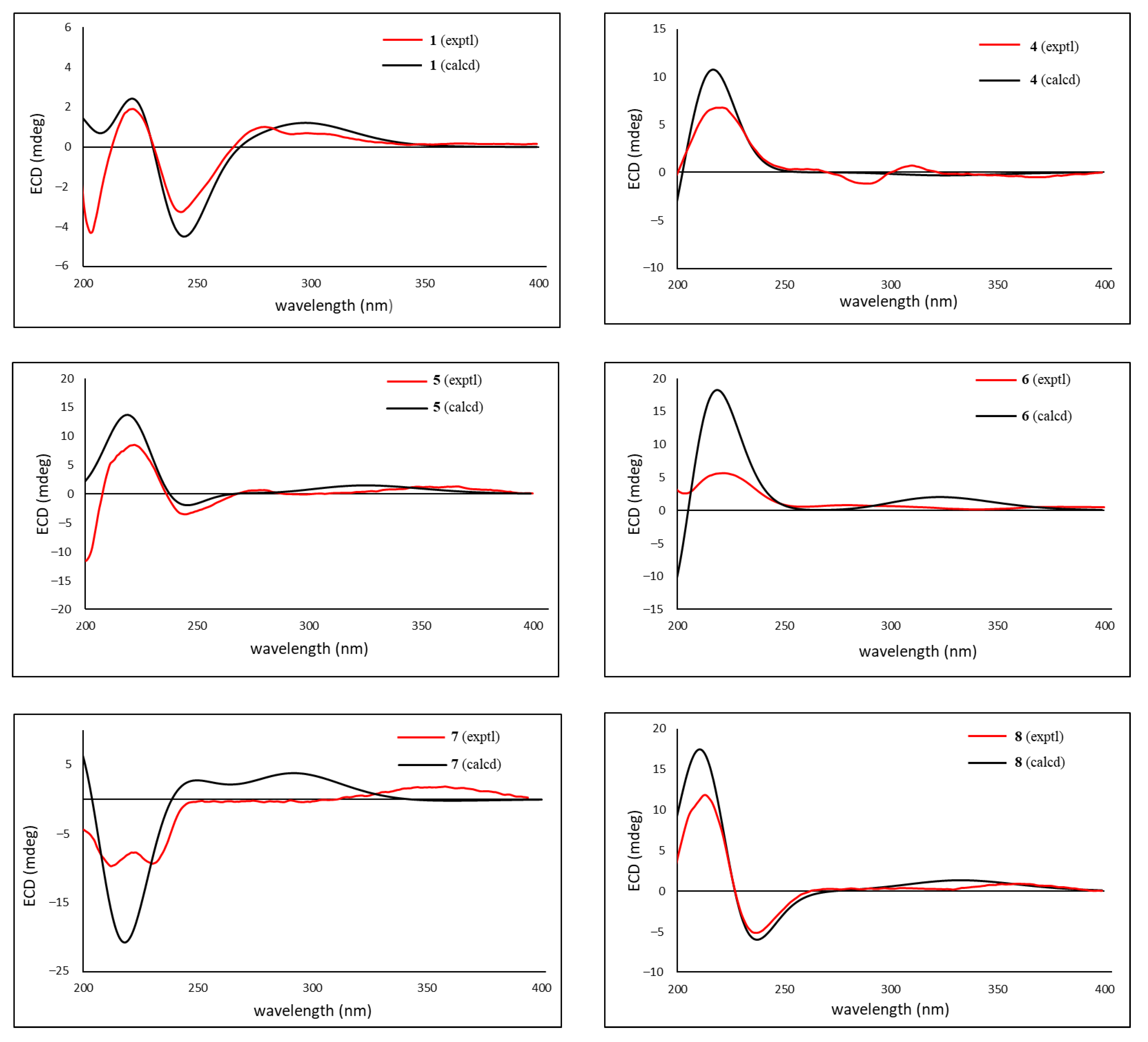
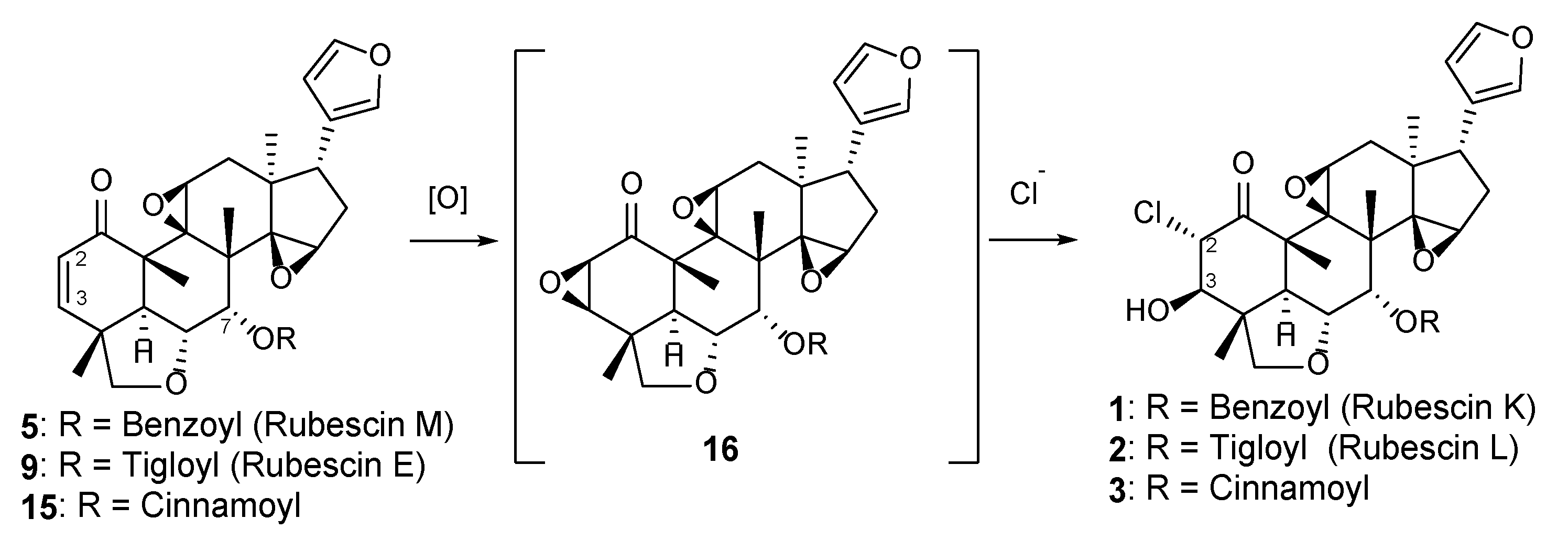
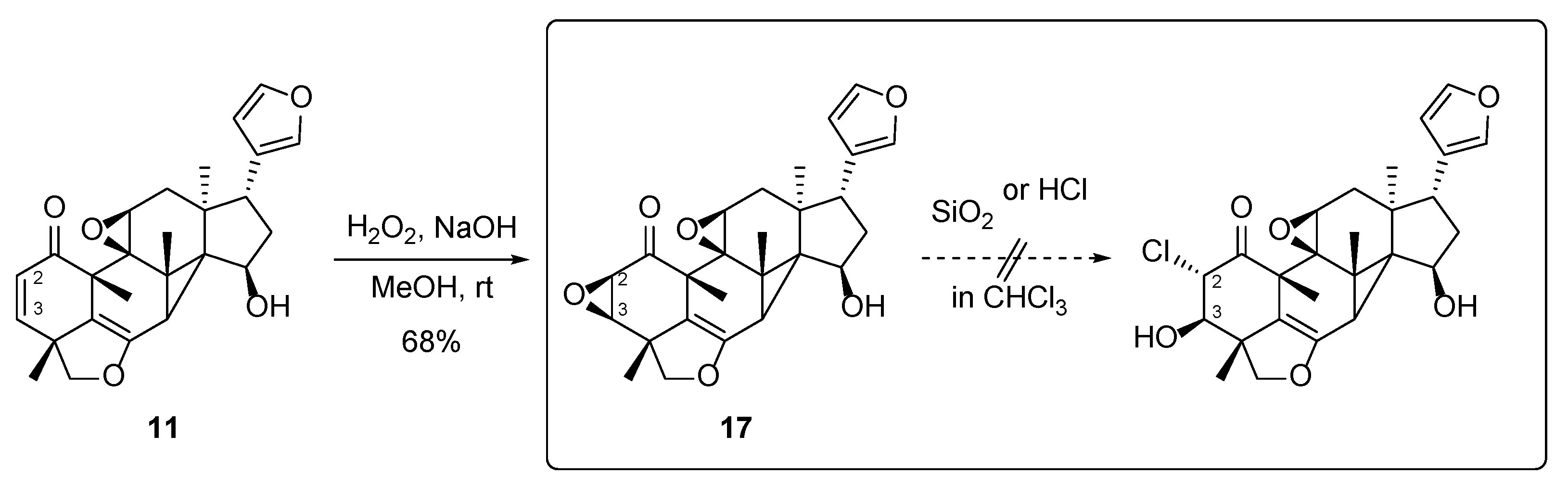
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Calculation of ECD Spectra
3.5. Antiproliferative Activity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luo, J.; Sun, Y.; Li, Q.; Kong, L. Research progress of meliaceous limonoids from 2011 to 2021. Nat. Prod. Rep. 2022, 39, 1325–1365. [Google Scholar] [CrossRef]
- Gouvea, C.F.; Dornelas, M.C.; Rodriguez, A.P.M. Floral development in the tribe Cedreleae (Meliaceae, sub-family Swietenioideae): Cedrela and Toona. Ann. Bot. 2008, 101, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Passos, M.S.; Nogueira, T.S.R.; Azevedo, O.d.A.; Vieira, M.G.C.; Terra, W.d.S.; Braz-Filho, R.; Vieira, I.J.C. Limonoids from the genus Trichilia and biological activities: Review. Phytochem. Rev. 2021, 20, 1055–1086. [Google Scholar] [CrossRef]
- Hilmayanti, E.; Nurlelasari; Supratman, U.; Kabayama, K.; Shinoyama, A.; Kukase, K. Limonoids with anti-inflammatory activity: A review. Phytochemistry 2022, 204, 113469. [Google Scholar] [CrossRef] [PubMed]
- Tsamo, A.T.; Mkounga, P.; Njayou, N.; Manautou, J.; Hultin, P.G.; Nkengfack, A.E. Rubescins A, B and C: New Havanensin Type Limonoids from Root Bark of Trichilia rubescens (Meliaceae). Chem. Pharm. Bull. 2013, 61, 1178–1183. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Armelle, T.T.; Pamela, N.K.; Pierre, M.; Muller, I.B.; Marat, K.; Sass, G.; Nkengfack, A.E. Antiplasmodial Limonoids from Trichilia rubescens (Meliaceae). Med. Chem. 2016, 12, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Tsamo, A.T.; Pagna, J.I.M.; Nangmo, P.K.; Mkounga, P.; Laatsch, H.; Nkengfack, A.E. Rubescins F–H, new vilasinin-type limonoids from the leaves of Trichilia rubescens (Meliaceae). Z. Naturforsch. C J. Biosci. 2019, 74, 175–182. [Google Scholar] [CrossRef]
- Tsamo, A.T.; Melong, R.; Mkounga, P.; Nkengfack, A.E. Rubescins I and J, further limonoid derivatives from the stem bark of Trichilia rubescens (Meliaceae). Nat. Prod. Res. 2019, 33, 196–203. [Google Scholar] [CrossRef]
- De Carvalho, A.C.V.; Ndi, C.P.; Tsopmo, A.; Tane, P.; Ayafor, J.; Connolly, J.D.; Teem, J.L. A Novel Natural Product Compound Enhances cAMP-Regulated Chloride Conductance of Cells Expressing CFTR∆F508. Mol. Med. 2002, 8, 75–87. [Google Scholar] [CrossRef]
