An Investigation of the Influence of Tyrosine Local Interactions on Electron Hopping in a Model Protein
Abstract
1. Introduction

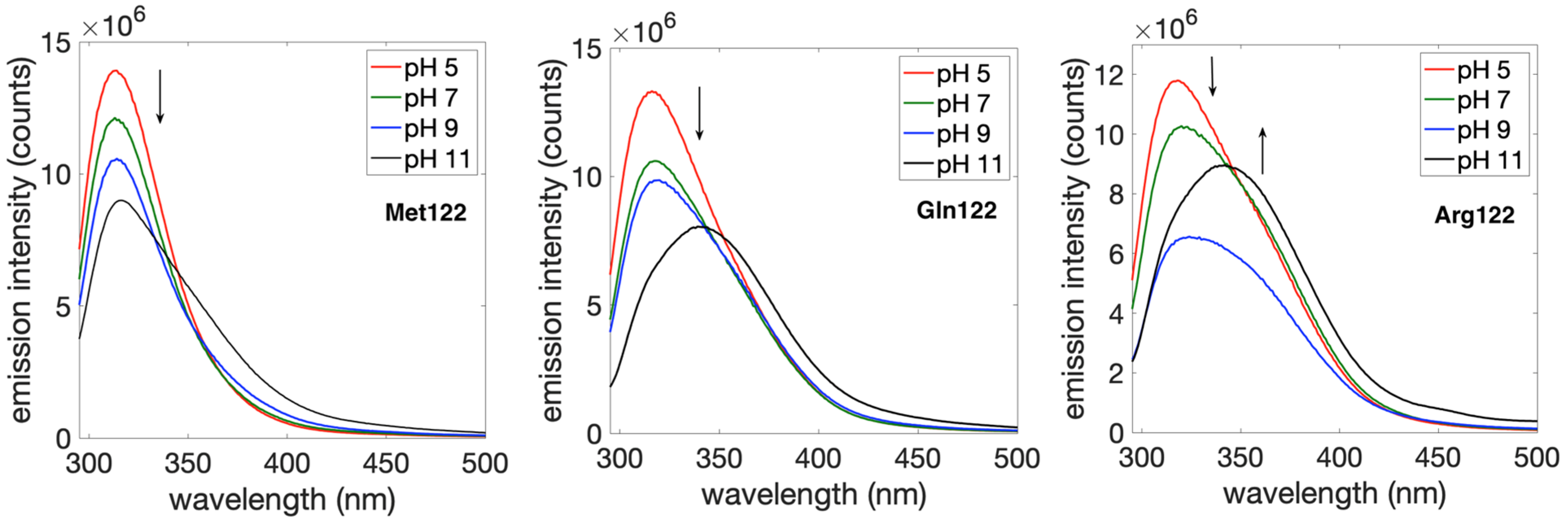
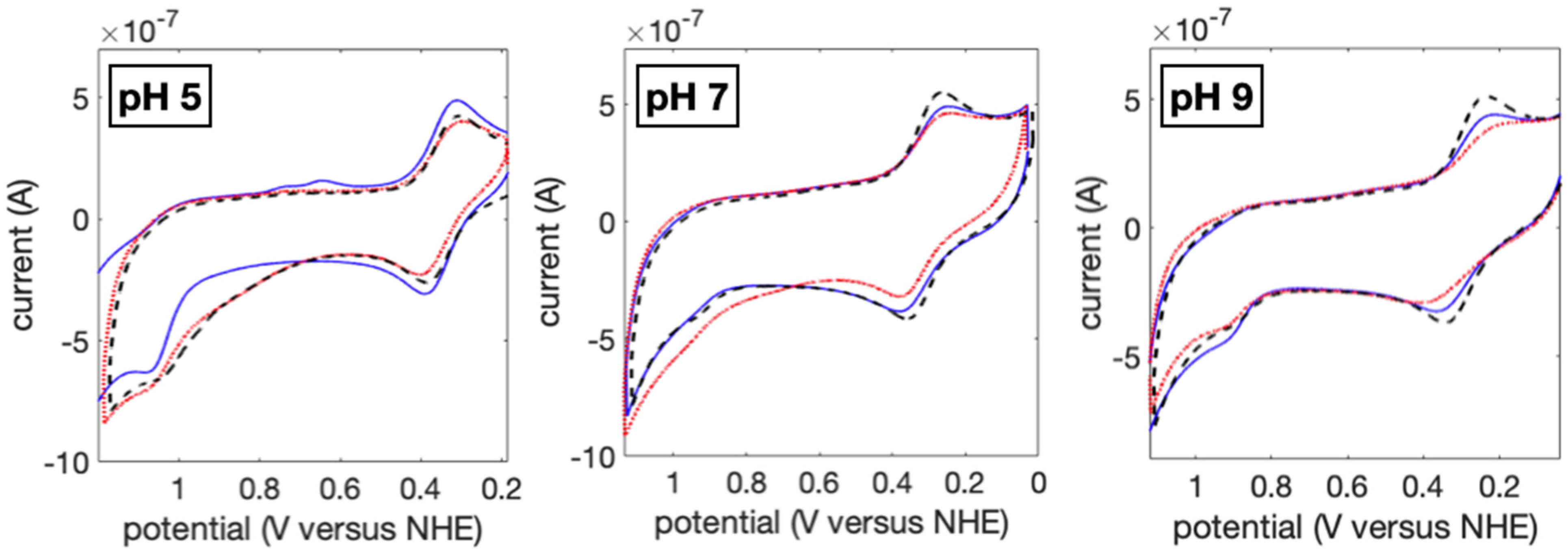
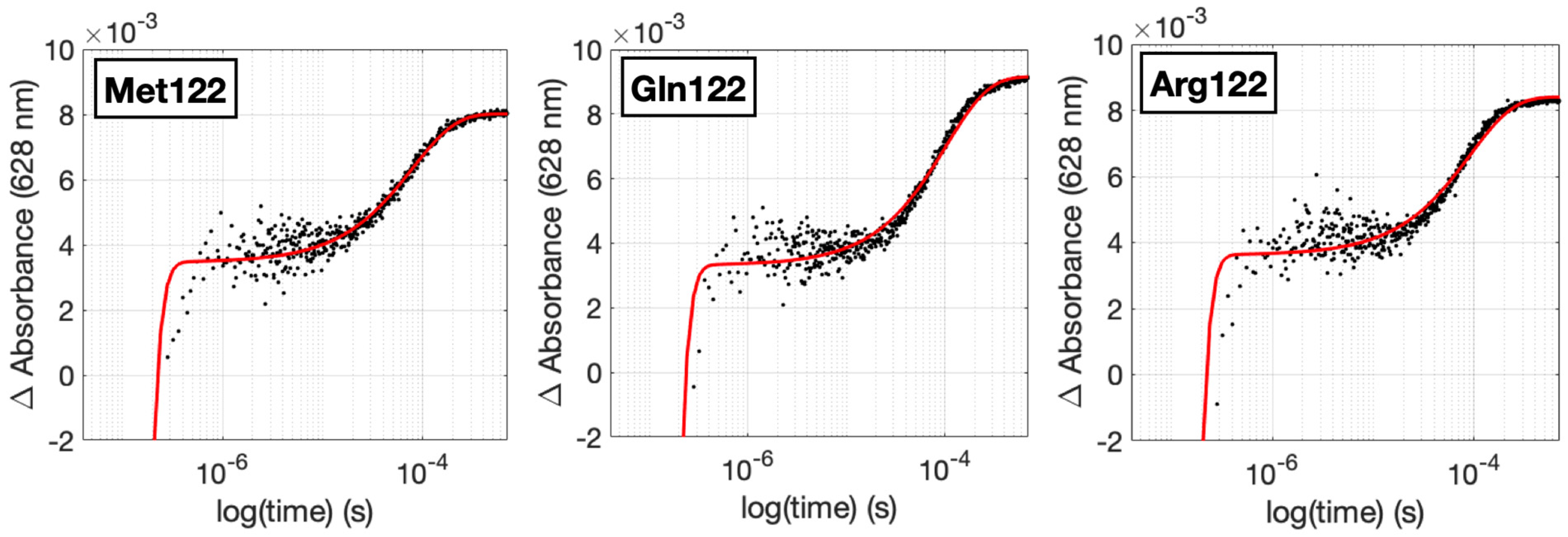
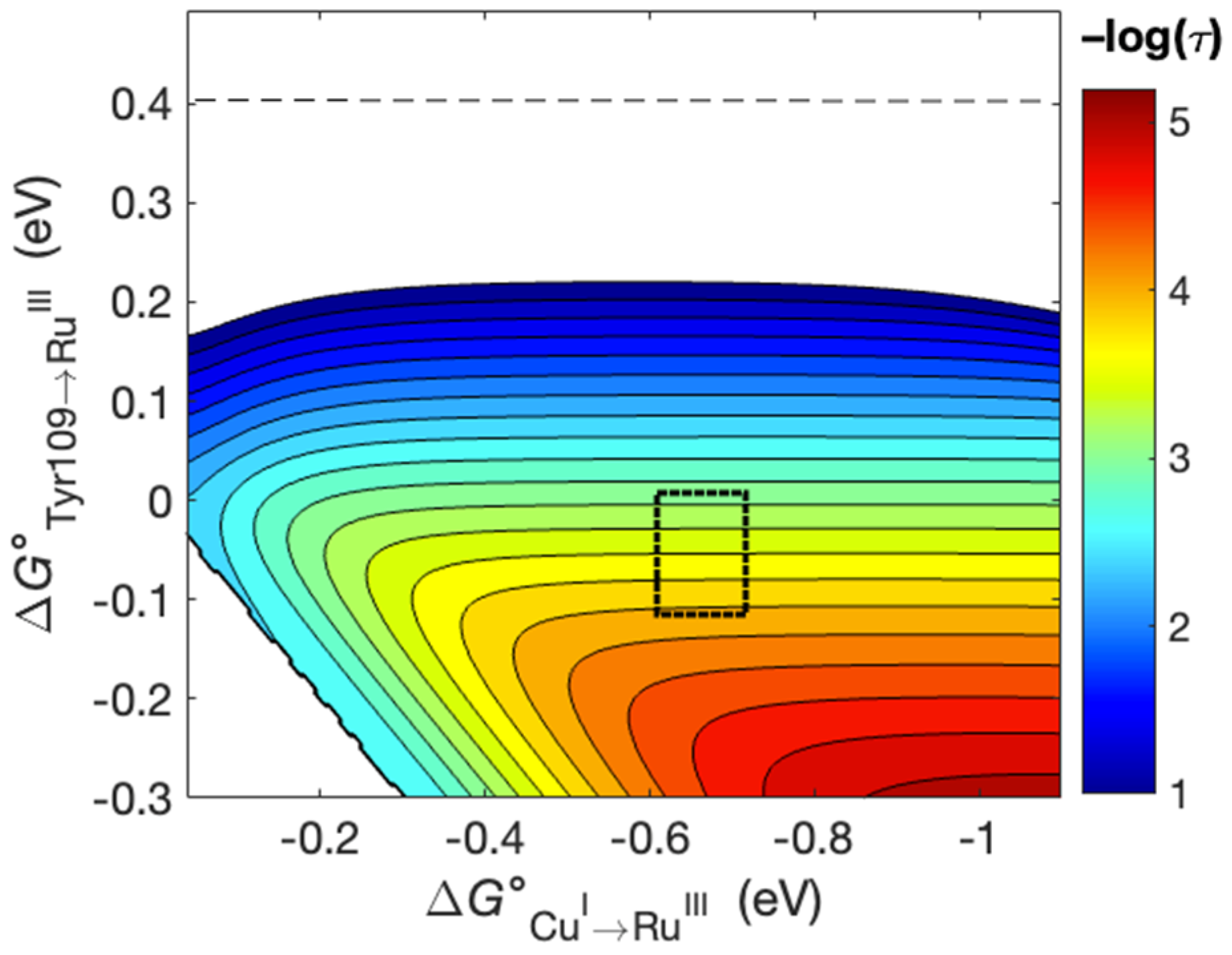
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lubitz, W.; Chrysina, M.; Cox, N. Water Oxidation in Photosystem II. Photosynth. Res. 2019, 142, 105–125. [Google Scholar] [CrossRef] [PubMed]
- Minnihan, E.C.; Nocera, D.G.; Stubbe, J. Reversible, Long-Range Radical Transfer in E. coli Class Ia Ribonucleotide Reductase. Acc. Chem. Res. 2013, 46, 2524–2535. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.L.; Urade, Y.; Jakobsson, P.-J. Enzymes of the Cyclooxygenase Pathways of Prostanoid Biosynthesis. Chem. Rev. 2011, 111, 5821–5865. [Google Scholar] [CrossRef] [PubMed]
- Ru, X.; Crane, B.R.; Zhang, P.; Beratan, D.N. Why Do Most Aromatics Fail to Support Hole Hopping in the Cytochrome c Peroxidase–Cytochrome c Complex? J. Phys. Chem. B 2021, 125, 7763–7773. [Google Scholar] [CrossRef] [PubMed]
- Yee, E.F.; Dzikovski, B.; Crane, B.R. Tuning Radical Relay Residues by Proton Management Rescues Protein Electron Hopping. J. Am. Chem. Soc. 2019, 141, 17571–17587. [Google Scholar] [CrossRef]
- Kathiresan, M.; English, A.M. LC-MS/MS Suggests That Hole Hopping in Cytochrome c Peroxidase Protects Its Heme from Oxidative Modification by Excess H2O2. Chem. Sci. 2017, 8, 1152–1162. [Google Scholar] [CrossRef]
- Kathiresan, M.; English, A.M. LC-MS/MS Proteoform Profiling Exposes Cytochrome c Peroxidase Self-Oxidation in Mitochondria and Functionally Important Hole Hopping from Its Heme. J. Am. Chem. Soc. 2018, 140, 12033–12039. [Google Scholar] [CrossRef]
- Gray, H.B.; Winkler, J.R. Hole Hopping through Tyrosine/Tryptophan Chains Protects Proteins from Oxidative Damage. Proc. Natl. Acad. Sci. USA 2015, 112, 10920–10925. [Google Scholar] [CrossRef]
