Gold/DNA-Cu2+ Complex Nanozyme-Based Aptamer Lateral Flow Assay for Highly Sensitive Detection of Kanamycin
Abstract
1. Introduction
2. Results
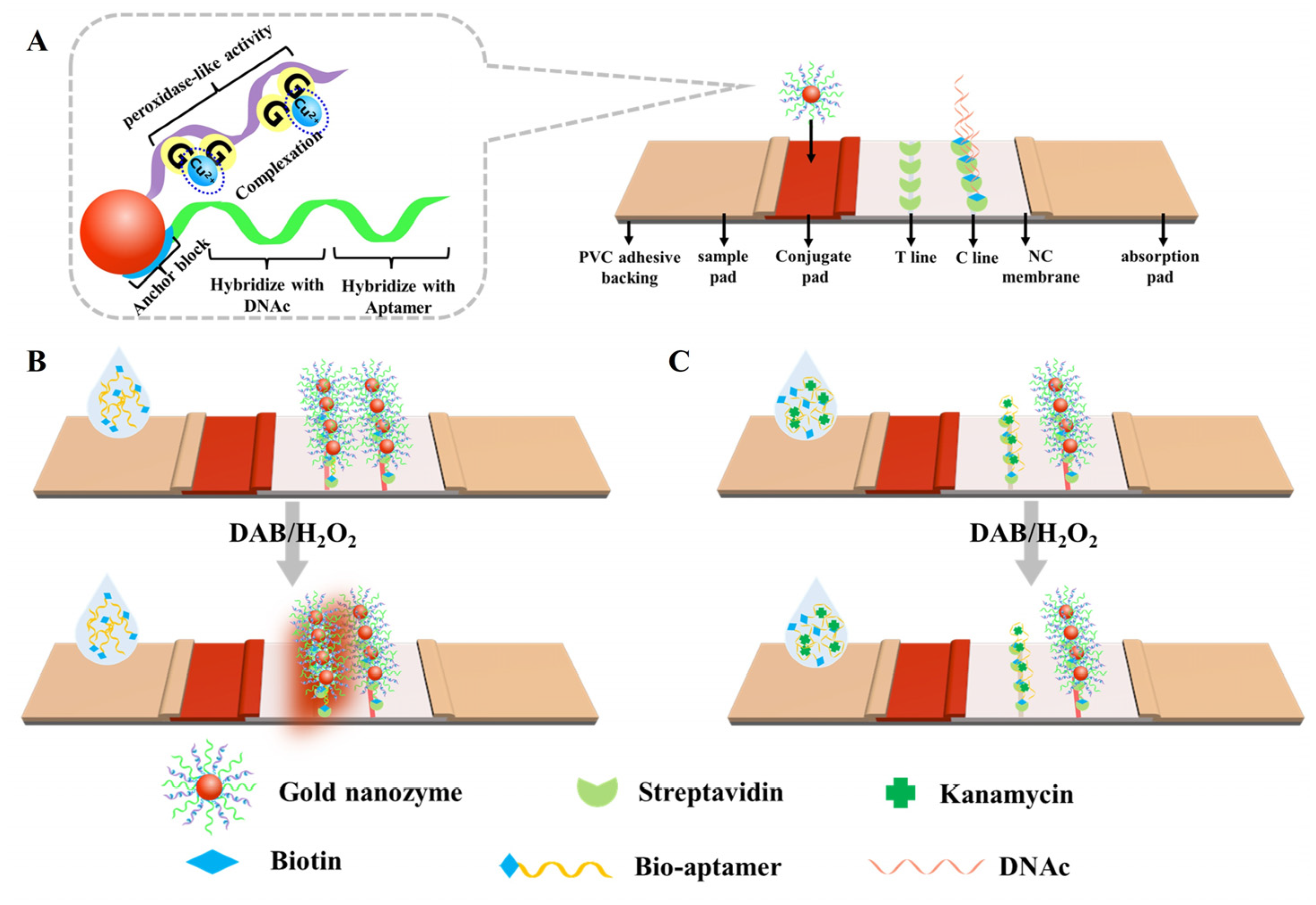
2.1. Principle of the Apt-LFA
2.2. Optimization of the Apt-LFA
2.2.1. Optimization of H2O2 and DAB Concentration
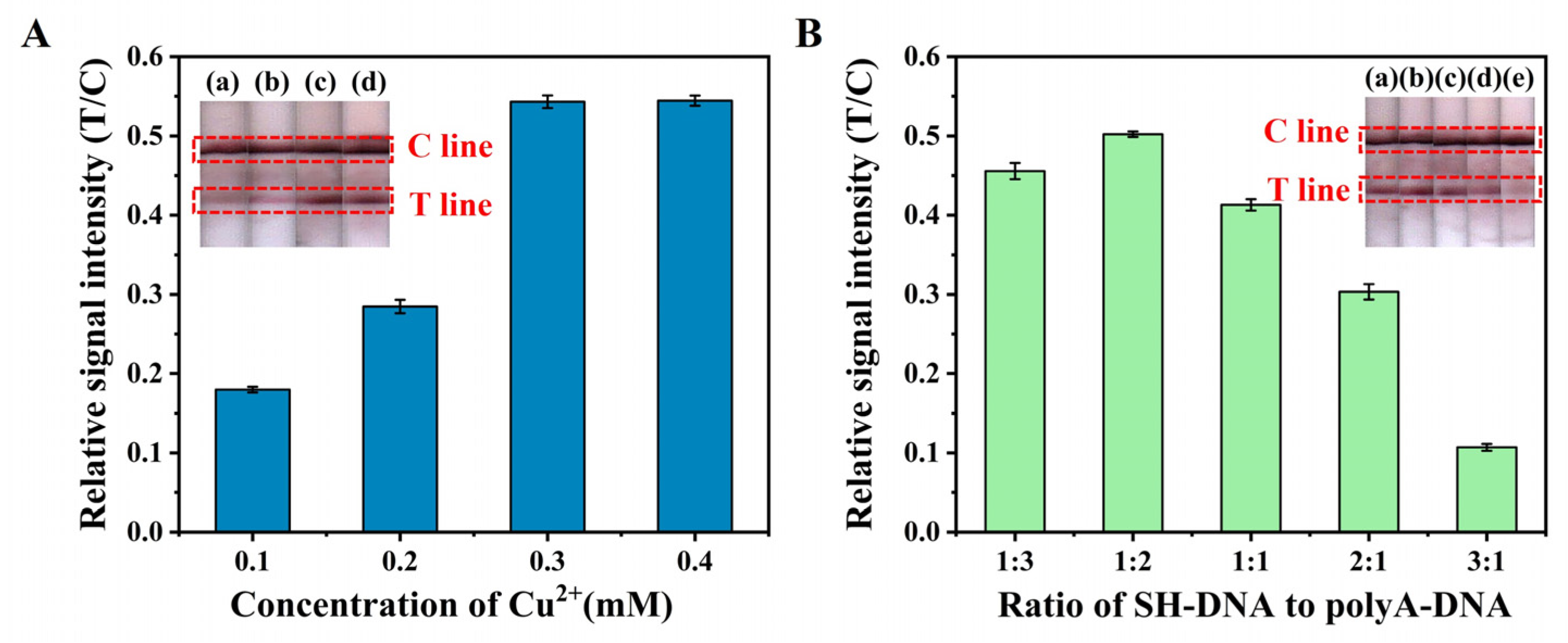
2.2.2. Optimization of Cu2+ Concentration
2.2.3. Optimization of the SH-DNA-to-polyA-DNA Ratio
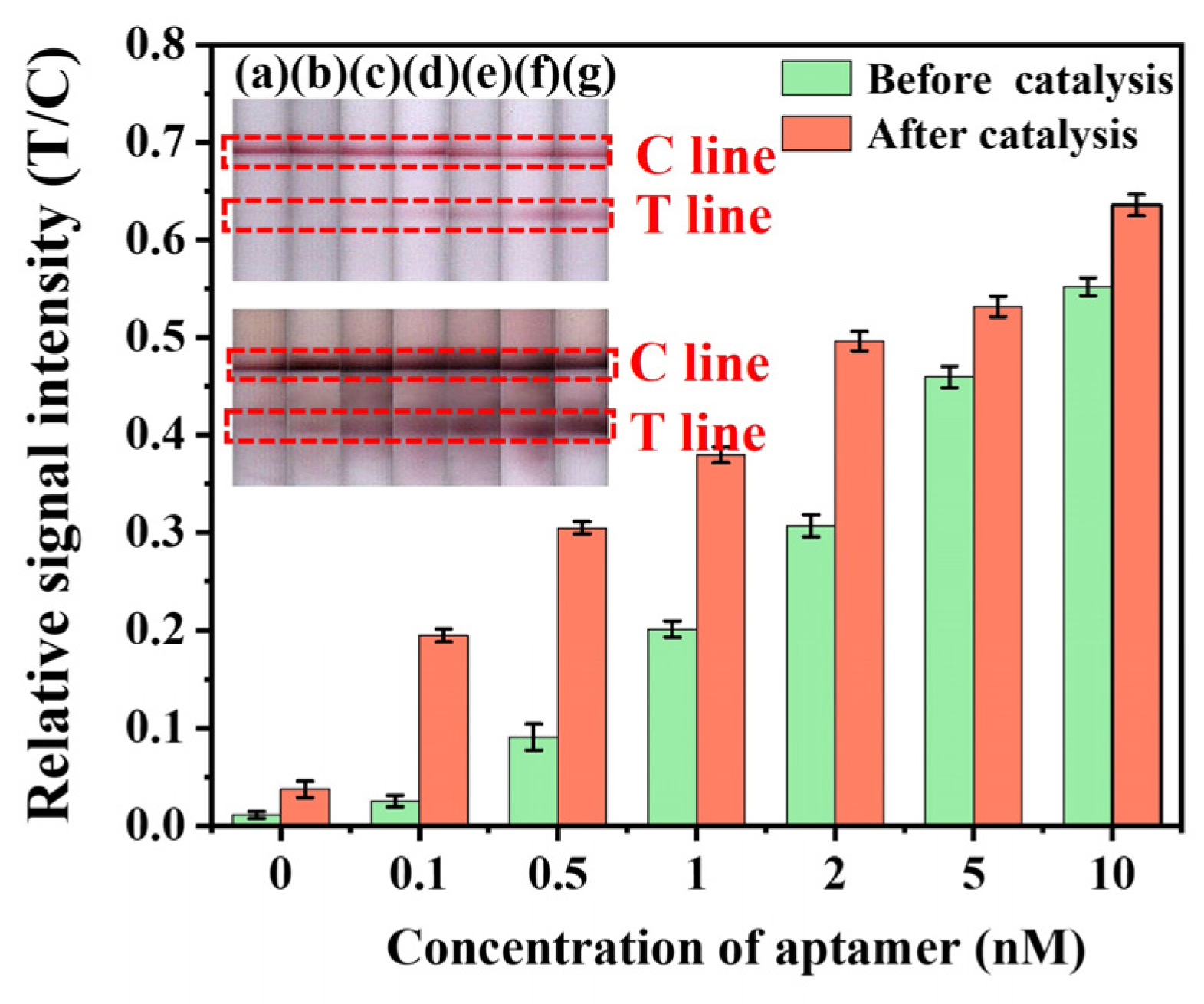
2.2.4. Optimization of Aptamer Concentration
2.3. Performance of Apt-LFA
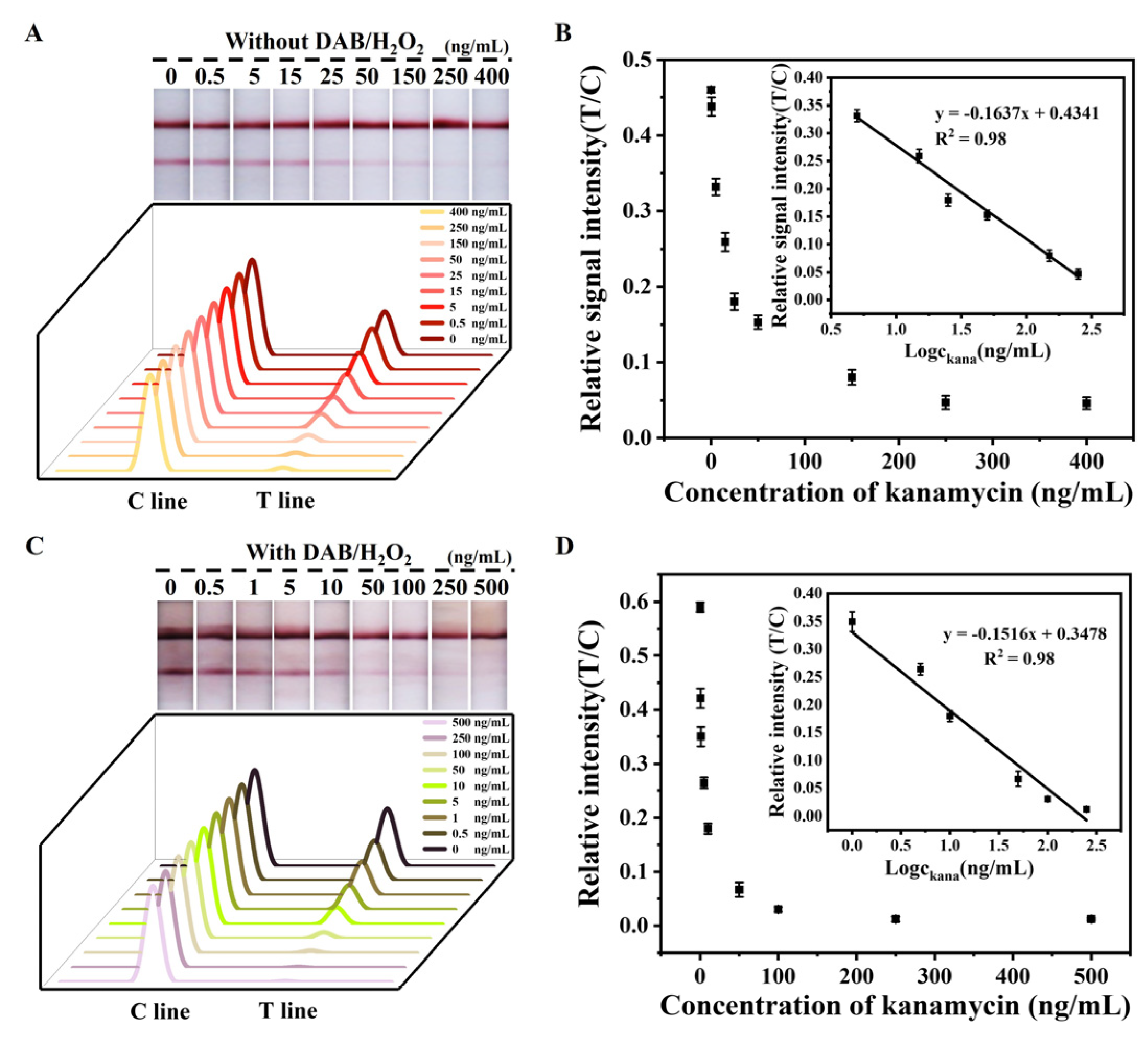
2.3.1. Detection of Kanamycin
2.3.2. Specificity Analysis
2.3.3. Real Sample Assay
3. Materials and Methods
3.1. Chemicals and Apparatus
3.2. Preparation of DNA-AuNPs Conjugates and AuNPs Nanozyme
3.3. Fabrication of the Apt-LFA Test Strip
3.4. Protocol of the Apt-LFA for Kanamycin
3.5. Sample Meaurements
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, W.; Shan, S.; Peng, J.; Liu, D.; Xia, J.; Shao, B.; Lai, W. Sensitive and hook effect–free lateral flow assay integrated with cascade signal transduction system. Sens. Actuators B Chem. 2020, 321, 128465. [Google Scholar] [CrossRef]
- Huang, L.; Tian, S.; Zhao, W.; Liu, K.; Ma, X.; Guo, J. Aptamer-based lateral flow assay on-site biosensors. Biosens. Bioelectron. 2021, 186, 113279. [Google Scholar] [CrossRef] [PubMed]
- Kasetsirikul, S.; Shiddiky, M.J.; Nguyen, N.-T. Challenges and perspectives in the development of paper-based lateral flow assays. Microfluid. Nanofluid 2020, 24, 1–18. [Google Scholar] [CrossRef]
