A Chemiluminescence Signal Amplification Method for MicroRNA Detection: The Combination of Molecular Aptamer Beacons with Enzyme-Free Hybridization Chain Reaction
Abstract
1. Introduction
2. Results
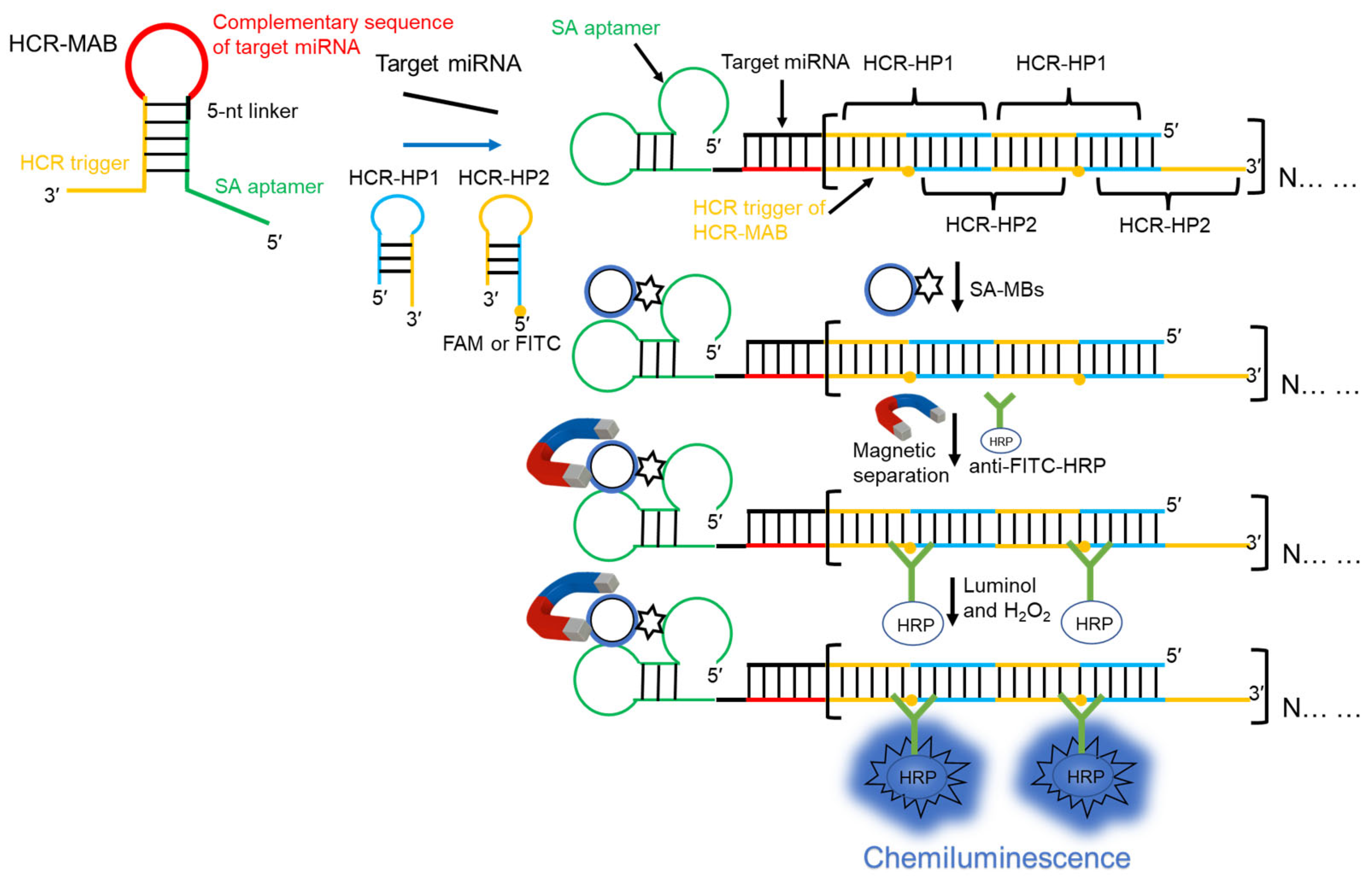
2.1. Principle of miRNA Detection
2.2. Verification of the Feasibility of the Detection System
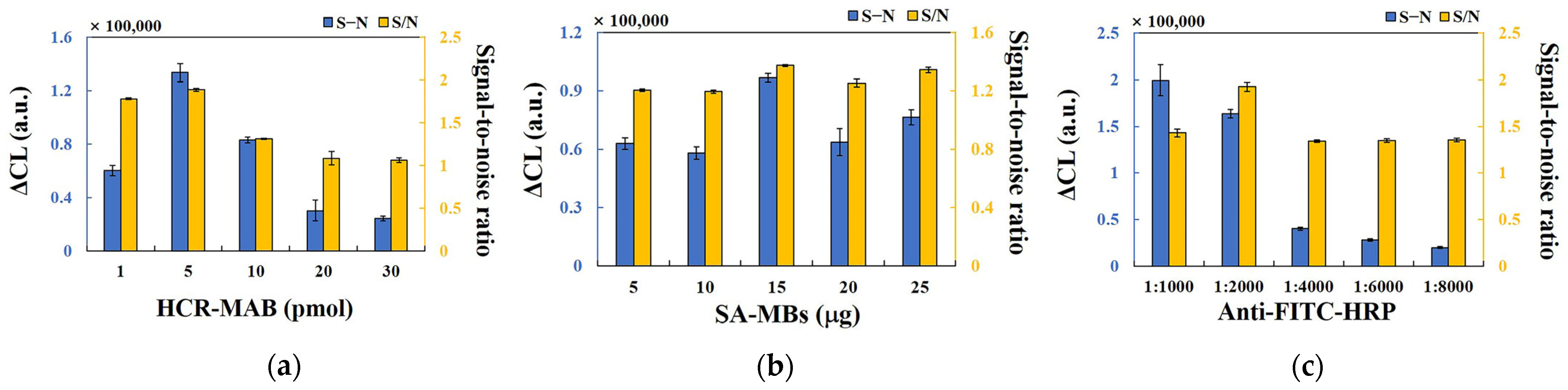
2.3. Optimization of Experimental Conditions
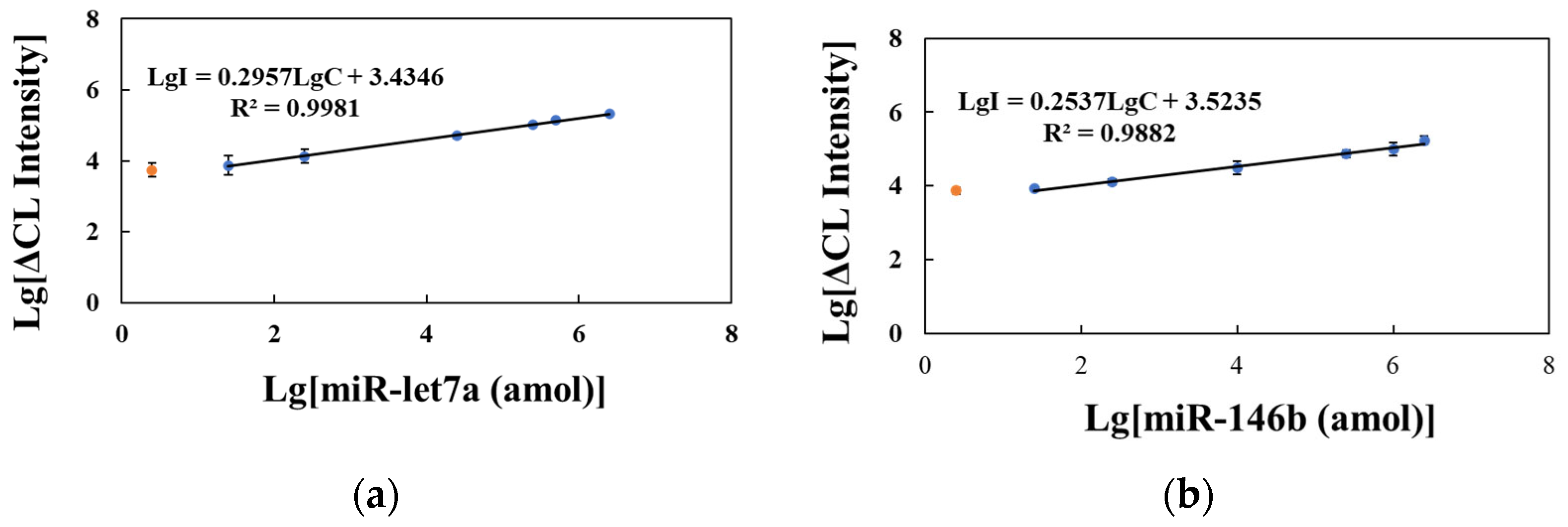
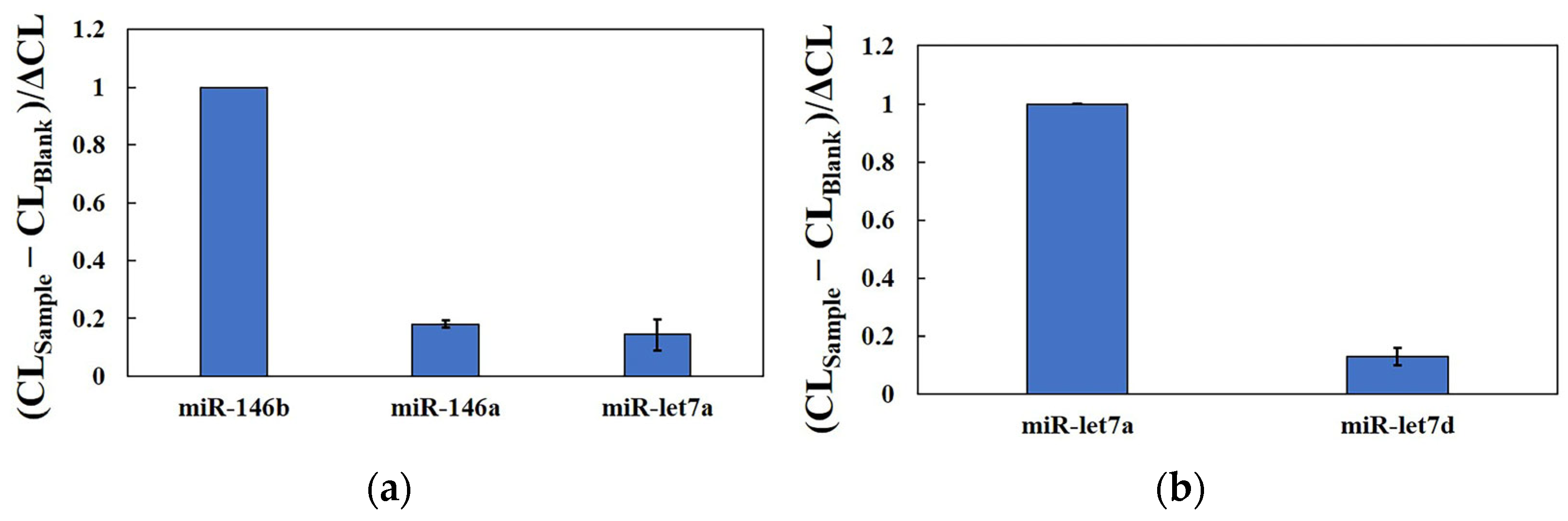
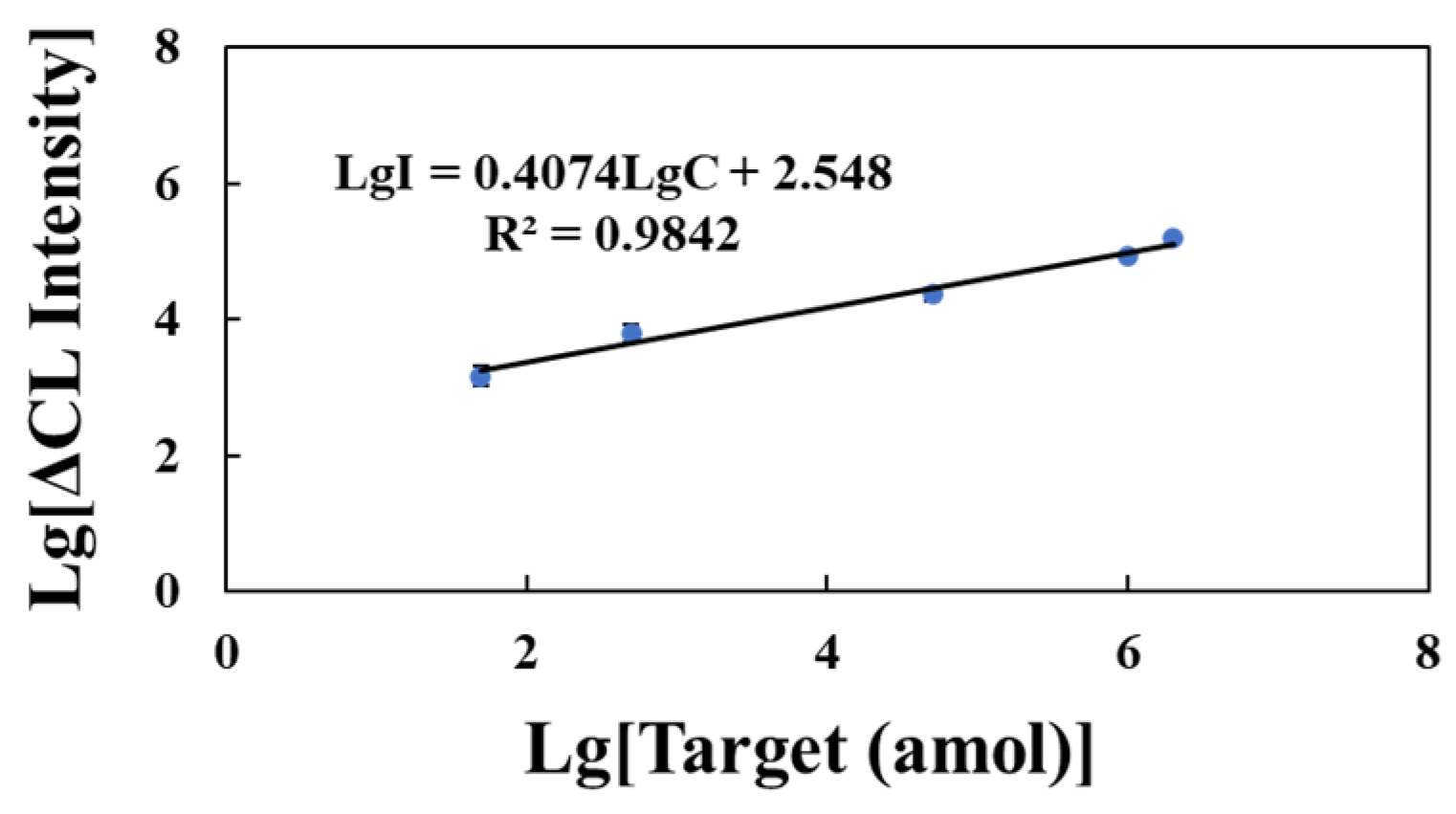
2.4. Performance of Detection Strategy and Selectivity
2.5. Testing of Actual Samples
3. Discussion
4. Materials and Methods
4.1. Reagents and Apparatus
4.2. Detection of Target miRNA
4.3. Agarose Gel Electrophoresis Analysis
4.4. Treatment of Healthy Human Blood and the Collection of Total RNA Samples from Cells
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Y.H.; Di, C.H.; Li, W.; Cai, W.B.; Tan, X.H.; Xu, L.W.; Yang, L.; Lou, G.Q.; Yan, Y.T. Oncomirs miRNA-221/222 and tumor suppressors miRNA-199a/195 are crucial miRNAs in liver cancer: A systematic analysis. Dig. Dis. Sci. 2016, 61, 2315–2327. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.D.; Li, Q.; Zhang, R.S.; Dai, X.L.; Chen, W.J.; Xing, D.M. Circulating microRNAs: Biomarkers of disease. Clin. Chim. Acta 2021, 516, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Song, C.; Chen, W.H.; Tang, L.X.; Ju, J.H.; Shen, W.; Kong, D.Z.; Tang, S. Duplex specific nuclease-enabled isothermal signal amplification combined with high-performance liquid chromatography for highly sensitive detection of dual-target microRNAs as disease biomarkers. Chin. J. Anal. Chem. 2021, 49, 216–225. [Google Scholar]