- Winkler, J.R.; Gray, H.B. Electron Flow through Biological Molecules: Does Hole Hopping Protect Proteins from Oxidative Damage? Q. Rev. Biophys. 2015, 48, 411–420. [Google Scholar] [CrossRef]
- Teo, R.D.; Wang, R.; Smithwick, E.R.; Migliore, A.; Therien, M.J.; Beratan, D.N. Mapping Hole Hopping Escape Routes in Proteins. Proc. Natl. Acad. Sci. USA 2019, 116, 15811–15816. [Google Scholar] [CrossRef]
- Cordes, M.; Köttgen, A.; Jasper, C.; Jacques, O.; Boudebous, H.; Giese, B. Influence of Amino Acid Side Chains on Long-Distance Electron Transfer in Peptides: Electron Hopping via “Stepping Stones”. Angew. Chem. Int. Ed. 2008, 47, 3461–3463. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.R.; Di Bilio, A.J.; Farrow, N.A.; Richards, J.H.; Gray, H.B. Electron Tunneling in Biological Molecules. Pure Appl. Chem. 1999, 71, 1753–1764. [Google Scholar] [CrossRef]
- Cordes, M.; Giese, B. Electron Transfer in Peptides and Proteins. Chem. Soc. Rev. 2009, 38, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Nag, L.; Sournia, P.; Myllykallio, H.; Liebl, U.; Vos, M.H. Identification of the TyrOH•+ Radical Cation in the Flavoenzyme TrmFO. J. Am. Chem. Soc. 2017, 139, 11500–11505. [Google Scholar] [CrossRef] [PubMed]
- Nag, L.; Sournia, P.; Myllykallio, H.; Liebl, U.; Vos, M.H. Correction to “Identification of the TyrOH●+ Radical Cation in the Flavoenzyme TrmFO”. J. Am. Chem. Soc. 2017, 139, 15554. [Google Scholar] [CrossRef] [PubMed]
- Pirisi, K.; Nag, L.; Fekete, Z.; Iuliano, J.N.; Tolentino Collado, J.; Clark, I.P.; Pécsi, I.; Sournia, P.; Liebl, U.; Greetham, G.M.; et al. Identification of the Vibrational Marker of Tyrosine Cation Radical Using Ultrafast Transient Infrared Spectroscopy of Flavoprotein Systems. Photochem. Photobiol. Sci. 2021, 20, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Costentin, C.; Louault, C.; Robert, M.; Savéant, J.-M. The Electrochemical Approach to Concerted Proton—Electron Transfers in the Oxidation of Phenols in Water. Proc. Natl. Acad. Sci. USA 2009, 106, 18143–18148. [Google Scholar] [CrossRef]