- Krief, S.; Martin, M.T.; Greller, P.; Kasenene, J.; Sevenet, T. Novel Antimalarial Compounds Isolated in a Survey of Self-Medicative Behavior of Wild Chimpanzees in Uganda. Antimicrob. Agents Chemother. 2004, 48, 3196–3199. [Google Scholar] [CrossRef]
- Nakatani, M.; Iwashita, T.; Mizukawa, K.; Hase, T. Trichilinin, a New Hexacyclic Limonoid from Trichilia roka. Heterocycles 1987, 26, 43–46. [Google Scholar] [CrossRef]
- Kowa, T.K.; Jansen, O.; Ledoux, A.; Mamede, L.; Wabo, H.K.; Tchinda, A.T.; Gregory, G.J.; Frederich, M. Bioassay-guided isolation of vilasinin–type limonoids and phenyl alkene from the leaves of Trichilia gilgiana and their antiplasmodial activities. Nat. Prod. Res. 2022, 36, 5039–5047. [Google Scholar] [CrossRef] [PubMed]
- El Sayed, K.A.; Hamann, M.T. A New Norcembranoid Dimer from the Red Sea Soft Coral Sinularia gardineri. J. Nat. Prod. 1996, 59, 687–689. [Google Scholar] [CrossRef] [PubMed]
- Oshima, Y.; Iwakawa, T.; Hikino, H. Alismol and alismoxide, sesquiterpenoids of Alisma rhizomes. Phytochemistry 1983, 22, 183–185. [Google Scholar] [CrossRef]
- Zhang, H.J.; Tan, G.T.; Santarsiero, B.D.; Mesecar, A.D.; Hung, N.V.; Cuong, N.M.; Soejarto, D.D.; Pezzuto, J.M.; Fong, H.S. New Sesquiterpenes from Litsea verticillate. J. Nat. Prod. 2003, 66, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, I.; Ikekawa, N.; Yagi, A.; Kawasaki, T.; Tsukamoto, T. Studies on the Plant Sterols and Triterpenes. II. Separation of Stigmasterol, β-Sitosterol and Campesterol, and about So-called “γ-Sitosterol.”. Chem. Pharm. Bull. 1965, 13, 379–384. [Google Scholar] [CrossRef][Green Version]
- Li, S.; Li, Y.; Xu, R.; Kong, L.Y.; Luo, J. New meliacarpin-type (C-seco) and C-ring intact limonoids from the fruits of Melia toosendan. Fitoterapia 2020, 144, 104605. [Google Scholar] [CrossRef]
- Liu, H.B.; Zhang, C.R.; Dong, S.H.; Dong, L.; Wu, Y.; Yue, J.M. Limonoids and Triterpenoids from the Seeds of Melia azedarach. Chem. Pharm. Bull. 2011, 59, 1003–1007. [Google Scholar] [CrossRef]
- Nakai, H.; Shiro, M.; Ochi, M. Ohchinolide A, a new limonoid from Melia azedarach L. var. japonica Makino. Acta Cryst. 1980, 36, 1698–1700. [Google Scholar] [CrossRef]
