Spectroscopic Relationship between XOD and TAOZHI Total Polyphenols Based on Chemometrics and Molecular Docking Techniques
Abstract
1. Introduction
2. Results
2.1. The Optimal Extraction Conditions for TZ-TPC
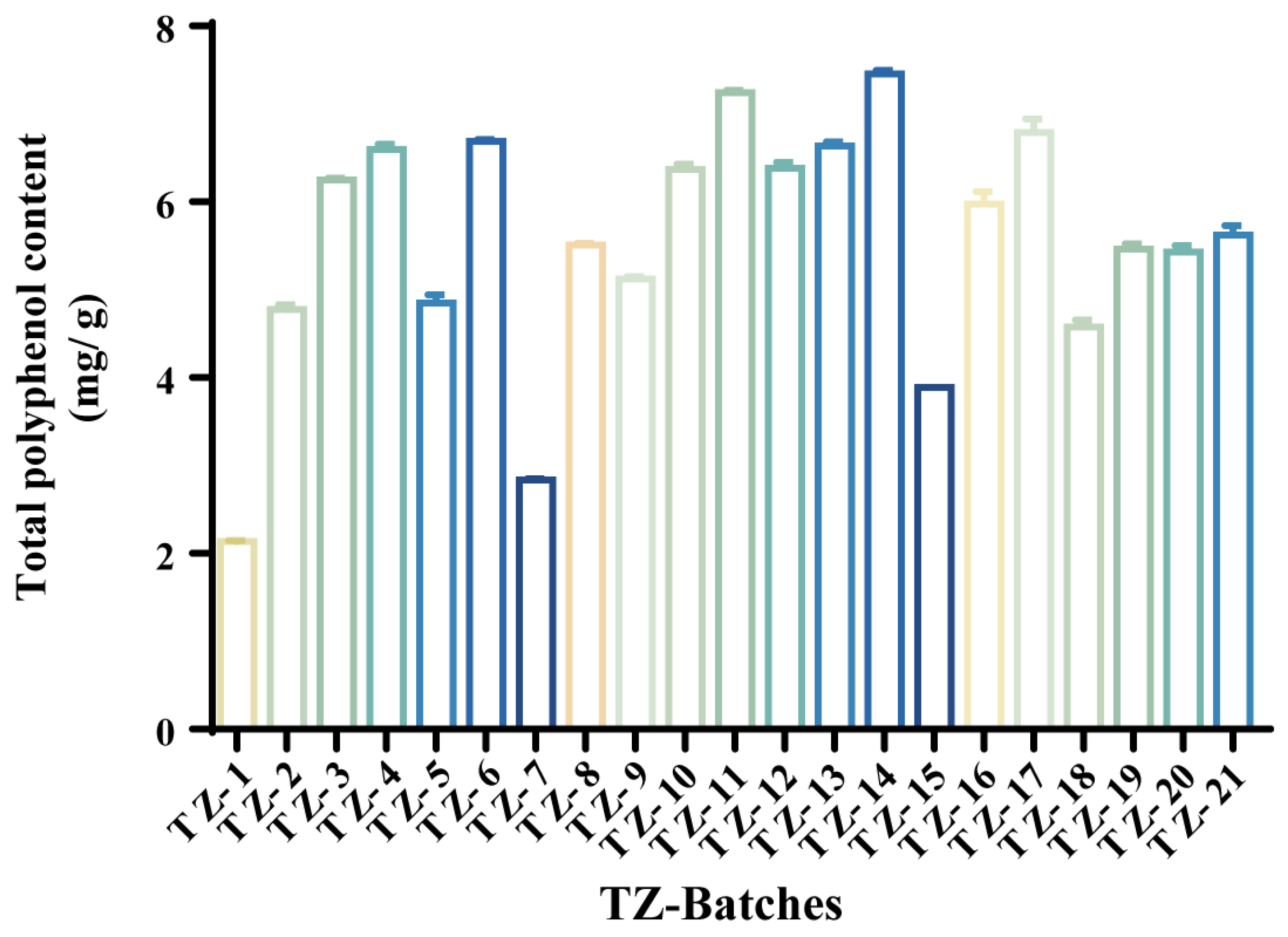
2.2. Extraction Rate of Different Batches of TZ–TPC
2.3. Optimal Reaction Conditions for XOD
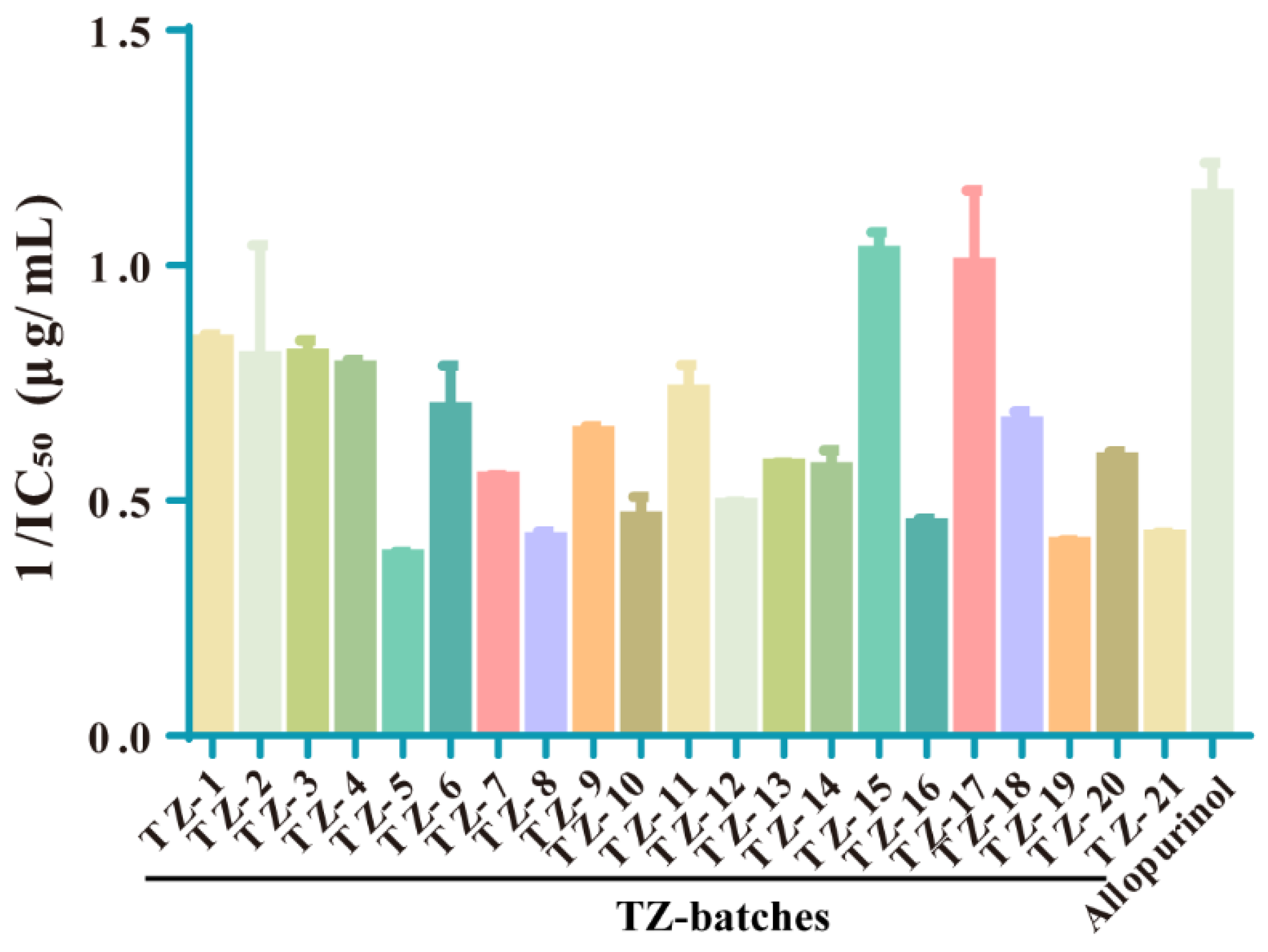
