Cross-Effects in Folding and Phase Transitions of hnRNP A1 and C9Orf72 RNA G4 In Vitro
Abstract
1. Introduction
2. Results and Discussion
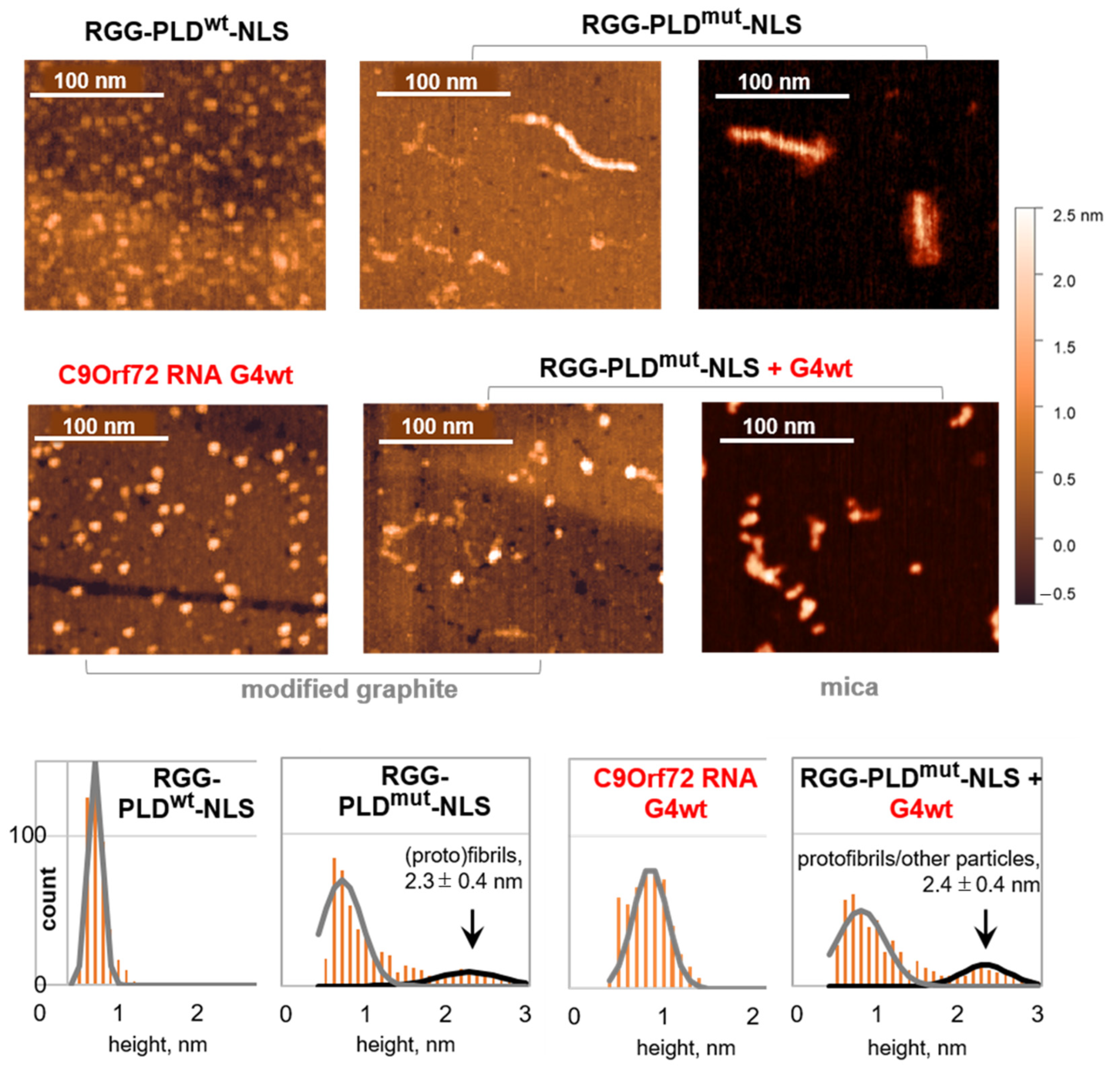
2.1. The RGG Motif Restricts the Spontaneous Fibrillation of the PLD-NLS Fragment of HnRNP A1
2.2. The C9Orf72 RNA G4 Modulates Fibrillation of RGG-PLD-NLS, Favoring Smaller Aggregates
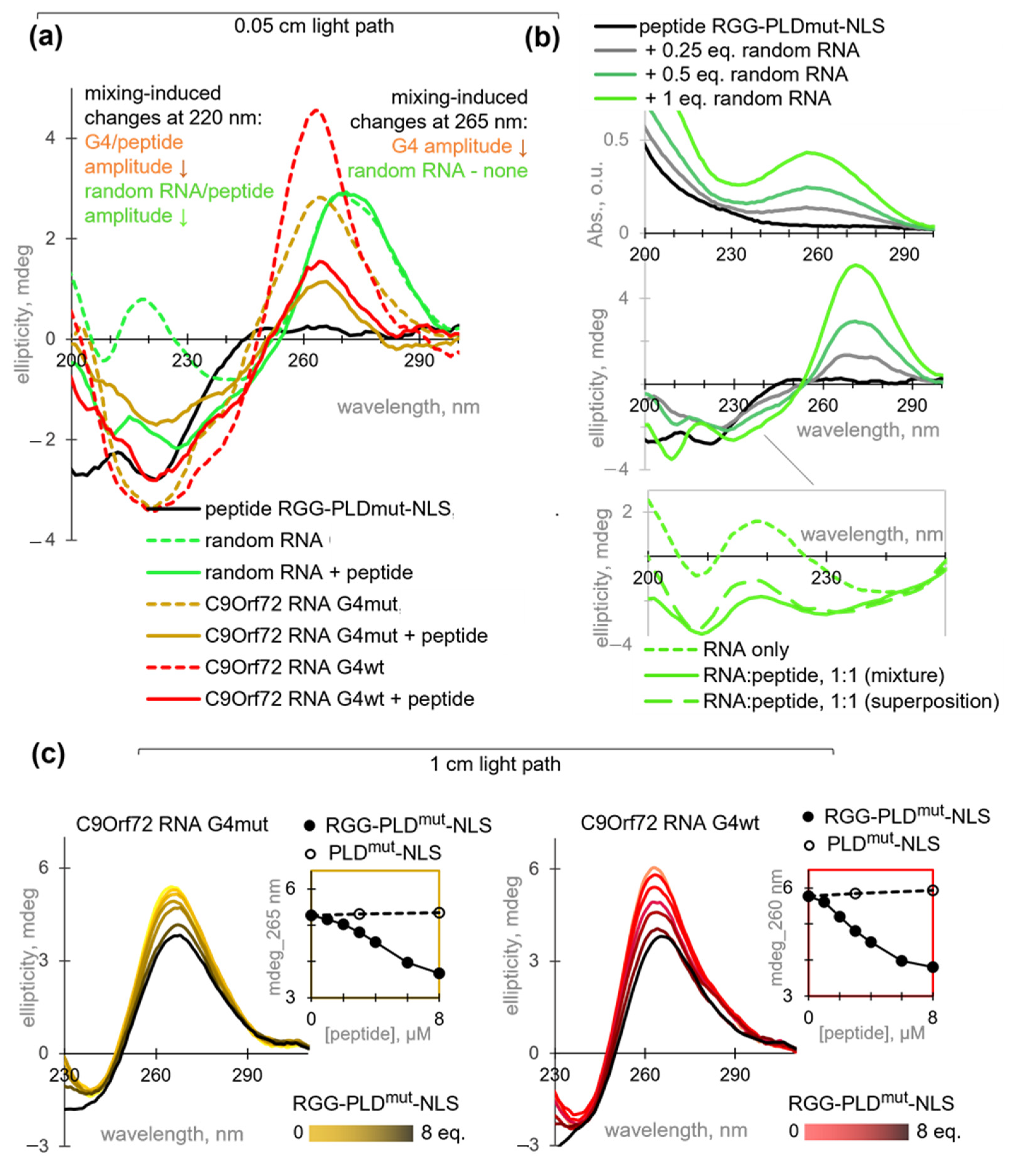
2.3. RGG-PLD-NLS Unfolds C9Orf72 RNA G4 and Its Mutant at Micromolar Concentrations
2.4. C9Orf72 RNA G4 Interferes with SR-Dependent Phase Separation of HnRNP A1 into Biocondensates
3. Materials and Methods
3.1. Peptides, Proteins, Oligonucleotides, and Reagents
3.2. Fibril Assembly and ThT Assays
3.3. Atomic Force Microscopy (AFM)
3.4. Circular Dichroism (CD) Spectroscopy
3.5. Biocondensate Assembly and Fluorescence Microscopy Imaging
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bayer, T.A. Proteinopathies, a Core Concept for Understanding and Ultimately Treating Degenerative Disorders? Eur. Neuropsychopharmacol. 2015, 25, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Hu, H. Sequestration of Cellular Interacting Partners by Protein Aggregates: Implication in a Loss-of-function Pathology. FEBS J. 2016, 283, 3705–3717. [Google Scholar] [CrossRef] [PubMed]
- Winklhofer, K.F.; Tatzelt, J.; Haass, C. The Two Faces of Protein Misfolding: Gain- and Loss-of-Function in Neurodegenerative Diseases. EMBO J. 2008, 27, 336–349. [Google Scholar] [CrossRef]
- Low, Y.-H.; Asi, Y.; Foti, S.C.; Lashley, T. Heterogeneous Nuclear Ribonucleoproteins: Implications in Neurological Diseases. Mol. Neurobiol. 2021, 58, 631–646. [Google Scholar] [CrossRef]
- Geuens, T.; Bouhy, D.; Timmerman, V. The HnRNP Family: Insights into Their Role in Health and Disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef]
- Tao, Y.; Zhang, Q.; Wang, H.; Yang, X.; Mu, H. Alternative Splicing and Related RNA Binding Proteins in Human Health and Disease. Signal Transduct. Target. Ther. 2024, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Lai, S.-K.; Sim, D.Y.; Ang, W.S.L.; Li, H.Y.; Roca, X. SRRM2 Organizes Splicing Condensates to Regulate Alternative Splicing. Nucleic Acids Res. 2022, 50, 8599–8614. [Google Scholar] [CrossRef]