- Akihisa, T.; Pan, X.; Nakamura, Y.; Kikuchi, T.; Takahashi, N.; Matsumoto, M.; Ogihara, E.; Fukatsu, M.; Koike, K.; Tokuda, H. Limonoids from the fruits of Melia azedarach and their cytotoxic activities. Phytochemistry 2013, 89, 59–70. [Google Scholar] [CrossRef]
- Zhu, G.Y.; Bai, L.P.; Liu, L.; Jiang, Z.H. Limonoids from the fruits of Melia toosendan and their NF-κB modulating activities. Phytochemistry 2014, 107, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, Y.; Nakaoka, M.; Yamamoto, T.; Takahshi, H.; Minami, H. Degraded and Oxetane-Bearing Limonoids from the Roots of Melia azedarach. Chem. Pharm. Bull. 2006, 54, 1219–1222. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.H.; Zhang, C.R.; He, X.F.; Liu, H.B.; Wu, Y.; Yue, J.M. Mesendanins A−J, Limonoids from the Leaves and Twigs of Melia toosendan. J. Nat. Prod. 2010, 73, 1344–1349. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, C.P.; Ke, C.Q.; Li, X.Q.; Xie, H.; Ye, Y. Limonoids from the fruits of Melia toosendan. Phytochemistry 2012, 73, 106–113. [Google Scholar] [CrossRef]
- Zeng, J.; Zhang, J. Chlorinated Natural Products and Related Halogenases. Isr. J. Chem. 2019, 59, 387–402. [Google Scholar] [CrossRef]
- Adak, S.; Moore, B.S. Cryptic halogenation reactions in natural product biosynthesis. Nat. Prod. Rep. 2021, 38, 1760–1774. [Google Scholar] [CrossRef]
- Gribble, G.W. A recent survey of naturally occurring organohalogen compounds. Environ. Chem. 2015, 12, 396–405. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, X.; Fredimoses, M.; Liao, S.; Liu, Y. Naturally occurring organoiodines. RSC Adv. 2014, 4, 57350–57376. [Google Scholar] [CrossRef]
- Bidleman, T.F.; Andersson, A.; Jantunen, L.M.; Kucklick, J.R.; Kylin, H.; Letcher, R.J.; Tysklind, M.; Wong, F. A review of halogenated natural products in Arctic, Subarctic and Nordic ecosystems. Emerg. Contam. 2019, 5, 89–115. [Google Scholar] [CrossRef]
- Wagner, C.; Omari, M.E.; Konig, G.M. Biohalogenation: Nature’s Way to Synthesize Halogenated Metabolites. J. Nat. Prod. 2009, 72, 540–553. [Google Scholar] [CrossRef]