2.4. The Inhibition of XOD Activity by Each Batch of TZ Was Determined, and the Enzyme Reaction Kinetics Was Judged
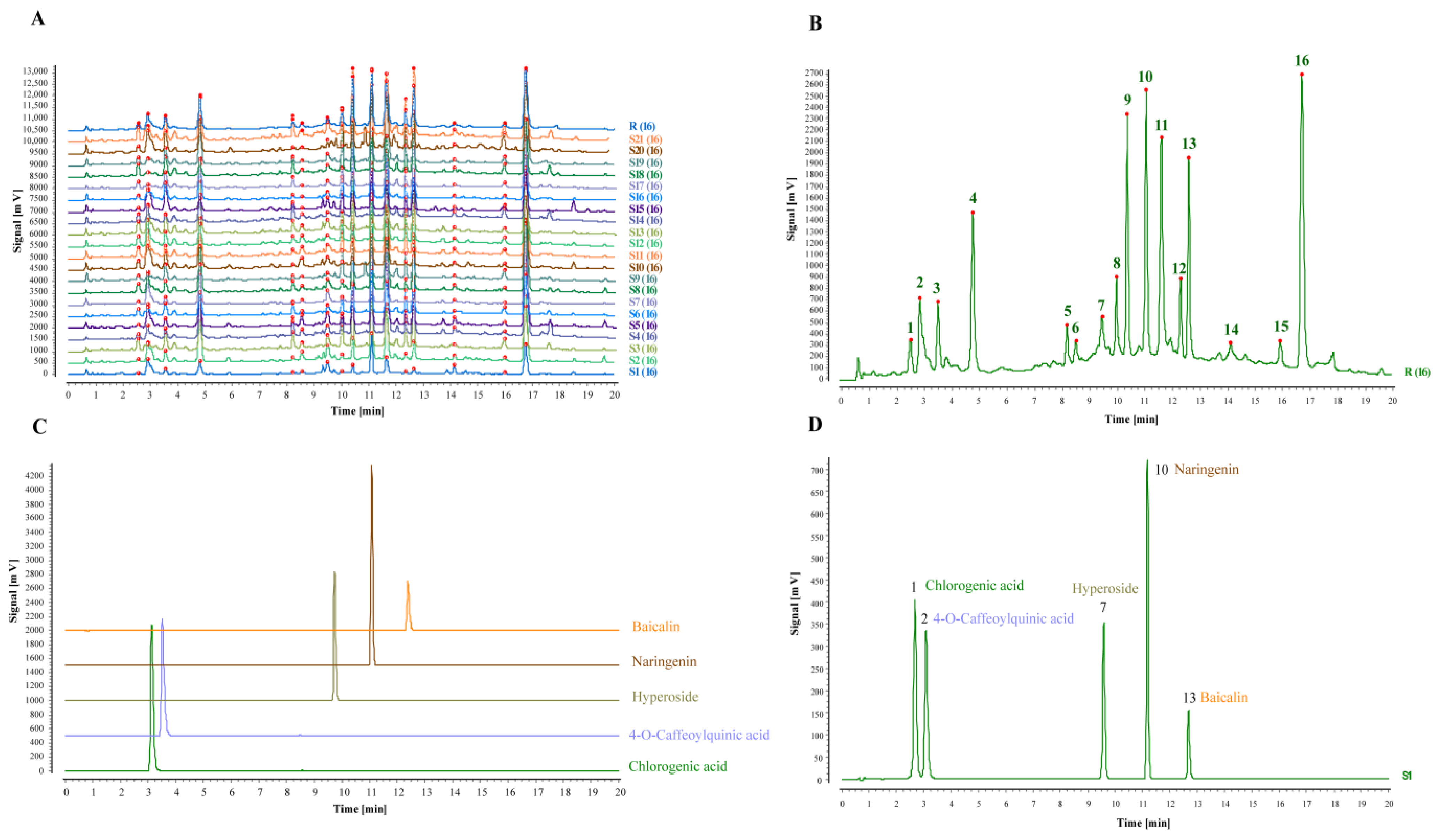
