STAT3 Decoy Oligodeoxynucleotides Suppress Liver Inflammation and Fibrosis in Liver Cancer Cells and a DDC-Induced Liver Injury Mouse Model
Abstract
1. Introduction
2. Results
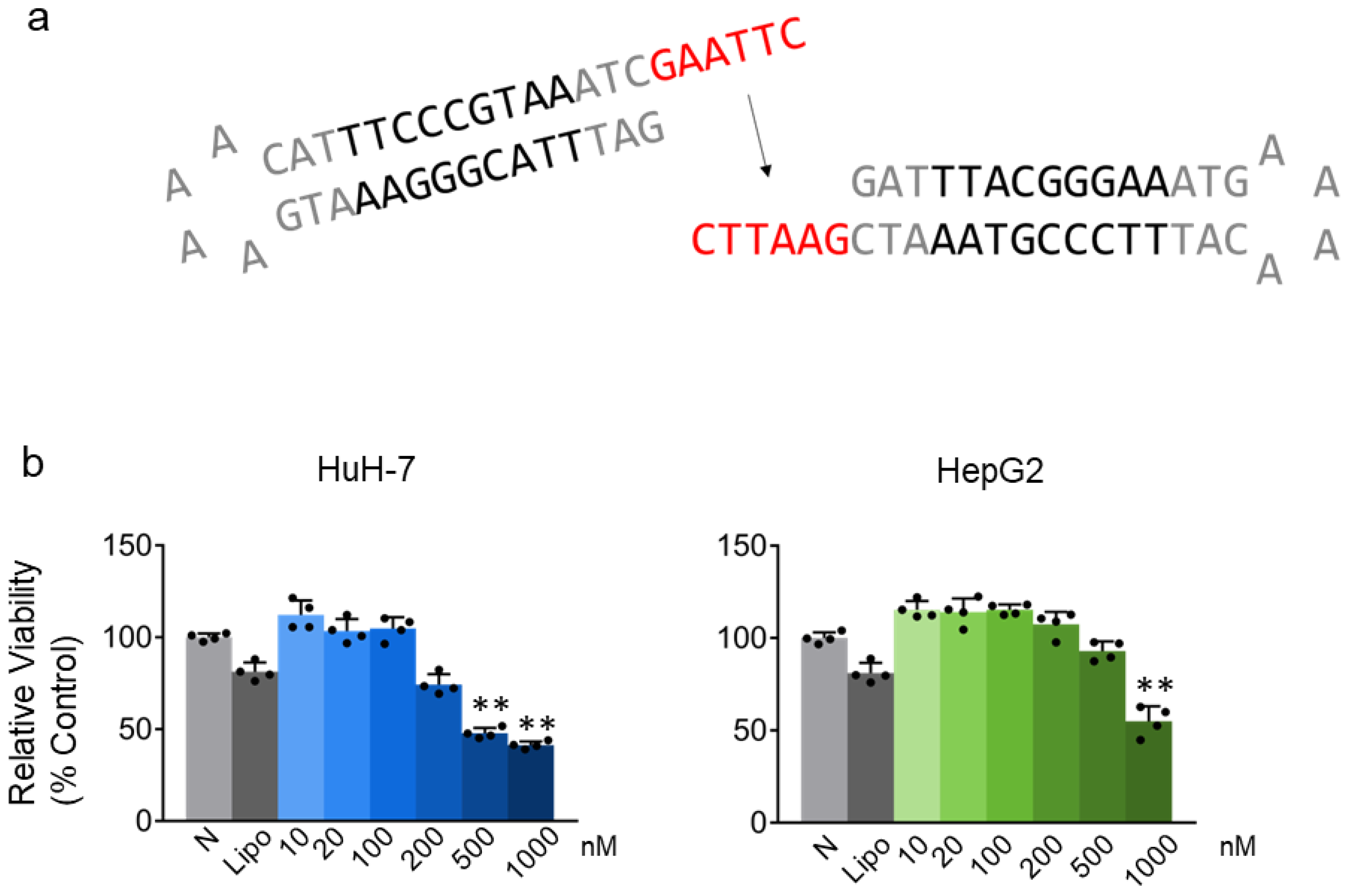
2.1. STAT3 Decoy ODN Inhibit the Proliferation of Hepatic Cancer HuH-7 and HepG2 Cells
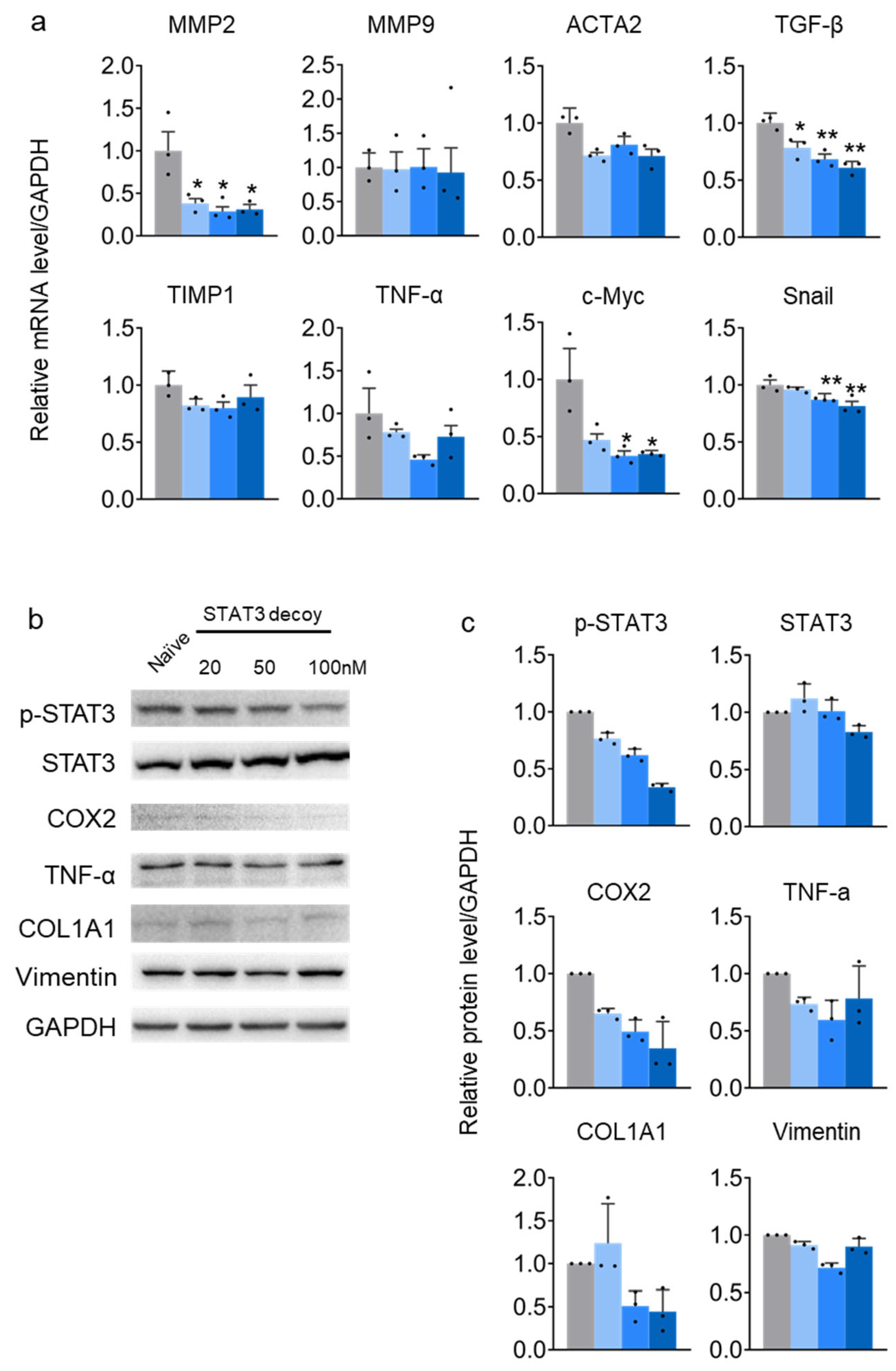
2.2. STAT3 Decoy ODN Suppress STAT3 Target Genes in HuH-7 Cells
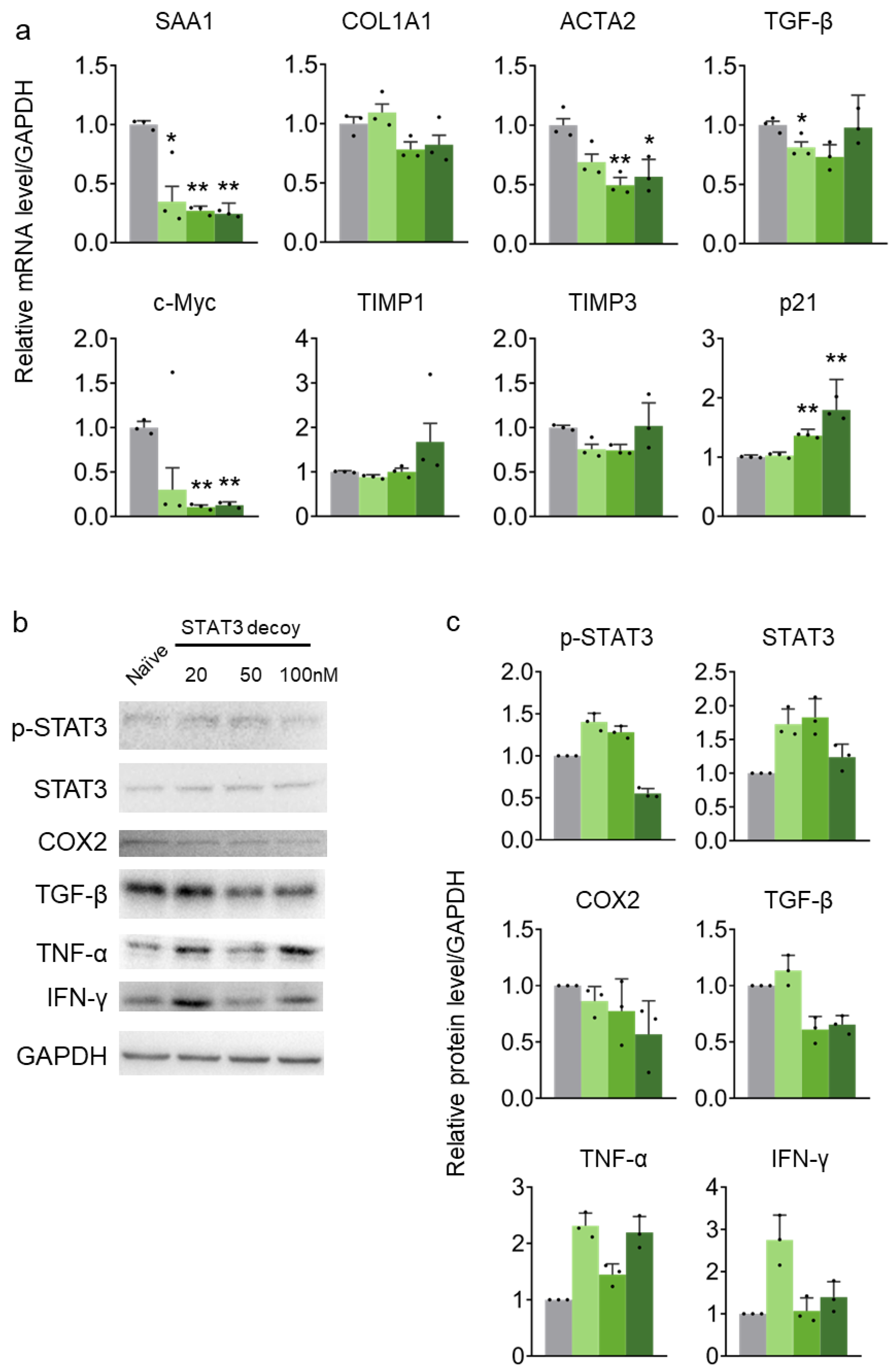
2.3. STAT3 Decoy ODN Suppress STAT3 Target Genes in HepG2 Cells
2.4. Treatment with Decoy ODN in DDC-Induced Liver Fibrosis Mice
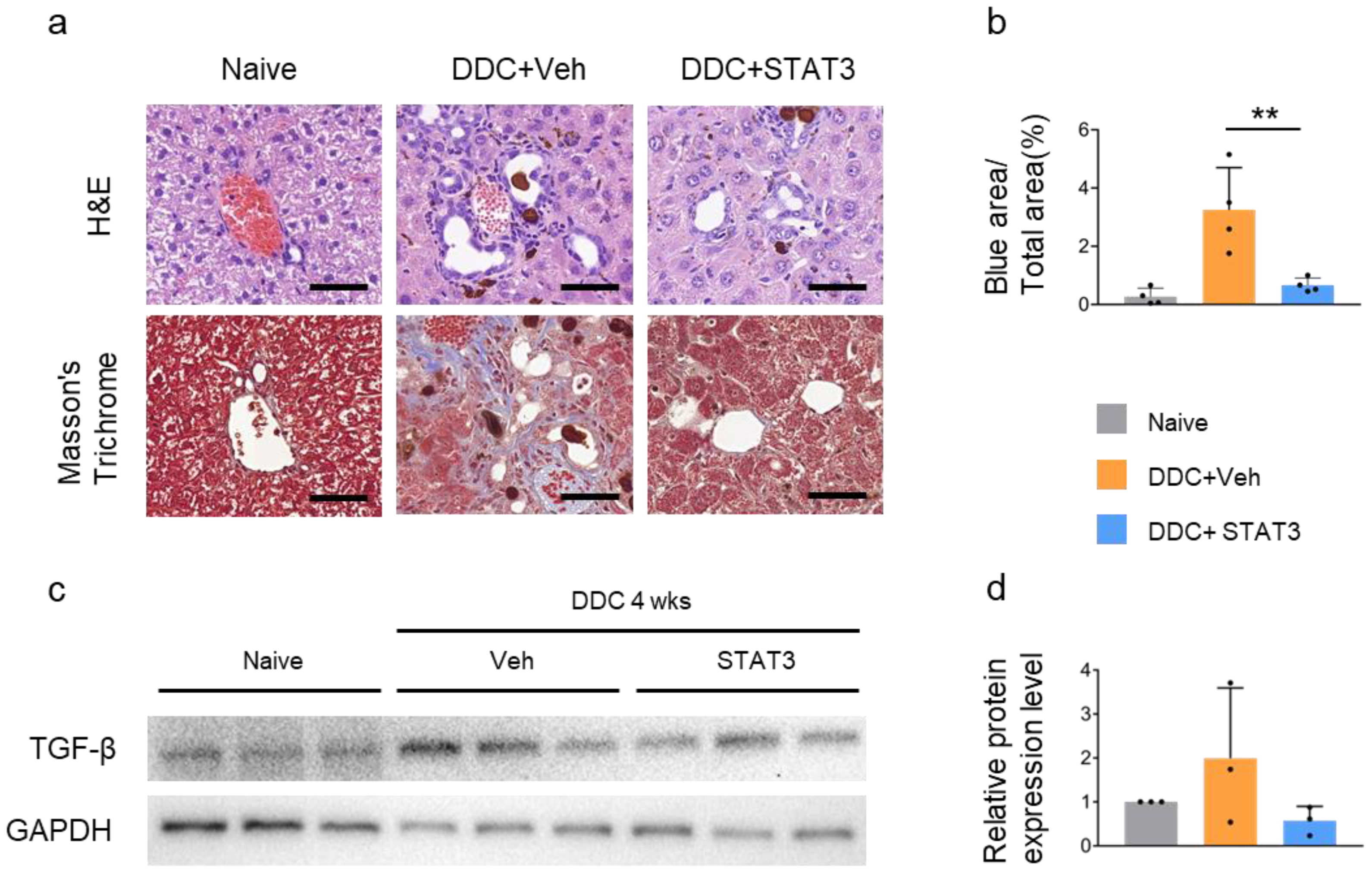
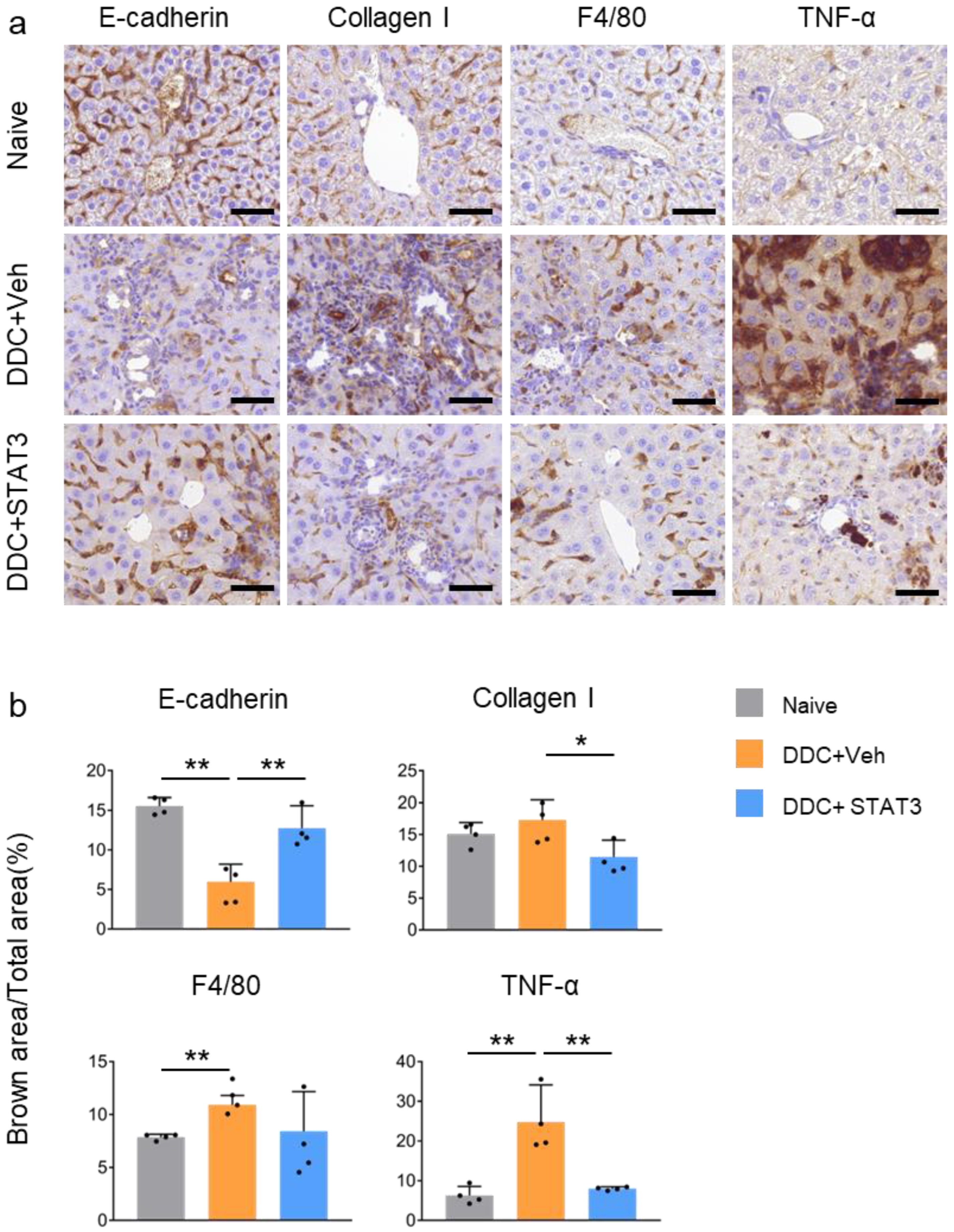
2.5. STAT3 Decoy ODN Ameliorate DDC-Induced Liver Fibrosis
2.6. STAT3 Decoy ODN Ameliorat DDC-Induced Fibrosis and Inflammation
3. Discussion
4. Materials and Methods
4.1. Decoy ODN Synthesis
4.2. Cell Culture
4.3. Cell Viability Assay
4.4. Animal Model
4.5. Histopathological Investigation
4.6. Immunohistochemical Staining
4.7. Western Blot Analysis
4.8. RNA Purification and qPCR Analysis
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wanless, I.R.; Nakashima, E.; Sherman, M. Regression of human cirrhosis: Morphologic features and the genesis of incomplete septal cirrhosis. Arch. Pathol. Lab. Med. 2000, 124, 1599–1607. [Google Scholar] [CrossRef]
- Benyon, R.C.; Iredale, J.P. Is liver fibrosis reversible? Gut 2000, 46, 443–446. [Google Scholar] [CrossRef]