- Nguyen, V.-T.; Song, S.; Park, S.; Joo, C. Recent advances in high-sensitivity detection methods for paper-based lateral-flow assay. Biosens. Bioelectron. 2020, 152, 112015. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 1992, 355, 850–852. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chen, L.; Chikkanna, A.; Chen, S.; Brusius, I.; Sbuh, N.; Veedu, R.N. Development of nucleic acid aptamer-based lateral flow assays: A robust platform for cost-effective point-of-care diagnosis. Theranostics 2021, 11, 5174. [Google Scholar] [CrossRef]
- Mao, M.; Xie, Z.; Ma, P.; Peng, C.; Wang, Z.; Wei, X.; Liu, G. Design and optimizing gold nanoparticle-cDNA nanoprobes for aptamer-based lateral flow assay: Application to rapid detection of acetamiprid. Biosens. Bioelectron. 2022, 207, 114114. [Google Scholar] [CrossRef]
- Chen, X.; Ding, L.; Huang, X.; Xiong, Y. Tailoring noble metal nanoparticle designs to enable sensitive lateral flow immunoassay. Theranostics 2022, 12, 574. [Google Scholar] [CrossRef]
- Chiu, T.-C.; Huang, C.-C. Aptamer-functionalized nano-biosensors. Sensors 2009, 9, 10356–10388. [Google Scholar] [CrossRef]
- Li, X.; Chen, X.; Mao, M.; Peng, C.; Wang, Z. Accelerated CRISPR/Cas12a-based small molecule detection using bivalent aptamer. Biosens. Bioelectron. 2022, 217, 114725. [Google Scholar] [CrossRef]
- Omidfar, K.; Khorsand, F.; Azizi, M.D. New analytical applications of gold nanoparticles as label in antibody based sensors. Biosens. Bioelectron. 