- Yokoi, A.; Matsuzaki, J.; Yamamoto, Y.; Yoneoka, Y.; Takahashi, K.; Shimizu, H.; Uehara, T.; Ishikawa, M.; Ikeda, S.; Sonoda, T.; et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat. Commun. 2018, 9, 4319. [Google Scholar] [CrossRef]
- Han, Y.; Hu, H.H.; Yu, L.S.; Zeng, S.; Min, J.Z.; Cai, S. A duplex-specific nuclease (DSN) and catalytic hairpin assembly (CHA)-mediated dual amplification method for miR-146b detection. Analyst 2023, 148, 556–561. [Google Scholar] [CrossRef]
- Talap, J.; AL-maskri, A.A.A.; Shen, M.Z.; Liu, H.; Jiang, X.F.; Xiao, G.Z.; Yu, L.S.; Zeng, S.; Jung, C.; Cai, S. Ultrasensitive detection of serum miRNA biomarkers related to papillary thyroid cancer using ligation-initiated phosphorothioated primer-based loop-mediated isothermal amplification. Sens. Actuator B-Chem. 2023, 374, 132785. [Google Scholar] [CrossRef]
- Yao, Y.Y.; Zhang, H.D.; Tian, T.T.; Liu, Y.X.; Zhu, R.D.; Ji, J.; Liu, B.H. Iodide-modified Ag nanoparticles coupled with DSN-assisted cycling amplification for label-free and ultrasensitive SERS detection of microRNA-21. Talanta 2021, 235, 122728. [Google Scholar] [CrossRef]
- Chen, C.F.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.H.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, J.; He, N.; Zhang, M.; Wu, L.; Chen, X.; Zhu, J.; Ran, F.; Chen, Q.; Zhang, H. CRISPR/Cas12a coupling with magnetic nanoparticles and cascaded strand displacement reaction for ultrasensitive fluorescence determination of exosomal miR-21. Molecules 2022, 27, 5338. [Google Scholar] [CrossRef]