- Iredale, J.P.; Benyon, R.C.; Pickering, J.; McCullen, M.; Northrop, M.; Pawley, S.; Hovell, C.; Arthur, M.J. Mechanisms of spontaneous resolution of rat liver fibrosis. Hepatic stellate cell apoptosis and reduced hepatic expression of metalloproteinase inhibitors. J. Clin. Investig. 1998, 102, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.J.; Rosenthal, N. Preparing the ground for tissue regeneration: From mechanism to therapy. Nat. Med. 2014, 20, 857–869. [Google Scholar] [CrossRef]
- Greuter, T.; Shah, V.H. Hepatic sinusoids in liver injury, inflammation, and fibrosis: New pathophysiological insights. J. Gastroenterol. 2016, 51, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Zhang, Z.; Wang, F.S. Liver fibrosis: Mechanisms of immune-mediated liver injury. Cell. Mol. Immunol. 2012, 9, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Houssein, H.S.H.; Mahato, R.I. Bioconjugation of oligonucleotides for treating liver fibrosis. Oligonucleotides 2007, 17, 349–404. [Google Scholar] [CrossRef] [PubMed]
- Eghbali-Fatourechi, G.; Sieck, G.C.; Prakash, Y.S.; Maercklein, P.; Gores, G.J.; Fitzpatrick, L.A. Type I procollagen production and cell proliferation is mediated by transforming growth factor-beta in a model of hepatic fibrosis. Endocrinology 1996, 137, 1894–1903. [Google Scholar] [CrossRef]
- Bataller, R.; Brenner, D.A. Liver fibrosis. J. Clin. Investig. 2005, 115, 209–218. [Google Scholar] [CrossRef]
- Gressner, A.M.; Weiskirchen, R.; Breitkopf, K.; Dooley, S. Roles of TGF-beta in hepatic fibrosis. Front. Biosci. Landmark 2002, 7, 793–807. [Google Scholar] [CrossRef]