- Galganski, L.; Urbanek, M.O.; Krzyzosiak, W.J. Nuclear Speckles: Molecular Organization, Biological Function and Role in Disease. Nucleic Acids Res. 2017, 45, 10350–10368. [Google Scholar] [CrossRef]
- Liao, S.E.; Regev, O. Splicing at the Phase-Separated Nuclear Speckle Interface: A Model. Nucleic Acids Res. 2021, 49, 636–645. [Google Scholar] [CrossRef]
- Chiou, N.-T.; Shankarling, G.; Lynch, K.W. HnRNP L and HnRNP A1 Induce Extended U1 SnRNA Interactions with an Exon to Repress Spliceosome Assembly. Mol. Cell 2013, 49, 972–982. [Google Scholar] [CrossRef]
- Bampton, A.; Gittings, L.M.; Fratta, P.; Lashley, T.; Gatt, A. The Role of HnRNPs in Frontotemporal Dementia and Amyotrophic Lateral Sclerosis. Acta Neuropathol. 2020, 140, 599–623. [Google Scholar] [CrossRef] [PubMed]
- Purice, M.D.; Taylor, J.P. Linking HnRNP Function to ALS and FTD Pathology. Front. Neurosci. 2018, 12, 326. [Google Scholar] [CrossRef] [PubMed]
- Kashima, T.; Rao, N.; David, C.J.; Manley, J.L. HnRNP A1 Functions with Specificity in Repression of SMN2 Exon 7 Splicing. Hum. Mol. Genet. 2007, 16, 3149–3159. [Google Scholar] [CrossRef]
- Berson, A.; Barbash, S.; Shaltiel, G.; Goll, Y.; Hanin, G.; Greenberg, D.S.; Ketzef, M.; Becker, A.J.; Friedman, A.; Soreq, H. Cholinergic-associated Loss of HnRNP-A/B in Alzheimer’s Disease Impairs Cortical Splicing and Cognitive Function in Mice. EMBO Mol. Med. 2012, 4, 730–742. [Google Scholar] [CrossRef]
- Singh, R.N.; Singh, N.N. Mechanism of Splicing Regulation of Spinal Muscular Atrophy Genes. In RNA Metabolism in Neurodegenerative Diseases. Advances in Neurobiology; Sattler, R., Donnelly, C., Eds.; Springer: Cham, Switzerland, 2018; Volume 20, pp. 31–61. ISBN 978-3-319-89688-5. [Google Scholar] [CrossRef]
- Clarke, J.P.; Thibault, P.A.; Salapa, H.E.; Levin, M.C. A Comprehensive Analysis of the Role of HnRNP A1 Function and Dysfunction in the Pathogenesis of Neurodegenerative Disease. Front. Mol. Biosci. 2021, 8, 659610. [Google Scholar] [CrossRef]
- Salapa, H.E.; Thibault, P.A.; Libner, C.D.; Ding, Y.; Clarke, J.-P.W.E.; Denomy, C.; Hutchinson, C.; Abidullah, H.M.; Austin Hammond, S.; Pastushok, L.; et al. HnRNP A1 Dysfunction Alters RNA Splicing and Drives Neurodegeneration in Multiple Sclerosis (MS). Nat. Commun. 2024, 15, 356. [Google Scholar] [CrossRef]