2013, 43, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Ren, M.; Li, Y.; Huang, Z.; Shu, M.; Yang, H.; Xiong, Y.; Xu, Y. Detection of aflatoxin B1 with immunochromatographic test strips: Enhanced signal sensitivity using gold nanoflowers. Talanta 2015, 142, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Ren, J.; Qu, X. Nanozymes: Classification, catalytic mechanisms, activity regulation, and applications. Chem. Rev. 2019, 119, 4357–4412. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, X.; Wang, Q.; Lou, Z.; Li, S.; Zhu, Y.; Qin, L.; Wei, H. Nanomaterials with enzyme-like characteristics (nanozymes): Next-generation artificial enzymes (II). Chem. Soc. Rev. 2019, 48, 1004–1076. [Google Scholar]
- Wei, D.; Zhang, X.; Chen, B.; Zeng, K. Using bimetallic Au@ Pt nanozymes as a visual tag and as an enzyme mimic in enhanced sensitive lateral-flow immunoassays: Application for the detection of streptomycin. Anal. Chim. Acta 2020, 1126, 106–113. [Google Scholar] [CrossRef]
- Baranwal, A.; Shukla, R.; Bansal, V. Nanozyme-enhanced paper-based biosensor technologies. TrAC Trends Anal. Chem. 2024, 172, 117573. [Google Scholar] [CrossRef]
- Tominaga, T. Enhanced sensitivity of lateral-flow test strip immunoassays using colloidal palladium nanoparticles and horseradish peroxidase. LWT 2017, 86, 566–570. [Google Scholar] [CrossRef]
- Han, J.; Zhang, L.; Hu, L.; Xing, K.; Lu, X.; Huang, Y.; Zhang, J.; Lai, W.; Chen, T. Nanozyme-based lateral flow assay for the sensitive detection of Escherichia coli O157: H7 in milk. J. Dairy Sci. 2018, 101, 5770–5779. [Google Scholar] [CrossRef]
- Liu, S.; Dou, L.; Yao, X.; Zhang, W.; Zhao, M.; Yin, X.; Sun, J.; Zhang, D.; Wang, J. Nanozyme amplification mediated on-demand multiplex lateral flow immunoassay with dual-readout and broadened detection range. Biosens. Bioelectron. 2020, 169, 112610. [Google Scholar] [CrossRef]