- Annoni, G.; Weiner, F.R.; Zern, M.A. Increased transforming growth factor-β1 gene expression in human liver disease. J. Hepatol. 1992, 14, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Scarpa, M.; Grillo, A.R.; Brun, P.; Macchi, V.; Stefani, A.; Signori, S.; Buda, A.; Fabris, P.; Giordani, M.T.; De Caro, R.; et al. Snail1 transcription factor is a critical mediator of hepatic stellate cell activation following hepatic injury. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G316–G326. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, Y.; Sone, M.; Nagahama, Y.; Kumagai, E.; Doi, Y.; Omori, Y.; Yoshioka, T.; Tokairin, T.; Yoshida, M.; Yamamoto, Y.; et al. Tumor necrosis factor-α promotes bile ductular transdifferentiation of mature rat hepatocytes in vitro. J. Cell. Biochem. 2013, 114, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.E., Jr.; Kerr, I.M.; Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef]
- Schindler, C. Cytokines and JAK–STAT signaling. Exp. Cell Res. 1999, 253, 7–14. [Google Scholar] [CrossRef]
- Darnell, J.E. Validating Stat3 in cancer therapy. Nat. Med. 2005, 11, 595–596. [Google Scholar] [CrossRef]
- Huynh, J.; Chand, A.; Gough, D.; Ernst, M. Therapeutically exploiting STAT3 activity in cancer—Using tissue repair as a road map. Nat. Rev. Cancer 2019, 19, 82–96. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Kortylewski, M.; Pardoll, D. Crosstalk between cancer and immune cells: Role of STAT3 in the tumour microenvironment. Nat. Rev. Immunol. 