- Deng, R.J.; Zhang, K.X.; Li, J.H. Isothermal amplification for microRNA detection: From the test tube to the cell. Acc. Chem. Res. 2017, 50, 1059–1068. [Google Scholar] [CrossRef]
- Kim, E.; Xu, J.X.; Kim, J.; Chun, H. Improving the robustness of a catalyzed hairpin assembly with a three-arm nanostructure for nonenzymatic signal amplification. Analyst 2022, 147, 1899–1905. [Google Scholar] [CrossRef] [PubMed]
- Bi, S.; Yue, S.Z.; Zhang, S.S. Hybridization chain reaction: A versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem. Soc. Rev. 2017, 46, 4281–4298. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.H.; Yang, K.K.; Tao, J.; Liu, Y.J.; Zhang, Q.; Habibi, S.; Nie, Z.H.; Xia, X.H. An enzyme-free signal amplification technique for ultrasensitive colorimetric assay of disease biomarkers. ACS Nano 2017, 11, 2052–2059. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Wang, G.; Zou, F.B.; Tang, S.; Chen, S.N.; Miao, P.; Jiao, J.W. Bifunctional probe-assisted hybridization chain reaction for circular RNA detection and live-cell imaging. Sens. Actuator B-Chem. 2023, 378, 133145. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, Y.L.; Ma, R.; Wang, W.J.; Zhang, L.; Li, J.; Sun, J.A.; Mao, X.Z. Aptasensing a class of small molecules based on split aptamers and hybridization chain reaction-assisted AuNPs nanozyme. Food Chem. 2023, 401, 134053. [Google Scholar] [CrossRef]
- Song, Z.H.; Zhou, Y.; Shen, M.Z.; Zhao, D.; Hu, H.H.; Zeng, S.; Sun, L.L.; Cai, S. MUC1 detection and in situ imaging method based on aptamer conformational switch and hybridization chain reaction. Talanta 2022, 239, 123129. [Google Scholar] [CrossRef]