- Corsi, A.; Bombieri, C.; Valenti, M.T.; Romanelli, M.G. Tau Isoforms: Gaining Insight into MAPT Alternative Splicing. Int. J. Mol. Sci. 2022, 23, 15383. [Google Scholar] [CrossRef]
- Guil, S.; Long, J.C.; Cáceres, J.F. HnRNP A1 Relocalization to the Stress Granules Reflects a Role in the Stress Response. Mol. Cell. Biol. 2006, 26, 5744–5758. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.-P.W.E.; Thibault, P.A.; Salapa, H.E.; Kim, D.E.; Hutchinson, C.; Levin, M.C. Multiple Sclerosis-Associated HnRNPA1 Mutations Alter HnRNPA1 Dynamics and Influence Stress Granule Formation. Int. J. Mol. Sci. 2021, 22, 2909. [Google Scholar] [CrossRef]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef]
- Linsenmeier, M.; Faltova, L.; Morelli, C.; Capasso Palmiero, U.; Seiffert, C.; Küffner, A.M.; Pinotsi, D.; Zhou, J.; Mezzenga, R.; Arosio, P. The Interface of Condensates of the HnRNPA1 Low-Complexity Domain Promotes Formation of Amyloid Fibrils. Nat. Chem. 2023, 15, 1340–1349. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Poudyal, M.; Dave, K.; Kadu, P.; Maji, S.K. Protein Misfolding and Amyloid Nucleation through Liquid–Liquid Phase Separation. Chem. Soc. Rev. 2024, 53, 4976–5013. [Google Scholar] [CrossRef] [PubMed]
- Nedelsky, N.B.; Taylor, J.P. Bridging Biophysics and Neurology: Aberrant Phase Transitions in Neurodegenerative Disease. Nat. Rev. Neurol. 2019, 15, 272–286. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, N.C.; Wang, Y.-D.; Scarborough, E.A.; Moore, J.; Diaz, Z.; MacLea, K.S.; Freibaum, B.; Li, S.; Molliex, A.; et al. Mutations in Prion-like Domains in HnRNPA2B1 and HnRNPA1 Cause Multisystem Proteinopathy and ALS. Nature 2013, 495, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Banerjee, S.; Savran, D.; Rajes, C.; Wiese, S.; Girdhar, A.; Schwierz, N.; Lee, C.; Shorter, J.; Schmidt, M.; et al. Cryo-EM Structure of the Full-Length HnRNPA1 Amyloid Fibril. J. Mol. Biol. 2023, 435, 168211. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, K.; Xia, W.; Feng, G.; Gu, J.; Ma, Y.; Gui, X.; Zhang, X.; Fang, Y.; Sun, B.; et al. The Nuclear Localization Sequence Mediates HnRNPA1 Amyloid Fibril Formation Revealed by CryoEM Structure. Nat. Commun. 2020, 11, 6349. [Google Scholar] [CrossRef]
- Nanaura, H.; Kawamukai, H.; Fujiwara, A.; Uehara, T.; Aiba, Y.; Nakanishi, M.; Shiota, T.; Hibino, M.; Wiriyasermkul, P.; Kikuchi, S.; et al. C9orf72-Derived Arginine-Rich Poly-Dipeptides Impede Phase Modifiers. Nat. Commun. 2021, 12, 5301. [Google Scholar] [CrossRef]