2007, 7, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Cen, L.; Kohout, J.; Hutzen, B.; Chan, C.; Hsieh, F.C.; Loy, A.; Huang, V.; Cheng, G.; Lin, J. Signal transducer and activator of transcription 3 activation is associated with bladder cancer cell growth and survival. Mol. Cancer 2008, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Leeman, R.J.; Lui, V.W.Y.; Grandis, J.R. STAT3 as a therapeutic target in head and neck cancer. Expert Opin. Biol. Ther. 2006, 6, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Yue, P.; Turkson, J. Targeting STAT3 in cancer: How successful are we? Expert Opin. Investig. Drugs 2009, 18, 45–56. [Google Scholar] [CrossRef]
- Leong, P.L.; Andrews, G.A.; Johnson, D.E.; Dyer, K.F.; Xi, S.; Mai, J.C.; Robbins, P.D.; Gadiparthi, S.; Burke, N.A.; Watkins, S.F.; et al. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc. Natl. Acad. Sci. USA 2003, 100, 4138–4143. [Google Scholar] [CrossRef]
- Lee, H.; Pal, S.K.; Reckamp, K.; Figlin, R.A.; Yu, H. STAT3: A target to enhance antitumor immune response. Cancer Immunol. Immunother. 2010, 344, 41–59. [Google Scholar] [CrossRef]
- Gao, B. Cytokines, STATs and liver disease. Cell. Mol. Immunol. 2005, 2, 92–100. [Google Scholar] [PubMed]
- Machida, K.; Tsukamoto, H.; Liu, J.C.; Han, Y.P.; Govindarajan, S.; Lai, M.M.; Akira, S.; Ou, J.H. C-Jun mediates hepatitis C virus hepatocarcinogenesis through signal transducer and activator of transcription 3 and nitric oxide–dependent impairment of oxidative DNA repair. Hepatology 2010, 52, 480–492. [Google Scholar] [CrossRef] [PubMed]
- Ruff-Jamison, S.; Chen, K.; Cohen, S. Induction by EGF and interferon-γ of tyrosine phosphorylated DNA binding proteins in mouse liver nuclei. Science 1993, 261, 1733–1736. [Google Scholar] [CrossRef]