- Crowe, C.; Molyneux, S.; Sharma, S.V.; Zhang, Y.; Gkotsi, D.S.; Connaris, H.; Goss, R.J.M. Halogenases: A palette of emerging opportunities for synthetic biology–synthetic chemistry and C–H functionalization. Chem. Soc. Rev. 2021, 50, 9443–9481. [Google Scholar] [CrossRef]
- Engvild, K.C. Chlorine-containing natural compounds in higher plants. Phytochemistry 1986, 25, 781–791. [Google Scholar] [CrossRef]
- Fehlberg, I.; Riberio, P.R.; dos Santos, I.B.F.; dos Sasntos, I.I.P.; Guedes, M.L.S.; Ferraz, C.G.; Cruz, F.G. Two new sesquiterpenoids and one new p-coumaroyl-triterpenoid derivative from Myrcia guianensis. Phytochem. Lett. 2022, 48, 5–10. [Google Scholar] [CrossRef]
- Ang, S.; Liu, C.; Hong, P.; Yang, L.; Hu, G.; Zheng, X.; Jin, J.; Wu, R.; Wong, W.L.; Zhang, K.; et al. Hirsutinolide-type sesquiterpenoids with anti-prostate cancer activity from Cyanthillium cinereum. Phytochemistry 2023, 216, 113887. [Google Scholar] [CrossRef]
- Li, H.; Liu, L.; Liu, G.; Li, J.; Aisa, H. A Chlorine-containing guaianolide sesquiterpenoids from Achillea millefolium L. with inhibitory effects against LPS-induced NO release in BV-2 microglial cells. Phytochemistry 2023, 207, 113567. [Google Scholar] [CrossRef]
- Yadav, P.A.; Kumar, C.P.; Siva, B.; Babu, K.S.; Allanki, A.D.; Sijwali, P.S.; Jain, N.; Rao, A.V. Synthesis and evaluation of anti-plasmodial and cytotoxic activities of epoxyazadiradione derivatives. Eur. J. Med. Chem. 2017, 134, 242–257. [Google Scholar] [CrossRef]
- Loughlin, W.A.; Jenkins, I.D.; Henderson, L.C.; Campitelli, M.R.; Healy, P. Total Synthesis of (±)-Hyphodermins A and D. J. Org. Chem. 2008, 73, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Lange, N.; Tontsa, A.T.; Wegscheid, C.; Mkounga, P.; Nkengfack, A.E.; Loscher, C.; Sass, G.; Tiegs, G. The Limonoids TS3 and Rubescin E Induce Apoptosis in Human Hepatoma Cell Lines and Interfere with NF-κB Signaling. PLoS ONE 2016, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Mohamad, K.; Hirasawa, Y.; Litaudon, M.; Awang, K.; Hadi, A.A.H.; Takeya, K.; Ekasari, W.; Widyawaruyanti, A.; Zaini, N.C.; Morita, H. Ceramicines B–D, new antiplasmodial limonoids from Chisocheton ceramicus. Bioorg. Med. Chem. 2009, 17, 727–730. [Google Scholar] [CrossRef] [PubMed]