- Mori, K.; Gotoh, S.; Ikeda, M. Aspects of Degradation and Translation of the Expanded C9orf72 Hexanucleotide Repeat RNA. J. Neurochem. 2023, 166, 156–171. [Google Scholar] [CrossRef]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef]
- Lin, Y.; Mori, E.; Kato, M.; Xiang, S.; Wu, L.; Kwon, I.; McKnight, S.L. Toxic PR Poly-Dipeptides Encoded by the C9orf72 Repeat Expansion Target LC Domain Polymers. Cell 2016, 167, 789–802.e12. [Google Scholar] [CrossRef]
- Lee, K.-H.; Zhang, P.; Kim, H.J.; Mitrea, D.M.; Sarkar, M.; Freibaum, B.D.; Cika, J.; Coughlin, M.; Messing, J.; Molliex, A.; et al. C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell 2016, 167, 774–788.e17. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, B.; Robberecht, W.; Van Den Bosch, L. RNA Toxicity in Non-coding Repeat Expansion Disorders. EMBO J. 2020, 39, e101112. [Google Scholar] [CrossRef] [PubMed]
- Solomon, D.A.; Smikle, R.; Reid, M.J.; Mizielinska, S. Altered Phase Separation and Cellular Impact in C9orf72-Linked ALS/FTD. Front. Cell. Neurosci. 2021, 15, 664151. [Google Scholar] [CrossRef]
- Bose, K.; Maity, A.; Ngo, K.H.; Vandana, J.J.; Shneider, N.A.; Phan, A.T. Formation of RNA G-Wires by G4C2 Repeats Associated with ALS and FTD. Biochem. Biophys. Res. Commun. 2022, 610, 113–118. [Google Scholar] [CrossRef]
- Wang, S.; Xu, Y. RNA Structure Promotes Liquid-to-Solid Phase Transition of Short RNAs in Neuronal Dysfunction. Commun. Biol. 2024, 7, 137. [Google Scholar] [CrossRef]
- Raguseo, F.; Wang, Y.; Li, J.; Petrić Howe, M.; Balendra, R.; Huyghebaert, A.; Vadukul, D.M.; Tanase, D.A.; Maher, T.E.; Malouf, L.; et al. The ALS/FTD-Related C9orf72 Hexanucleotide Repeat Expansion Forms RNA Condensates through Multimolecular G-Quadruplexes. Nat. Commun. 2023, 14, 8272. [Google Scholar] [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Liu, C.; Cai, Q.; Luo, Z.; Miao, H.; Shi, X.; Xu, N.; Fung, C.P.; Choy, T.T.; Yan, B.; et al. Crystal Structure of Parallel G-Quadruplex Formed by the Two-Repeat ALS- and FTD-Related GGGGCC Sequence. Nucleic Acids Res. 2021, 49, 5881–5890. [Google Scholar] [CrossRef]