- He, G.; Yu, G.Y.; Temkin, V.; Ogata, H.; Kuntzen, C.; Sakurai, T.; Sieghart, W.; Peck-Radosavljevic, M.; Leffert, H.L.; Karin, M. Hepatocyte IKKβ/NF-κB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell 2010, 17, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.J.; Dzau, V.J. Therapeutic applications of transcription factor decoy oligonucleotides. J. Clin. Investig. 2000, 106, 1071–1075. [Google Scholar] [CrossRef]
- Bielinska, A.; Shivdasani, R.A.; Zhang, L.Q.; Nabel, G.J. Regulation of gene expression with double-stranded phosphorothioate oligonucleotides. Science 1990, 250, 997–1000. [Google Scholar] [CrossRef]
- Morishita, R.; Gibbons, G.H.; Horiuchi, M.; Ellison, K.E.; Nakama, M.; Zhang, L.; Kaneda, Y.; Ogihara, T.; Dzau, V.J. A gene therapy strategy using a transcription factor decoy of the E2F binding site inhibits smooth muscle proliferation in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 5855–5859. [Google Scholar] [CrossRef]
- Clerbaux, L.A.; Van Hul, N.; Gouw, A.S.H.; Manco, R.; Español-Suñer, R.; Leclercq, I.A. Relevance of the CDE and DDC Mouse Models to Study Ductular Reaction in Chronic Human Liver Diseases. In Experimental Animal Models of Human Diseases—An Effective Therapeutic Strategy; IntechOpen: London, UK, 2018; pp. 1–17. [Google Scholar] [CrossRef]