- McCloud, T.G. High Throughput Extraction of Plant, Marine and Fungal Specimens for Preservation of Biologically Active Molecules. Molecules 2010, 15, 4526–4563. [Google Scholar] [CrossRef] [PubMed]
- Pascitelli, G.; Bruhn, T. Good Computational Practice in the Assignment of Absolute Configurations by TDDFT Calculations of ECD Spectra. Chirality 2016, 28, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Hirazawa, S.; Saito, Y.; Sagano, M.; Goto, M.; Nakagawa-Goto, K. Chemical Space Expansion of Flavonoids: Induction of Mitotic Inhibition by Replacing Ring B with a 10π-Electron System, Benzo[b]thiophene. J. Nat. Prod. 2022, 85, 136–147. [Google Scholar] [CrossRef] [PubMed]
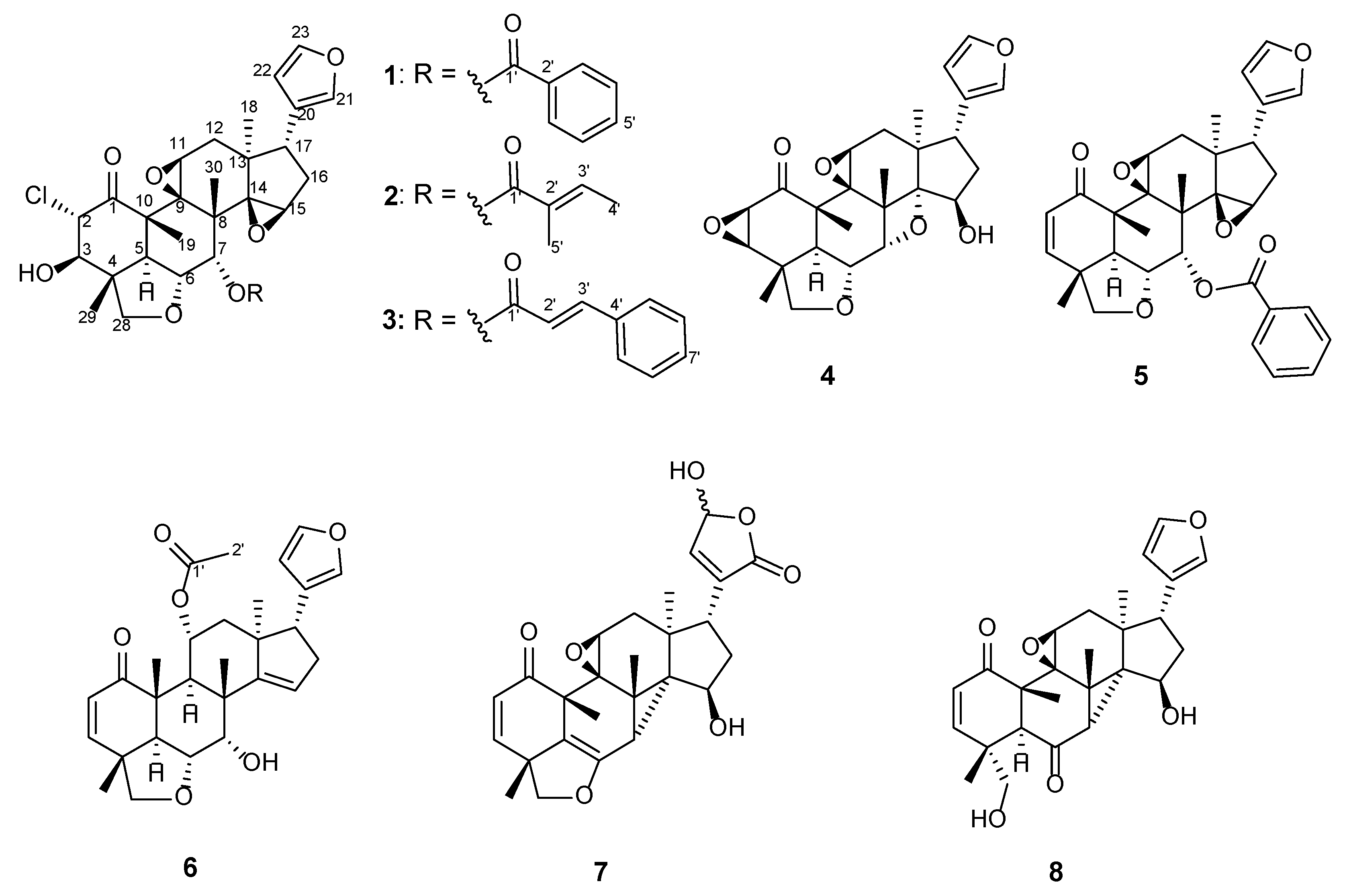
| Position | 1 | 2 | 3 | ||
|---|---|---|---|---|---|
| δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | |||
| 2 | 4.74, d (10.1) | 4.72, d (10.3) | 4.70, d (10.1) | ||
| 3 | 3.87, d (10.1) | 3.77, d (10.3) | 3.84, d (10.1) | ||
| 5 | 2.54, d (11.9) | 2.38, d (11.9) | 2.43, d (11.9) | ||
| 6 | 4.59, dd (11.9, 3.8) | 4.51, dd (11.9, 3.8) | 4.54, dd (11.9, 3.9) | ||
| 7 | 5.66, d (3.8) | 5.47, d (3.8) | 5.48, d (3.8) | ||
| 11 | 4.68, dd (6.9, 1.4) | 4.65, dd (6.9, 1.4) | 4.65, dd (6.8, 1.3) | ||
| 12a | 1.97, dd (13.7, 6.9) | 1.98, dd (13.3, 6.9) | 2.03, dd (13.5, 6.8) | ||
| 12b | 1.84, dd (13.7, 1.4) | 1.83, overlap | 1.87, d (13.5) | ||
| 15 | 3.53, brs a | 3.46, brs a | 3.45, brs a | ||
| 16a | 2.07, ddd (13.3, 6.5, 1.0) | 2.11, ddd (13.4, 6.5, 0.7) | 2.12, ddd (13.4, 6.4, 1.0) | ||
| 16b | 1.42, dd (13.3, 11.3) | 1.54, overlap | 1.59, dd (13.4, 11.3) | ||
| 17 | 2.47, dd (11.3, 6.5) | 2.51, dd (11.3, 6.5) | 2.53, dd (11.3, 6.4) | ||
| 18 | 0.61, s | 0.62, s | 0.77, s | ||
| 19 | 1.54, s | 1.50, s | 1.51, s | ||
| 21 | 6.88, dd (1.6, 0.9) | 7.03, brs | 7.01, dd (1.6, 0.9) | ||
| 22 | 5.87, dd (1.6, 0.9) | 6.07, brs | 6.07, dd (1.6, 0.9) | ||
| 23 | 7.24, t (1.6) | 7.35, t (1.7) | 7.31, t (1.6) | ||
| 28a | 3.85, d (7.3) | 3.91, d (7.9) | 3.92, d (7.8) | ||
| 28b | 3.48, d (7.3) | 3.52, d (7.9) | 3.54, d (7.8) | ||
| 29 | 1.40, s | 1.39, s | 1.38, s | ||
| 30 | 1.26, s | 1.20, s | 1.22, s | ||
| OH-3 | 2.68, brs a | 2.61, brs a | 2.58, brs a | ||
| Ester moiety at C-7 | |||||
| position | δH (J in Hz) | position | δH (J in Hz) | position | δH (J in Hz) |
| 2′ | 2′ | 2′ | 6.37, d (15.9) | ||