- Bonin, J.; Costentin, C.; Louault, C.; Robert, M.; Routier, M.; Savéant, J.-M. Intrinsic Reactivity and Driving Force Dependence in Concerted Proton–Electron Transfers to Water Illustrated by Phenol Oxidation. Proc. Natl. Acad. Sci. USA 2010, 107, 3367–3372. [Google Scholar] [CrossRef]
- Warren, J.J.; Herrera, N.; Hill, M.G.; Winkler, J.R.; Gray, H.B. Electron Flow through Nitrotyrosinate in Pseudomonas aeruginosa Azurin. J. Am. Chem. Soc. 2013, 135, 11151–11158. [Google Scholar] [CrossRef]
- Giese, B.; Napp, M.; Jacques, O.; Boudebous, H.; Taylor, A.M.; Wirz, J. Multistep Electron Transfer in Oligopeptides: Direct Observation of Radical Cation Intermediates. Angew. Chem. Int. Ed. 2005, 117, 4141–4143. [Google Scholar] [CrossRef]
- Cordes, M.; Jacques, O.; Köttgen, A.; Jasper, C.; Boudebous, H.; Giese, B. Development of a Model System for the Study of Long Distance Electron Transfer in Peptides. Adv. Synth. Catal. 2008, 350, 1053–1062. [Google Scholar] [CrossRef]
- Gao, J.; Müller, P.; Wang, M.; Eckhardt, S.; Lauz, M.; Fromm, K.M.; Giese, B. Electron Transfer in Peptides: The Influence of Charged Amino Acids. Angew. Chem. Int. Ed. 2011, 50, 1926–1930. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Kasanmascheff, M.; Huynh, M.; Quartararo, A.; Costentin, C.; Bejenke, I.; Nocera, D.G.; Bennati, M.; Tommos, C.; Stubbe, J. Properties of Site-Specifically Incorporated 3-Aminotyrosine in Proteins To Study Redox-Active Tyrosines: Escherichia coli Ribonucleotide Reductase as a Paradigm. Biochemistry 2018, 57, 3402–3415. [Google Scholar] [CrossRef] [PubMed]
- Minnihan, E.C.; Young, D.D.; Schultz, P.G.; Stubbe, J. Incorporation of Fluorotyrosines into Ribonucleotide Reductase Using an Evolved, Polyspecific Aminoacyl-tRNA Synthetase. J. Am. Chem. Soc. 2011, 133, 15942–15945. [Google Scholar] [CrossRef]