- Fickert, P.; Stöger, U.; Fuchsbichler, A.; Moustafa, T.; Marschall, H.U.; Weiglein, A.H.; Tsybrovskyy, O.; Jaeschke, H.; Zatloukal, K.; Denk, H.; et al. A new xenobiotic-induced mouse model of sclerosing cholangitis and biliary fibrosis. Am. J. Pathol. 2007, 171, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Calabro, S.R.; Maczurek, A.E.; Morgan, A.J.; Tu, T.; Wen, V.W.; Yee, C.; Mridha, A.; Lee, M.; d’Avigdor, W.; Locarnini, S.A.; et al. Hepatocyte produced matrix metalloproteinases are regulated by CD147 in liver fibrogenesis. PLoS ONE 2014, 9, e90571. [Google Scholar] [CrossRef]
- Wang, Z.; Li, J.; Xiao, W.; Long, J.; Zhang, H. The STAT3 inhibitor S3I-201 suppresses fibrogenesis and angiogenesis in liver fibrosis. Lab. Investig. 2018, 98, 1600–1613. [Google Scholar] [CrossRef]
- Kim, J.Y.; An, H.J.; Kim, W.H.; Park, Y.Y.; Park, K.D.; Park, K.K. Apamin suppresses biliary fibrosis and activation of hepatic stellate cells. Int. J. Mol. Med. 2017, 39, 1188–1194. [Google Scholar] [CrossRef]
- Kim, J.Y.; Jang, H.J.; Leem, J.; Kim, G.M. Protective effects of bee venom-derived phospholipase A2 against cholestatic liver disease in mice. Biomedicines 2021, 9, 992. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, J.; Wang, J.; Jiang, R.; Zhao, T. Gut microbiota combined with metabolomics reveal the mechanism of curcumol on liver fibrosis in mice. Biomed. Pharmacother. 2022, 152, 113204. [Google Scholar] [CrossRef]
- Fabregat, I.; Moreno-Càceres, J.; Sánchez, A.; Dooley, S.; Dewidar, B.; Giannelli, G.; Ten Dijke, P.; IT-LIVER Consortium. TGF-β signalling and liver disease. FEBS J. 2016, 283, 2219–2232. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.Y.; Heller, M.; Meng, Z.; Yu, L.R.; Tang, Y.; Zhou, M.; Zhang, Y.E. Transforming growth factor-β (TGF-β) directly activates the JAK1-STAT3 axis to induce hepatic fibrosis in coordination with the SMAD pathway. J. Biol. Chem. 2017, 292, 4302–4312. [Google Scholar] [CrossRef]