- Choi, H.M.T.; Chang, J.Y.; Trinh, L.A.; Padilla, J.E.; Fraser, S.E.; Pierce, N.A. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 2010, 28, 1208–1212. [Google Scholar] [CrossRef]
- Wang, J.Y.; Li, H.X.; Du, C.Y.; Li, Y.; Ma, X.Y.; Yang, C.Y.; Xu, W.T.; Sun, C.Y. Structure-switching aptamer triggering signal amplification strategy for tobramycin detection based on hybridization chain reaction and fluorescence synergism. Talanta 2022, 243, 123318. [Google Scholar] [CrossRef]
- Kachwala, M.J.; Smith, C.W.; Nandu, N.; Yigit, M.V. Reprogrammable gel electrophoresis detection assay using crispr-cas12a and hybridization chain reaction. Anal. Chem. 2021, 93, 1934–1938. [Google Scholar] [CrossRef]
- Mei, S.Y.; Liu, B.R.; Xiong, X.L.; Hun, X. One-step fabrication of trimetallic alloy nanozyme catalyzer for luminol-H2O2 chemiluminescence and its application for miRNA-21 detection coupled with miRNA walking machine. J. Pharm. Biomed. Anal. 2020, 186, 113280. [Google Scholar] [CrossRef]
- Cai, S.; Ye, J.W.; AL-maskri, A.A.A.; Sun, L.L.; Zeng, S. A conformational switch-based aptasensor for the chemiluminescence detection of microRNA. Luminescence 2019, 34, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.T.; Xu, W.; Guo, J.B.; Ouyang, J.; Na, N. Chemiluminescence resonance energy transfer-based mesoporous silica nanosensors for the detection of miRNA. ACS Sens. 2020, 5, 2800–2805. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Wei, Q.X.; Yang, L.Y.; Li, J.Y.; Lu, T.C.; Liu, Z.J.; Zhong, G.X.; Weng, X.H.; Xu, X.W. Multimodal ochratoxin A-aptasensor using 3′-FAM-enhanced exonuclease I tool and magnetic microbead carrier. Anal. Chem. 2022, 94, 10921–10929. [Google Scholar] [CrossRef] [PubMed]