- Liu, C.; Zhou, B.; Xu, N.; Fung, C.P.; Yan, B.; Suen, M.C.; Huang, Z.; Zhu, G. The Parallel Tetrameric DNA G-Quadruplex Formed by the Two-Repeat C9orf72 GGGGCC Sequence in Solution. Magn. Reson. Lett. 2022, 2, 196–204. [Google Scholar] [CrossRef]
- Zhou, B.; Liu, C.; Geng, Y.; Zhu, G. Topology of a G-Quadruplex DNA Formed by C9orf72 Hexanucleotide Repeats Associated with ALS and FTD. Sci. Rep. 2015, 5, 16673. [Google Scholar] [CrossRef]
- Wang, J.; Choi, J.-M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174, 688–699.e16. [Google Scholar] [CrossRef]
- Conlon, E.G.; Lu, L.; Sharma, A.; Yamazaki, T.; Tang, T.; Shneider, N.A.; Manley, J.L. The C9ORF72 GGGGCC Expansion Forms RNA G-Quadruplex Inclusions and Sequesters HnRNP H to Disrupt Splicing in ALS Brains. Elife 2016, 5, e17820. [Google Scholar] [CrossRef]
- Paramasivam, M.; Membrino, A.; Cogoi, S.; Fukuda, H.; Nakagama, H.; Xodo, L.E. Protein HnRNP A1 and Its Derivative Up1 Unfold Quadruplex DNA in the Human KRAS Promoter: Implications for Transcription. Nucleic Acids Res. 2009, 37, 2841–2853. [Google Scholar] [CrossRef]
- Fujino, Y.; Ueyama, M.; Ishiguro, T.; Ozawa, D.; Ito, H.; Sugiki, T.; Murata, A.; Ishiguro, A.; Gendron, T.; Mori, K.; et al. FUS Regulates RAN Translation through Modulating the G-Quadruplex Structure of GGGGCC Repeat RNA in C9orf72-Linked ALS/FTD. Elife 2023, 12, RP84338. [Google Scholar] [CrossRef]
- Shorter, J.; Taylor, J.P. Disease Mutations in the Prion-like Domains of HnRNPA1 and HnRNPA2/B1 Introduce Potent Steric Zippers That Drive Excess RNP Granule Assembly. Rare Dis. 2013, 1, e25200. [Google Scholar] [CrossRef]
- Li, Y.R.; King, O.D.; Shorter, J.; Gitler, A.D. Stress Granules as Crucibles of ALS Pathogenesis. J. Cell Biol. 2013, 201, 361–372. [Google Scholar] [CrossRef]
- Fontana, E.; Bongianni, M.; Benussi, A.; Bronzato, E.; Scialo, C.; Sacchetto, L.; Cagnin, A.; Castriciano, S.; Buratti, E.; Gardoni, F.; et al. Detection of TDP-43 Seeding Activity in the Olfactory Mucosa from Patients with Frontotemporal Dementia. Alzheimer’s Dement. 2024, 20, 1156–1165. [Google Scholar] [CrossRef]
- Dubrovin, E.V. Atomic Force Microscopy-Based Approaches for Single-Molecule Investigation of Nucleic Acid–Protein Complexes. Biophys. Rev. 2023, 15, 1015–1033. [Google Scholar] [CrossRef]