- Cheng, N.; Song, Y.; Zeinhom, M.M.; Chang, Y.-C.; Sheng, L.; Li, H.; Du, D.; Li, L.; Zhu, M.-J.; Luo, Y. Nanozyme-mediated dual immunoassay integrated with smartphone for use in simultaneous detection of pathogens. ACS Appl. Mater. Interfaces 2017, 9, 40671–40680. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, M.; Fu, Q.; Ouyang, H.; Wen, W.; Song, Y.; Zhu, C.; Lin, Y.; Du, D. A nanozyme-and ambient light-based smartphone platform for simultaneous detection of dual biomarkers from exposure to organophosphorus pesticides. Anal. Chem. 2018, 90, 7391–7398. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Wang, X.; Liu, B.; Liu, J. Conjugation of antibodies and aptamers on nanozymes for developing biosensors. Biosens. Bioelectron. 2020, 168, 112537. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Kumar, A.; Sachan, M.; Gupta, S.; Nara, S. Aptamer-gold nanozyme based competitive lateral flow assay for rapid detection of CA125 in human serum. Biosens. Bioelectron. 2020, 165, 112368. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, G.; Pan, Q.; Xue, F.; Wang, Z.; Peng, C. Rapid and ultra-sensitive lateral flow assay for pathogens based on multivalent aptamer and magnetic nanozyme. Biosens. Bioelectron. 2024, 250, 116044. [Google Scholar] [CrossRef]
- Kim, B.-H.; Yoon, I.S.; Lee, J.-S. Masking nanoparticle surfaces for sensitive and selective colorimetric detection of proteins. Anal. Chem. 2013, 85, 10542–10548. [Google Scholar] [CrossRef]