- Moutsiopoulou, A.; Broyles, D.; Dikici, E.; Daunert, S.; Deo, S.K. Molecular aptamer beacons and their applications in sensing, imaging, and diagnostics. Small 2019, 15, 1902248. [Google Scholar] [CrossRef] [PubMed]
- Moutsiopoulou, A.; Broyles, D.; Joda, H.; Dikici, E.; Kaur, A.; Kaifer, A.; Daunert, S.; Deo, S.K. Bioluminescent protein-inhibitor pair in the design of a molecular aptamer beacon biosensing system. Anal. Chem. 2022, 92, 7393–7398. [Google Scholar] [CrossRef]
- Han, C.; Li, Q.; Ji, H.S.; Xing, W.P.; Zhang, L.M.; Zhang, L.Y. Aptamers: The powerful molecular tools for virus detection. Chem. Asian J. 2021, 16, 1298–1306. [Google Scholar] [CrossRef]
- Bing, T.; Yang, X.J.; Mei, H.C.; Cao, Z.H.; Shangguan, D.H. Conservative secondary structure motif of streptavidin-binding aptamers generated by different laboratories. Bioorg. Med. Chem. 2010, 18, 1798–1805. [Google Scholar] [CrossRef]
- Jiang, H.; Peng, Z.; Lv, X.F.; Liu, Y.; Li, X.Q.; Deng, Y.L. Hybrid chain reaction nanoscaffold-based functional nucleic acid nanomaterial cascaded with rolling circle amplification for signal enhanced miRNA let-7a detection. Microchim. Acta 2024, 191, 533. [Google Scholar] [CrossRef]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. [Google Scholar] [CrossRef]
- Mo, L.T.; Liang, D.L.; Mo, M.X.; Yang, C.; Lin, W.Y. Dual-detection of miRNAs in living cells via hybridization chain reaction on DNA tetrahedron. Sens. Actuator B-Chem. 2023, 375, 132955. [Google Scholar]