- Dubrovin, E.V.; Klinov, D.V.; Schäffer, T.E. Evidence of (Anti)Metamorphic Properties of Modified Graphitic Surfaces Obtained in Real Time at a Single-Molecule Level. Colloids Surf. B 2020, 193, 111077. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Cao, Q.; Hughes, M.P.; Sawaya, M.R.; Boyer, D.R.; Cascio, D.; Eisenberg, D.S. CryoEM Structure of the Low-Complexity Domain of HnRNPA2 and Its Conversion to Pathogenic Amyloid. Nat. Commun. 2020, 11, 4090. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.K.-P.; Obi, I.; Sabouri, N. The RGG Domain in the C-Terminus of the DEAD Box Helicases Dbp2 and Ded1 Is Necessary for G-Quadruplex Destabilization. Nucleic Acids Res. 2021, 49, 8339–8354. [Google Scholar] [CrossRef]
- Corrêa, D.H.A.; Ramos, C.H.I. The Use of Circular Dichroism Spectroscopy to Study Protein Folding, Form and Function. Afr. J. Biochem. Res. 2009, 3, 164–173. [Google Scholar]
- Gładysz, M.; Andrałojć, W.; Czapik, T.; Gdaniec, Z.; Kierzek, R. Thermodynamic and Structural Contributions of the 6-Thioguanosine Residue to Helical Properties of RNA. Sci. Rep. 2019, 9, 4385. [Google Scholar] [CrossRef]
- Larsen, A.T.; Fahrenbach, A.C.; Sheng, J.; Pian, J.; Szostak, J.W. Thermodynamic Insights into 2-Thiouridine-Enhanced RNA Hybridization. Nucleic Acids Res. 2015, 43, 7675–7687. [Google Scholar] [CrossRef]
- Liu, X.; Ishizuka, T.; Bao, H.L.; Wada, K.; Takeda, Y.; Iida, K.; Nagasawa, K.; Yang, D.; Xu, Y. Structure-Dependent Binding of hnRNPA1 to Telomere RNA. J. Am. Chem. Soc. 2017, 139, 7533–7539. [Google Scholar] [CrossRef]
- Zhang, Q.-S.; Manche, L.; Xu, R.-M.; Krainer, A.R. HnRNP A1 Associates with Telomere Ends and Stimulates Telomerase Activity. RNA 2006, 12, 1116–1128. [Google Scholar] [CrossRef]
- Cogoi, S.; Rapozzi, V.; Cauci, S.; Xodo, L.E. Critical Role of HnRNP A1 in Activating KRAS Transcription in Pancreatic Cancer Cells: A Molecular Mechanism Involving G4 DNA. Biochim. Biophys. Acta 2017, 1861, 1389–1398. [Google Scholar] [CrossRef]
- Mendoza, O.; Bourdoncle, A.; Boulé, J.-B.; Brosh, R.M.; Mergny, J.-L. G-Quadruplexes and Helicases. Nucleic Acids Res. 2016, 44, 1989–2006. [Google Scholar] [CrossRef]
- Ding, J.; Hayashi, M.K.; Zhang, Y.; Manche, L.; Krainer, A.R.; Xu, R.-M. Crystal Structure of the Two-RRM Domain of HnRNP A1 (UP1) Complexed with Single-Stranded Telomeric DNA. Genes Dev. 1999, 13, 1102–1115. [Google Scholar] [CrossRef]