- Ravichandran, K.R.; Zong, A.B.; Taguchi, A.T.; Nocera, D.G.; Stubbe, J.; Tommos, C. Formal Reduction Potentials of Difluorotyrosine and Trifluorotyrosine Protein Residues: Defining the Thermodynamics of Multistep Radical Transfer. J. Am. Chem. Soc. 2017, 139, 2994–3004. [Google Scholar] [CrossRef] [PubMed]
- Oyala, P.H.; Ravichandran, K.R.; Funk, M.A.; Stucky, P.A.; Stich, T.A.; Drennan, C.L.; Britt, R.D.; Stubbe, J. Biophysical Characterization of Fluorotyrosine Probes Site-Specifically Incorporated into Enzymes: E. coli Ribonucleotide Reductase As an Example. J. Am. Chem. Soc. 2016, 138, 7951–7964. [Google Scholar] [CrossRef]
- Warren, J.J.; Shafaat, O.S.; Winkler, J.R.; Gray, H.B. Proton-Coupled Electron Hopping in Ru-Modified P. aeruginosa Azurin. J. Biol. Inorg. Chem. 2016, 21, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Rayner, D.M.; Krajcarski, D.T.; Szabo, A.G. Excited State Acid–Base Equilibrium of Tyrosine. Can. J. Chem. 1978, 56, 1238–1245. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluoresence Spectroscopy, 2nd ed.; Kleuwer Academic/Plenum: New York, NY, USA, 1999. [Google Scholar]
- Pascher, T.; Karlsson, B.G.; Nordling, M.; Malmström, B.G.; Vänngård, T. Reduction Potentials and Their pH Dependence in Site-Directed-Mutant Forms of Azurin from Pseudomonas aeruginosa. Eur. J. Biochem. 1993, 212, 289–296. [Google Scholar] [CrossRef]
- St. Clair, C.S.; Ellis, W.R.; Gray, H.B. Spectroelectrochemistry of Blue Copper Proteins: pH and Temperature Dependences of the Reduction Potentials of Five Azurins. Inorg. Chim. Acta 1992, 191, 149–155. [Google Scholar] [CrossRef]
- Berry, B.W.; Martinez-Rivera, M.C.; Tommos, C. Reversible Voltammograms and a Pourbaix Diagram for a Protein Tyrosine Radical. Proc. Natl. Acad. Sci. USA 2012, 109, 9739–9743. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Rivera, M.C.; Berry, B.W.; Valentine, K.G.; Westerlund, K.; Hay, S.; Tommos, C. Electrochemical and Structural Properties of a Protein System Designed To Generate Tyrosine Pourbaix Diagrams. J. Am Chem. Soc. 2011, 133, 17786–17795. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Langen, R.; Chang, I.-J.; Germanas, J.P.; Richards, J.H.; Winkler, J.R.; Gray, H.B. Electron Tunneling in Proteins: Coupling Through a β Strand. Science 1995, 268, 1733–1735. [Google Scholar] [CrossRef] [PubMed]