- Wang, Y.H.; Feng, H.; Huang, K.; Quan, J.F.; Liu, X.H.; Jiang, H.; Wang, X.M. Target-triggered hybridization chain reaction for ultrasensitive dual-signal miRNA detection. Biosens. Bioelectron. 2022, 215, 114572. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Chen, J.; Yang, H.H.; Yin, W.; Li, C.R.; Xu, Y.Z.; Liu, S.Y.; Dai, Z.; Zou, X.Y. Light-controlled recruitable hybridization chain reaction on exosome vehicles for highly sensitive microRNA imaging in living cells. Anal. Chem. 2022, 94, 9665–9673. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Hu, J.; Li, Z.; Xu, Q.; Wang, C.S.; Jia, H.M.; Zhou, H.; Lin, P.; Zhao, W.W. Hybridization chain reaction for regulating surface capacitance of organic photoelectrochemical transistor toward sensitive miRNA detection. Biosens. Bioelectron. 2022, 209, 114224. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Pan, M.; Zou, Z.Q.; Fan, L.; Liu, X.Q. Hybridization chain reaction-mediated luminescent silver nanocluster system for amplified detection of miRNA-21. Chin. J. Anal. Chem. 2020, 48, 1193–1201. [Google Scholar]
- Ding, L.H.; Liu, H.Y.; Zhang, L.N.; Li, L.; Yu, J.H. Label-free detection of microRNA based on the fluorescence quenching of silicon nanoparticles induced by catalyzed hairpin assembly coupled with hybridization chain reaction. Sens. Actuator B-Chem. 2018, 254, 370–376. [Google Scholar] [CrossRef]
- Xu, J.; Lin, B.; Wang, Y.; Xu, Z.A.; Cheng, G.Y.; Zhang, W. Label-free fluorescent DNA detection strategy based on hybridization chain reaction and vacant site-binding molecule. Chin. J. Anal. Chem. 2018, 46, 1095–1101. [Google Scholar]