| 3′/7′ | 7.92, dd (8.3, 1.3) | 3′ | 6.72, dd (7.2, 1.4) | 3′ | 7.79, d (15.9) |
| 4′/6′ | 7.47, dd (8.3, 8.3) | 4′ | 1.83, d (7.2) | 4′ | - |
| 5′ | 7.62, m | 5′ | 1.86, brs | 5′/9′ | 7.53, dd (7.8, 2.2) |
| 6′/8′ | 7.44, m | ||||
| 7′ | 7.43, m | ||||
| Position | 1 | 2 | 3 | ||
|---|---|---|---|---|---|
| δC | δC | δC | |||
| 1 | 199.4 | 199.3 | 199.5 | ||
| 2 | 68.8 | 68.7 | 68.7 | ||
| 3 | 79.4 | 79.5 | 79.2 | ||
| 4 | 45.4 | 45.3 | 45.2 | ||
| 5 | 48.2 | 48.1 | 47.8 | ||
| 6 | 71.3 | 71.3 | 71.3 | ||
| 7 | 74.9 | 74.1 | 74.1 | ||
| 8 | 45.1 | 45.2 | 45.0 | ||
| 9 | 64.9 | 64.9 | 64.9 | ||
| 10 | 50.1 | 50.1 | 50.0 | ||
| 11 | 60.2 | 60.2 | 60.2 | ||
| 12 | 35.1 | 35.3 | 35.2 | ||
| 13 | 41.1 | 41.1 | 41.2 | ||
| 14 | 68.4 | 68.3 | 68.4 | ||
| 15 | 55.6 | 55.6 | 55.5 | ||
| 16 | 31.1 | 31.1 | 31.1 | ||
| 17 | 38.6 | 38.8 | 38.7 | ||
| 18 | 21.7 | 21.4 | 18.1 | ||
| 19 | 18.1 | 18.2 | 21.2 | ||
| 20 | 122.7 | 123.0 | 122.9 | ||
| 21 | 139.3 | 139.4 | 139.4 | ||
| 22 | 110.8 | 110.9 | 110.9 | ||
| 23 | 142.8 | 142.9 | 143.0 | ||
| 28 | 83.0 | 82.9 | 83.0 | ||
| 29 | 14.3 | 14.4 | 14.3 | ||
| 30 | 23.0 | 22.8 | 22.9 | ||
| Ester moiety at C-7 | |||||
| position | δC | position | δC | position | δC |
| 1′ | 165.9 | 1′ | 167.2 | 1′ | 165.9 |
| 2′ | 129.8 | 2′ | 128.7 | 2′ | 116.7 |
| 3′/7′ | 128.7 | 3′ | 138.5 | 3′ | 147.1 |
| 4′/6′ | 129.6 | 4′ | 14.6 | 4′ | 133.9 |
| 5′ | 133.6 | 5′ | 12.7 | 5′/9′ | 128.3 |
| 6′/8′ | 129.1 | ||||
| 7′ | 130.9 | ||||
| Position | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|
| δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | |
| 2 | 3.15, d (2.8) | 5.99, d (9.6) | 5.87, d (9.6) | 5.85, d (9.8) | 5.90, d (10.3) |
| 3 | 3.57, d (2.8) | 7.10, d (9.6) | 6.94, d (9.6) | 6.68, d (9.8) | 6.64, d (10.3) |
| 5 | 3.12, d (12.6) | 3.05, d (12.7) | 2.66, d (12.4) | 3.67, s | |
| 6 | 4.00, dd (12.6, 4.1) | 4.49, dd (12.7, 4.1) | 4.34, dd (12.4, 3.1) | ||
| 7 | 5.02, d (4.1) | 5.68, d (4.1) | 4.09, d (3.1) | 1.66, s | 2.00, s |
| 9 | 2.44, d (3.7) | ||||
| 11 | 3.18, dd (6.5, 4.7) | 3.98, dd (6.5, 1.4) | 6.23, dd (8.0, 3.7) | 2.69, dd (6.3, 4.3) | 3.04, dd (6.5, 3.6) |
| 12a | 2.21, dd (13.8, 6.5) | 1.90, dd (13.6, 6.5) | 2.50, dd (14.6, 8.0) | 2.60, dd (13.5, 6.3) | 2.27, dd (13.8, 6.5) |