- Regan, J.J.; Di Bilio, A.J.; Langen, R.; Skov, L.K.; Winkler, J.R.; Gray, H.B.; Onuchic, J.N. Electron Tunneling in Azurin: The Coupling across a β-Sheet. Chem. Biol. 1995, 2, 489–496. [Google Scholar] [CrossRef]
- Faham, S.; Day, M.W.; Connick, W.B.; Crane, B.R.; Bilio, A.J.D.; Schaefer, W.P.; Rees, D.C.; Gray, H.B. Structures of Ruthenium-Modified Pseudomonas Aeruginosa Azurin and [Ru(2,2′-Bipyridine)2(Imidazole)2]SO4·10H2O. Acta Crystallogr. D 1999, 55, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Chang, I.J.; Gray, H.B.; Winkler, J.R. High-Driving-Force Electron Transfer in Metalloproteins: Intramolecular Oxidation of Ferrocytochrome c by Ru(2,2’-Bpy)2(Im)(His-33)3+. J. Am. Chem. Soc. 1991, 113, 7056–7057. [Google Scholar] [CrossRef]
- Krishtalik, L.I. pH-Dependent Redox Potential: How to Use It Correctly in the Activation Energy Analysis. Biochim. Biophys. Acta 2003, 1604, 13–21. [Google Scholar] [CrossRef]
- Fedoretz-Maxwell, B.P.; Shin, C.H.; MacNeil, G.A.; Worrall, L.J.; Park, R.; Strynadka, N.C.J.; Walsby, C.J.; Warren, J.J. The Impact of Second Coordination Sphere Methionine-Aromatic Interactions in Copper Proteins. Inorg. Chem. 2022, 61, 5563–5571. [Google Scholar] [CrossRef]
- Shih, C.; Museth, A.K.; Abrahamsson, M.; Blanco-Rodriguez, A.M.; Di Bilio, A.J.; Sudhamsu, J.; Crane, B.R.; Ronayne, K.L.; Towrie, M.; Vlcek, A.; et al. Tryptophan-Accelerated Electron Flow Through Proteins. Science 2008, 320, 1760–1762. [Google Scholar] [CrossRef]
- Warren, J.J.; Ener, M.E.; Vlček, A.; Winkler, J.R.; Gray, H.B. Electron Hopping through Proteins. Coord. Chem. Rev. 2012, 256, 2478–2487. [Google Scholar] [CrossRef]