- Zhang, Z.K.; Zhang, L.; Liu, Y.M.; Hu, C.X.; Liu, Q.J. Sensitive DNA detection using a branched DNA as a sensor coupled with hybridization chain reaction. ChemistrySelect 2023, 7, e202201891. [Google Scholar] [CrossRef]

| Added (fmol) | Found (fmol) | Recovery (%) | RSD (%) | Mean Recovery (%) |
|---|---|---|---|---|
| 1000 | 981.00 ± 83.27 | 98.10 | 8.49 | 103.32 |
| 500 | 559.33 ± 52.34 | 111.87 | 9.36 | |
| 250 | 249.99 ± 31.13 | 99.99 | 12.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.; Li, J.; Li, M.; An, R.; Zhang, X.; Cai, S. A Chemiluminescence Signal Amplification Method for MicroRNA Detection: The Combination of Molecular Aptamer Beacons with Enzyme-Free Hybridization Chain Reaction. Molecules 2024, 29, 5782. https://doi.org/10.3390/molecules29235782
Han Y, Li J, Li M, An R, Zhang X, Cai S. A Chemiluminescence Signal Amplification Method for MicroRNA Detection: The Combination of Molecular Aptamer Beacons with Enzyme-Free Hybridization Chain Reaction. Molecules. 2024; 29(23):5782. https://doi.org/10.3390/molecules29235782
Chicago/Turabian StyleHan, Yu, Jialin Li, Man Li, Ran An, Xu Zhang, and Sheng Cai. 2024. "A Chemiluminescence Signal Amplification Method for MicroRNA Detection: The Combination of Molecular Aptamer Beacons with Enzyme-Free Hybridization Chain Reaction" Molecules 29, no. 23: 5782. https://doi.org/10.3390/molecules29235782
APA StyleHan, Y., Li, J., Li, M., An, R., Zhang, X., & Cai, S. (2024). A Chemiluminescence Signal Amplification Method for MicroRNA Detection: The Combination of Molecular Aptamer Beacons with Enzyme-Free Hybridization Chain Reaction. Molecules, 29(23), 5782. https://doi.org/10.3390/molecules29235782