- Weerathunge, P.; Ramanathan, R.; Shukla, R.; Sharma, T.K.; Bansal, V. Aptamer-controlled reversible inhibition of gold nanozyme activity for pesticide sensing. Anal. Chem. 2014, 86, 11937–11941. [Google Scholar] [CrossRef]
- Weerathunge, P.; Ramanathan, R.; Torok, V.A.; Hodgson, K.; Xu, Y.; Goodacre, R.; Behera, B.K.; Bansal, V. Ultrasensitive colorimetric detection of murine norovirus using NanoZyme aptasensor. Anal. Chem. 2019, 91, 3270–3276. [Google Scholar] [CrossRef]
- Sharma, T.K.; Ramanathan, R.; Weerathunge, P.; Mohammadtaheri, M.; Daima, H.K.; Shukla, R.; Bansal, V. Aptamer-mediated ‘turn-off/turn-on’nanozyme activity of gold nanoparticles for kanamycin detection. Chem. Commun. 2014, 50, 15856–15859. [Google Scholar] [CrossRef]
- Santangelo, M.G.; Antoni, P.M.; Spingler, B.; Jeschke, G. Can Copper (II) Mediate Hoogsteen Base-Pairing in a Left-Handed DNA Duplex? A Pulse EPR Study. ChemPhysChem 2010, 11, 599–606. [Google Scholar] [CrossRef]
- Chang, Y.; Zhang, Z.; Hao, J.; Yang, W.; Tang, J. A simple label free colorimetric method for glyphosate detection based on the inhibition of peroxidase-like activity of Cu (Ⅱ). Sens. Actuators B Chem. 2016, 228, 410–415. [Google Scholar] [CrossRef]
- Li, W.; Zhao, X.; Zhang, J.; Fu, Y. Cu (II)-coordinated GpG-duplex DNA as peroxidase mimetics and its application for label-free detection of Cu2+ ions. Biosens. Bioelectron. 2014, 60, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Loomans, E.E.; Van Wiltenburg, J.; Koets, M.; Van Amerongen, A. Neamin as an immunogen for the development of a generic ELISA detecting gentamicin, kanamycin, and neomycin in milk. J. Agric. Food Chem. 2003, 51, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Qi, T.; Jin, Y.; Liang, L.; Zhao, J. A signal-on fluorescent aptasensor based on gold nanoparticles for kanamycin detection. RSC Adv. 2021, 11, 10054–10060. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhan, L.; Qin, Z.; Sackrison, J.; Bischof, J.C. Ultrasensitive and highly specific lateral flow assays for point-of-care diagnosis. ACS Nano 2021, 15, 3593–3611. [Google Scholar] [CrossRef]
- Ou, Y.; Jin, X.; Liu, J.; Tian, Y.; Zhou, N. Visual detection of kanamycin with DNA-functionalized gold nanoparticles probe in aptamer-based strip biosensor. Anal. Biochem. 2019, 587, 113432. [Google Scholar] [CrossRef]
- Frens, G. Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nat. Phys. Sci. 1973, 241, 20–22. [Google Scholar] [CrossRef]
- Grabar, K.C.; Freeman, R.G.; Hommer, M.B.; Natan, M.J. Preparation and characterization of Au colloid monolayers. Anal. Chem. 1995, 67, 735–743. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, B.; Dave, N.; Servos, M.R.; Liu, J. Instantaneous attachment of an ultrahigh density of nonthiolated DNA to gold nanoparticles and its applications. Langmuir 2012, 28, 17053–17060. [Google Scholar] [CrossRef]
- Liao, L.; Li, X.; Jiang, B.; Zhou, W.; Yuan, R.; Xiang, Y. Cascaded and nonlinear DNA assembly amplification for sensitive and aptamer-based detection of kanamycin. Anal. Chim. Acta 2022, 1204, 339730. [Google Scholar] [CrossRef]
- Umrao, S.; Anusha, S.; Jain, V.; Chakraborty, B.; Roy, R. Smartphone-based kanamycin sensing with ratiometric FRET. RSC Adv. 2019, 9, 6143–6151. [Google Scholar] [CrossRef]