- Warren, J.J.; Winkler, J.R.; Gray, H.B. Hopping Maps for Photosynthetic Reaction Centers. Coord. Chem. Rev. 2013, 257, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.B.; Cho, M.P.; Wishart, J.F.; Emge, T.J.; Isied, S.S. Cis-Bis(Bipyridine)Ruthenium Imidazole Derivatives: A Spectroscopic, Kinetic, and Structural Study. Inorg. Chem. 1996, 35, 7241–7245. [Google Scholar] [CrossRef] [PubMed]
- Marcus, R.A.; Sutin, N. Electron Transfers in Chemistry and Biology. Biochim. Biophys. Acta 1985, 811, 265–322. [Google Scholar] [CrossRef]
- Gibbs, C.A.; Fedoretz-Maxwell, B.P.; MacNeil, G.A.; Walsby, C.J.; Warren, J.J. Proximal Methionine Amino Acid Residue Affects the Properties of Redox-Active Tryptophan in an Artificial Model Protein. ACS Omega 2023, 8, 19798–19806. [Google Scholar] [CrossRef]
- Tommos, C. Insights into the Thermodynamics and Kinetics of Amino-Acid Radicals in Proteins. Annu. Rev. Biophys. 2022, 51, 453–471. [Google Scholar] [CrossRef] [PubMed]
- Tommos, C.; Skalicky, J.J.; Pilloud, D.L.; Wand, A.J.; Dutton, P.L. De Novo Proteins as Models of Radical Enzymes. Biochemistry 1999, 38, 9495–9507. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Baumann, U.; Reymond, J.-L. An Efficient One-Step Site-Directed and Site-Saturation Mutagenesis Protocol. Nucleic Acids Res. 2004, 32, e115. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chakraborty, S.; Hosseinzadeh, P.; Yu, Y.; Tian, S.; Petrik, I.; Bhagi, A.; Lu, Y. Metalloproteins Containing Cytochrome, Iron–Sulfur, or Copper Redox Centers. Chem. Rev. 2014, 114, 4366–4469. [Google Scholar] [CrossRef]
- Wang, M.; Gao, J.; Müller, P.; Giese, B. Electron Transfer in Peptides with Cysteine and Methionine as Relay Amino Acids. Angew. Chem. Int. Ed. 2009, 48, 4232–4234. [Google Scholar] [CrossRef]
- Brunelle, P.; Rauk, A. One-Electron Oxidation of Methionine in Peptide Environments: The Effect of Three-Electron Bonding on the Reduction Potential of the Radical Cation. J. Phys. Chem. A 2004, 108, 11032–11041. [Google Scholar] [CrossRef]
- Brunelle, P.; Schöneich, C.; Rauk, A. One-Electron Oxidation of Methionine Peptides—Stability of the Three-Electron S—N(Amide) Bond. Can. J. Chem. 2006, 84, 893–904. [Google Scholar] [CrossRef]