| 12b | 1.31, overlap | 1.85, d (13.6) | 1.64, dd (14.6, 3.7) | 1.47, dd (13.5, 4.3) | 1.51, dd (13.8, 3.6) |
| 15 | 4.66, brs a | 3.50, brs a | 5.67, brs a | 3.90, brs a | 4.00, brs a |
| 16a | 2.08, ddd (13.9, 13.0, 2.9) | 2.07, dd (13.2, 6.5) | 2.53, ddd (15.6, 11.3, 1.7) | 2.17, ddd (13.0, 11.6, 3.6) | 2.20, ddd (13.8, 12.6, 3.4) |
| 16b | 1.85, ddd (13.0, 5.3, 1.0) | 1.41, dd (13.2, 11.3) | 2.45, ddd (15.6, 7.6, 3.4) | 1.95/1.93, dd (11.6, 5.6) | 1.95, ddd (12.6, 4.8, 0.9) |
| 17 | 3.12, dd (13.9, 5.3) | 2.47, dd (11.3, 6.5) | 2.89, dd (11.3, 7.6) | 3.34/3.32, dd (13.0, 5.6) | 3.43, dd (13.8, 4.8) |
| 18 | 0.76, s | 0.64, s | 0.82, s | 0.66/0.63, s | 0.67, s |
| 19 | 1.48, s | 1.46, s | 1.35, s | 1.64/1.63, s | 1.67, s |
| 21 | 7.17, dd (1.6, 0.8) | 6.84, brs a | 7.25, brs a | 7.21, dd (1.7, 0.8) | |
| 22 | 6.20, dd (1.6, 0.8) | 5.87, brs a | 6.30, brs a | 6.85, s | 6.23, dd (1.7, 0.8) |
| 23 | 7.37, t (1.6) | 7.22, t (1.7) | 7.37, t (1.7) | 6.12, brs | 7.21, t (1.7) |
| 28a | 3.96, d (7.7) | 3.70, d (7.4) | 3.79, d (7.2) | 4.29, d (9.0) | 4.20, dd (10.2, 7.1) |
| 28b | 3.92, d (7.7) | 3.54, d (7.4) | 3.62, d (7.2) | 4.07/4.05, d (9.0) | 3.45, dd (10.2, 7.1) |
| 29 | 1.26, s | 1.35, s | 1.36, s | 1.36, s | 1.20, s |
| 30 | 1.31, s | 1.27, s | 1.46, s | 1.68/1.67, s | 1.67, s |
| OH-7 | 2.09, s | ||||
| OH-15 | 1.42, d (2.9) | 1.55, overlap | 1.56, d (2.8) | ||
| OH-28 | 1.64, d (7.1) | ||||
| 2′ | 2.04, s | ||||
| 3′/7′ | 7.94, dd (7.9, 1.4) | ||||
| 4′/6′ | 7.46, dd (7.9, 7.9) | ||||
| 5′ | 7.59, m |
| Position | 4 | 5 | 6 | 7 a | 8 |
|---|---|---|---|---|---|
| δC | δC | δC | δC | δC | |
| 1 | 202.4 | 200.3 | 202.1 | 197.1 | 196.7 |
| 2 | 51.9 | 131.3 | 130.0 | 128.54/128.53 | 127.6 |
| 3 | 60.4 | 151.7 | 150.8 | 152.4/152.3 | 155.4 |
| 4 | 39.0 | 42.8 | 41.9 | 44.8 | 42.1 |
| 5 | 51.8 | 50.8 | 48.0 | 118.2 | 55.4 |
| 6 | 71.8 | 71.3 | 73.8 | 149.7 | 207.1 |
| 7 | 80.0 | 75.5 | 75.4 | 29.1 | 44.4 |
| 8 | 47.9 | 44.8 | 45.9 | 22.7 | 29.3 |
| 9 | 61.8 | 64.6 | 39.4 | 60.5 | 61.3 |
| 10 | 47.8 | 47.6 | 47.3 | 48.2 | 49.3 |
| 11 | 59.4 | 60.4 | 71.0 | 58.7 | 58.1 |
| 12 | 37.2 | 35.3 | 44.2 | 40.9/40.7 | 39.9 |
| 13 | 45.6 | 41.4 | 45.5 | 43.8/43.7 | 43.9 |
| 14 | 96.1 | 68.3 | 158.4 | 51.4, 51.3 | 55.2 |