- Xu, M.Y.; Hu, J.J.; Shen, J.; Wang, M.L.; Zhang, Q.Q.; Qu, Y.; Lu, L.G. Stat3 signaling activation crosslinking of TGF-β1 in hepatic stellate cell exacerbates liver injury and fibrosis. Biochim. Biophys. Acta 2014, 1842, 2237–2245. [Google Scholar] [CrossRef]
- Zhao, J.; Qi, Y.F.; Yu, Y.R. STAT3: A key regulator in liver fibrosis. Ann. Hepatol. 2021, 21, 100224. [Google Scholar] [CrossRef]
- Zheng, B.; Yang, Y.; Han, Q.; Yin, C.; Pan, Z.; Zhang, J. STAT3 directly regulates NKp46 transcription in NK cells of HBeAg-negative CHB patients. J. Leukoc. Biol. 2019, 106, 987–996. [Google Scholar] [CrossRef] [PubMed]

| Primer | Sequence (5′-3′) |
|---|---|
| hGAPDH_Fwd | GTCTCCTCTGACTTCAACAGCG |
| hGAPDH_Rvs | ACCACCCTGTTGCTGTAGCCAA |
| hMMP2_Fwd | AGCGAGTGGATGCCGCCTTTAA |
| hMMP2_Rvs | CATTCCAGGCATCTGCGATGAG |
| hMMP9_Fwd | GCCACTACTGTGCCTTTGAGTC |
| hMMP9_Rvs | CCCTCAGAGAATCGCCAGTACT |
| hACTA2_Fwd | CTATGCCTCTGGACGCACAACT |
| hACTA2_Rvs | CAGATCCAGACGCATGATGGCA |
| hTGF-β1_Fwd | TACCTGAACCCGTGTTGCTCTC |
| hTGF-β1_Rvs | GTTGCTGAGGTATCGCCAGGAA |
| hTIMP1_Fwd | GGAGAGTGTCTGCGGATACTTC |
| hTIMP1_Rvs | GCAGGTAGTGATGTGCAAGAGTC |
| hTIMP3_Fwd | TACCGAGGCTTCACCAAGATGC |
| hTIMP3_Rvs | CATCTTGCCATCATAGACGCGAC |
| hTNF-α_Fwd | CTCTTCTGCCTGCTGCACTTTG |
| hTNF-α_Rvs | ATGGGCTACAGGCTTGTCACTC |
| hc-Myc_Fwd | CCTGGTGCTCCATGAGGAGAC |
| hc-Myc_Rvs | CAGACTCTGACCTTTTGCCAGG |
| hSnail_Fwd | TTACACCTTTGCATACAGAACCC |