- Beratan, D.N. Why Are DNA and Protein Electron Transfer So Different? Annu. Rev. Phys. Chem. 2019, 70, 71–97. [Google Scholar] [CrossRef] [PubMed]
- Beratan, D.N.; Liu, C.; Migliore, A.; Polizzi, N.F.; Skourtis, S.S.; Zhang, P.; Zhang, Y. Charge Transfer in Dynamical Biosystems, or The Treachery of (Static) Images. Acc. Chem. Res. 2015, 48, 474–481. [Google Scholar] [CrossRef]
- Beratan, D.N.; Skourtis, S.S.; Balabin, I.A.; Balaeff, A.; Keinan, S.; Venkatramani, R.; Xiao, D. Steering Electrons on Moving Pathways. Acc. Chem. Res. 2009, 42, 1669–1678. [Google Scholar] [CrossRef] [PubMed]
- Zahler, C.T.; Shaw, B.F. What Are We Missing by Not Measuring the Net Charge of Proteins? Chem. Eur. J. 2019, 25, 7581–7590. [Google Scholar] [CrossRef] [PubMed]
- Zahler, C.T.; Zhou, H.; Abdolvahabi, A.; Holden, R.L.; Rasouli, S.; Tao, P.; Shaw, B.F. Direct Measurement of Charge Regulation in Metalloprotein Electron Transfer. Angew. Chem. Int. Ed. 2018, 57, 5364–5368. [Google Scholar] [CrossRef] [PubMed]
- Markle, T.F.; Tronic, T.A.; DiPasquale, A.G.; Kaminsky, W.; Mayer, J.M. Effect of Basic Site Substituents on Concerted Proton-Electron Transfer in Hydrogen-Bonded Pyridyl-Phenols. J. Phys. Chem. A 2012, 116, 12249–12259. [Google Scholar] [CrossRef]
- Markle, T.F.; Tenderholt, A.L.; Mayer, J.M. Probing Quantum and Dynamic Effects in Concerted Proton-Electron Transfer Reactions of Phenol-Base Compounds. J. Phys. Chem. B 2011, 116, 571–584. [Google Scholar] [CrossRef]
- Irebo, T.; Reece, S.Y.; Sjoedin, M.; Nocera, D.G.; Hammarström, L. Proton-Coupled Electron Transfer of Tyrosine Oxidation: Buffer Dependence and Parallel Mechanisms. J. Am. Chem. Soc. 2007, 129, 15462–15464. [Google Scholar] [CrossRef]
- Wenger, O.S. Proton-Coupled Electron Transfer with Photoexcited Metal Complexes. Acc. Chem. Res. 2013, 46, 1517–1526. [Google Scholar] [CrossRef]
| pH = | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|