| 15 | 76.1 | 55.2 | 121.6 | 79.0 | 80.3 |
| 16 | 37.1 | 31.1 | 34.3 | 37.5/37.4 | 38.6 |
| 17 | 43.0 | 38.7 | 52.3 | 45.0/44.9 | 46.0 |
| 18 | 19.9 | 21.8 | 22.5 | 19.4 | 22.3 |
| 19 | 15.9 | 17.4 | 16.3 | 21.9 | 21.0 |
| 20 | 123.9 | 122.8 | 124.1 | 138.8/138.9 | 124.1 |
| 21 | 139.5 | 139.3 | 139.8 | 171.3/171.0 | 139.6 |
| 22 | 111.1 | 110.9 | 110.9 | 144.5/144.9 | 110.9 |
| 23 | 142.9 | 142.7 | 142.8 | 96.7/96.3 | 143.1 |
| 28 | 79.4 | 79.7 | 80.1 | 81.4 | 70.3 |
| 29 | 16.8 | 21.1 | 20.1 | 28.1 | 17.4 |
| 30 | 21.6 | 22.8 | 27.8 | 19.2 | 18.9 |
| 1′ | 166.1 | 169.7 | |||
| 2′ | 128.6 | 21.8 | |||
| 3′/7′ | 129.9 | ||||
| 4′/6′ | 129.8 | ||||
| 5′ | 133.4 |
| Cell Lines a (IC50 μM) | |||||
|---|---|---|---|---|---|
| Compounds | A549 | MDA-MB-231 | MCF-7 | KB | KB-VIN |
| 1 | 2.53 | 5.72 | 1.34 | 3.38 | 4.02 |
| 5 | 3.17 | 5.45 | 2.85 | 4.48 | 2.09 |
| 6 | 3.39 | 17.6 | 4.56 | 6.83 | 8.07 |
| 8 | 37.4 | >40 | >40 | >40 | >40 |
| 9 | 2.14 | 5.88 | 0.96 | 3.68 | 4.35 |
| 11 | 1.60 | 20.9 | 8.46 | 6.83 | 8.09 |
| 13 | 1.42 | 6.76 | 8.07 | 4.38 | 3.28 |
| 14 | 0.56 | 2.06 | 0.75 | 0.57 | 0.54 |
| PXL b (nM) | 2.57 | 12.41 | 2.71 | 6.76 | 2939.33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amuti, S.; Saito, Y.; Fukuyoshi, S.; Miyake, K.; Newman, D.J.; O’Keefe, B.R.; Lee, K.-H.; Nakagawa-Goto, K. Unusual Vilasinin-Class Limonoids from Trichilia rubescens. Molecules 2024, 29, 651. https://doi.org/10.3390/molecules29030651
Amuti S, Saito Y, Fukuyoshi S, Miyake K, Newman DJ, O’Keefe BR, Lee K-H, Nakagawa-Goto K. Unusual Vilasinin-Class Limonoids from Trichilia rubescens. Molecules. 2024; 29(3):651. https://doi.org/10.3390/molecules29030651
Chicago/Turabian StyleAmuti, Saidanxia, Yohei Saito, Shuichi Fukuyoshi, Katsunori Miyake, David J. Newman, Barry R. O’Keefe, Kuo-Hsiung Lee, and Kyoko Nakagawa-Goto. 2024. "Unusual Vilasinin-Class Limonoids from Trichilia rubescens" Molecules 29, no. 3: 651. https://doi.org/10.3390/molecules29030651
APA StyleAmuti, S., Saito, Y., Fukuyoshi, S., Miyake, K., Newman, D. J., O’Keefe, B. R., Lee, K.-H., & Nakagawa-Goto, K. (2024). Unusual Vilasinin-Class Limonoids from Trichilia rubescens. Molecules, 29(3), 651. https://doi.org/10.3390/molecules29030651