- Fei, J.; Jadaliha, M.; Harmon, T.S.; Li, I.T.S.; Hua, B.; Hao, Q.; Holehouse, A.S.; Reyer, M.; Sun, Q.; Freier, S.M.; et al. Quantitative Analysis of Multilayer Organization of Proteins and RNA in Nuclear Speckles at Super Resolution. J. Cell. Sci. 2017, 130, 4180–4192. [Google Scholar] [CrossRef]
- André, A.A.M.; Yewdall, N.A.; Spruijt, E. Crowding-Induced Phase Separation and Gelling by Co-Condensation of PEG in NPM1-RRNA Condensates. Biophys. J. 2023, 122, 397–407. [Google Scholar] [CrossRef] [PubMed]



| Code 1 | Sequence | Position (Protein) |
|---|---|---|
| zipper motifmut | SYNVFG | 259–264 (hnRNP A1) |
| PLDmut-NLS | GGGYGGSGDGYNGFGNDGSNFGGGGSYNVFGN YNNQSSN | 234–272 (hnRNP A1) |
| RGG-PLDmut-NLS | RGGNFSGRGGFGGSRGGGGYGGSGDGYNGFGN DGSNFGGGGSYNVFGNYNNQSSN | 218–272 (hnRNP A1) |
| RGG-PLDwt-NLS | RGGNFSGRGGFGGSRGGGGYGGSGDGYNGFGN DGSNFGGGGSYNDFGNYNNQSSN | 218–272 (hnRNP A1) |
| SRSFfr | PRSPSYGRSRSRSRSRSRSRSRSNSRSRSYSP | 197–228 (SRSF1) |
| C9Orf72 RNA G4wt | GGGGCCGGGGCCGGGGCCGGGG | N/A |
| C9Orf72 RNA G4mut | GGGCCGGGCCGGGCCGGG | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vedekhina, T.; Svetlova, J.; Pavlova, I.; Barinov, N.; Alieva, S.; Malakhova, E.; Rubtsov, P.; Shtork, A.; Klinov, D.; Varizhuk, A. Cross-Effects in Folding and Phase Transitions of hnRNP A1 and C9Orf72 RNA G4 In Vitro. Molecules 2024, 29, 4369. https://doi.org/10.3390/molecules29184369
Vedekhina T, Svetlova J, Pavlova I, Barinov N, Alieva S, Malakhova E, Rubtsov P, Shtork A, Klinov D, Varizhuk A. Cross-Effects in Folding and Phase Transitions of hnRNP A1 and C9Orf72 RNA G4 In Vitro. Molecules. 2024; 29(18):4369. https://doi.org/10.3390/molecules29184369
Chicago/Turabian StyleVedekhina, Tatiana, Julia Svetlova, Iuliia Pavlova, Nikolay Barinov, Sabina Alieva, Elizaveta Malakhova, Pavel Rubtsov, Alina Shtork, Dmitry Klinov, and Anna Varizhuk. 2024. "Cross-Effects in Folding and Phase Transitions of hnRNP A1 and C9Orf72 RNA G4 In Vitro" Molecules 29, no. 18: 4369. https://doi.org/10.3390/molecules29184369
APA StyleVedekhina, T., Svetlova, J., Pavlova, I., Barinov, N., Alieva, S., Malakhova, E., Rubtsov, P., Shtork, A., Klinov, D., & Varizhuk, A. (2024). Cross-Effects in Folding and Phase Transitions of hnRNP A1 and C9Orf72 RNA G4 In Vitro. Molecules, 29(18), 4369. https://doi.org/10.3390/molecules29184369