| His107Tyr109Met122 | 313 | 313 | 314 | 313 | 314 | 314 | 314 | 315 |
| His107Tyr109Gln122 | 316 | 316 | 316 | 317 | 317 | 318 | 319 | 340 |
| His107Tyr109Arg122 | 317 | 317 | 318 | 319 | 321 | 321 | 324 | 341 |
| pH = | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|
| Cu | 0.348 | 0.339 | 0.312 | 0.292 | 0.288 |
| Tyr109Met122 | 1.04 | 0.975 | 0.930 | 0.877 | 0.876 |
| Tyr109Gln122 | 1.02 | 0.962 | 0.927 | 0.898 | 0.893 |
| Tyr109Arg122 | 1.02 | 0.980 | 0.912 | 0.880 | 0.876 |
| pH | Met122 | Gln122 | Arg122 | Lys122 b |
|---|---|---|---|---|
| 6 | (6.8 ± 0.5) × 103 3.83 | (6.3 ± 0.6) × 103 3.80 | (5.4 ± 1.1) × 103 3.73 | (5.7 ± 0.3) × 103 3.76 |
| 7 | (1.2 ± 0.1) × 104 4.09 | (1.1 ± 0.1) × 104 4.03 | (1.1 ± 0.1) × 104 4.05 | (1.2 ± 0.2) × 104 4.08 |
| 8 | (1.8 ± 0.1) × 104 4.26 | (1.6 ± 0.1) × 104 4.19 | (1.6 ± 0.1) × 104 4.19 | (1.7 ± 0.2) × 104 4.23 |
| 9 | (2.6 ± 0.1) × 104 4.42 | (2.1 ± 0.1) × 104 4.33 | (2.2 ± 0.1) × 104 4.32 | (1.8 ± 0.2) × 104 4.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gibbs, C.A.; Ghazi, N.; Tao, J.; Warren, J.J. An Investigation of the Influence of Tyrosine Local Interactions on Electron Hopping in a Model Protein. Molecules 2024, 29, 350. https://doi.org/10.3390/molecules29020350
Gibbs CA, Ghazi N, Tao J, Warren JJ. An Investigation of the Influence of Tyrosine Local Interactions on Electron Hopping in a Model Protein. Molecules. 2024; 29(2):350. https://doi.org/10.3390/molecules29020350
Chicago/Turabian StyleGibbs, Curtis A., Nikta Ghazi, Jody Tao, and Jeffrey J. Warren. 2024. "An Investigation of the Influence of Tyrosine Local Interactions on Electron Hopping in a Model Protein" Molecules 29, no. 2: 350. https://doi.org/10.3390/molecules29020350
APA StyleGibbs, C. A., Ghazi, N., Tao, J., & Warren, J. J. (2024). An Investigation of the Influence of Tyrosine Local Interactions on Electron Hopping in a Model Protein. Molecules, 29(2), 350. https://doi.org/10.3390/molecules29020350