- Tang, Y.; Hu, Y.; Zhou, P.; Wang, C.; Tao, H.; Wu, Y. Colorimetric detection of kanamycin residue in foods based on the aptamer-enhanced peroxidase-mimicking activity of layered WS2 nanosheets. J. Agric. Food Chem. 2021, 69, 2884–2893. [Google Scholar] [CrossRef]
- Liu, J.; Zeng, J.; Tian, Y.; Zhou, N. An aptamer and functionalized nanoparticle-based strip biosensor for on-site detection of kanamycin in food samples. Analyst 2018, 143, 182–189. [Google Scholar] [CrossRef]

| Spiked (ng mL−1) | Detected (ng mL−1) | Recovery (%) | RSD (%) |
|---|---|---|---|
| 0 | ND a | - | - |
| 50 | 48 | 95.3 | 5.5 |
| 500 | 528 | 105.5 | 8.1 |
| 1000 | 1046 | 104.6 | 6.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Chang, R.; Tai, S.; Mao, M.; Peng, C. Gold/DNA-Cu2+ Complex Nanozyme-Based Aptamer Lateral Flow Assay for Highly Sensitive Detection of Kanamycin. Molecules 2024, 29, 4569. https://doi.org/10.3390/molecules29194569
Li X, Chang R, Tai S, Mao M, Peng C. Gold/DNA-Cu2+ Complex Nanozyme-Based Aptamer Lateral Flow Assay for Highly Sensitive Detection of Kanamycin. Molecules. 2024; 29(19):4569. https://doi.org/10.3390/molecules29194569
Chicago/Turabian StyleLi, Xiuping, Rui Chang, Shengmei Tai, Minxin Mao, and Chifang Peng. 2024. "Gold/DNA-Cu2+ Complex Nanozyme-Based Aptamer Lateral Flow Assay for Highly Sensitive Detection of Kanamycin" Molecules 29, no. 19: 4569. https://doi.org/10.3390/molecules29194569
APA StyleLi, X., Chang, R., Tai, S., Mao, M., & Peng, C. (2024). Gold/DNA-Cu2+ Complex Nanozyme-Based Aptamer Lateral Flow Assay for Highly Sensitive Detection of Kanamycin. Molecules, 29(19), 4569. https://doi.org/10.3390/molecules29194569