| hSnail_Rvs | TTTACGATTACACCCAGACTGC |
| hSAA1_Fwd | TCGTTCCTTGGCGAGGCTTTTG |
| hSAA1_Rvs | AGGTCCCCTTTTGGCAGCATCA |
| hCOL1A1_Fwd | GATTCCCTGGACCTAAAGGTGC |
| hCOL1A1_Rvs | AGCCTCTCCATCTTTGCCAGCA |
| hp21_Fwd | AGGTGGACCTGGAGACTCTCAG |
| hp21_Rvs | TCCTCTTGGAGAAGATCAGCCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, H.J.; Kim, Y.-A.; Ryu, J.; Park, K.-K.; Lee, S.-J.; Kim, B.S.; Song, J.-E.; Kim, J.D. STAT3 Decoy Oligodeoxynucleotides Suppress Liver Inflammation and Fibrosis in Liver Cancer Cells and a DDC-Induced Liver Injury Mouse Model. Molecules 2024, 29, 593. https://doi.org/10.3390/molecules29030593
Choi HJ, Kim Y-A, Ryu J, Park K-K, Lee S-J, Kim BS, Song J-E, Kim JD. STAT3 Decoy Oligodeoxynucleotides Suppress Liver Inflammation and Fibrosis in Liver Cancer Cells and a DDC-Induced Liver Injury Mouse Model. Molecules. 2024; 29(3):593. https://doi.org/10.3390/molecules29030593
Chicago/Turabian StyleChoi, Hye Jin, Young-Ah Kim, Junghwa Ryu, Kwan-Kyu Park, Sun-Jae Lee, Byung Seok Kim, Jeong-En Song, and Joo Dong Kim. 2024. "STAT3 Decoy Oligodeoxynucleotides Suppress Liver Inflammation and Fibrosis in Liver Cancer Cells and a DDC-Induced Liver Injury Mouse Model" Molecules 29, no. 3: 593. https://doi.org/10.3390/molecules29030593
APA StyleChoi, H. J., Kim, Y.-A., Ryu, J., Park, K.-K., Lee, S.-J., Kim, B. S., Song, J.-E., & Kim, J. D. (2024). STAT3 Decoy Oligodeoxynucleotides Suppress Liver Inflammation and Fibrosis in Liver Cancer Cells and a DDC-Induced Liver Injury Mouse Model. Molecules, 29(3), 593. https://doi.org/10.3390/molecules29030593