2.5. Fingerprinting of TZ Batches and Methodological Validation
2.5.1. Establishment of TZ Characteristic Fingerprint and Similarity Evaluation
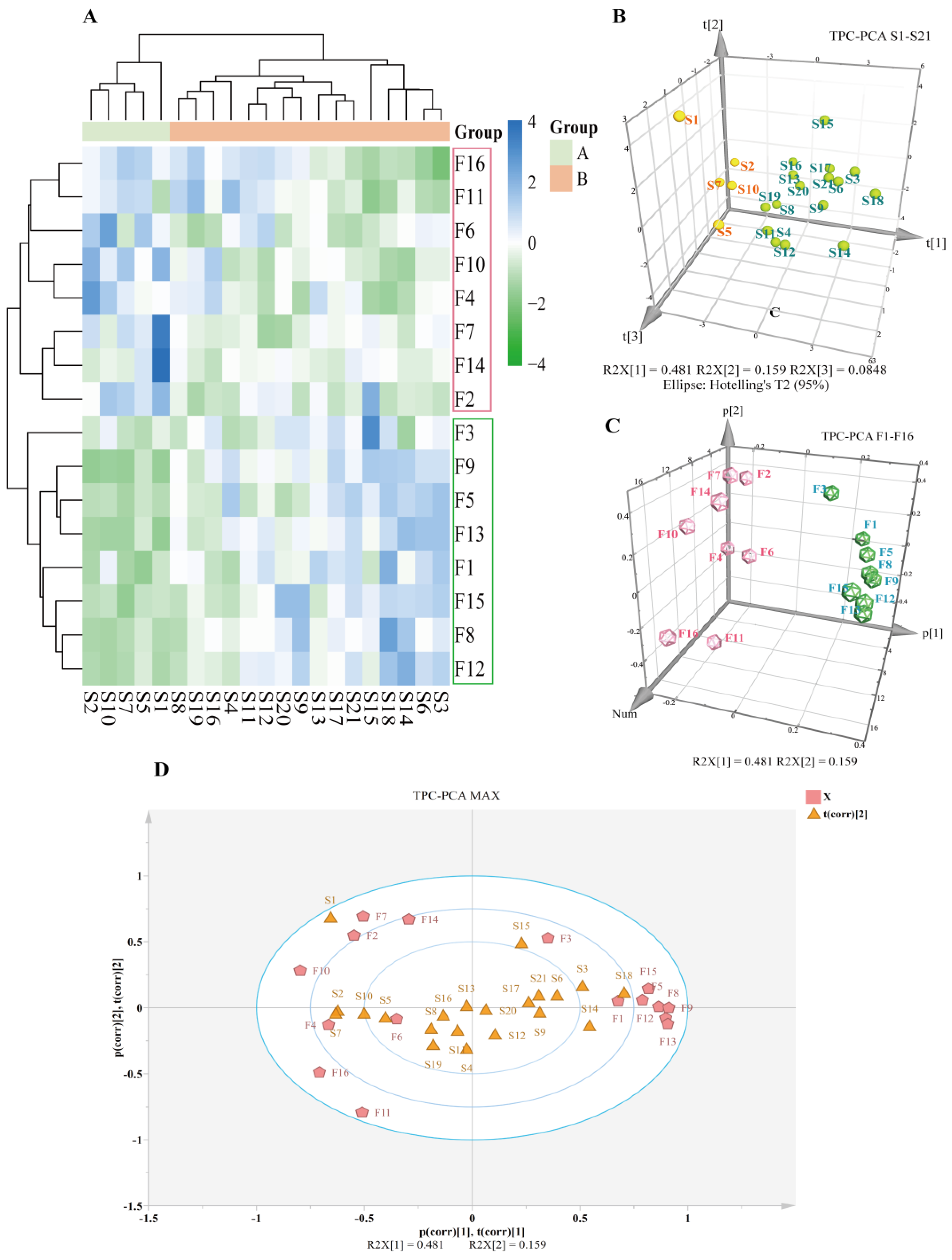
2.5.2. CA and PCA Analyses
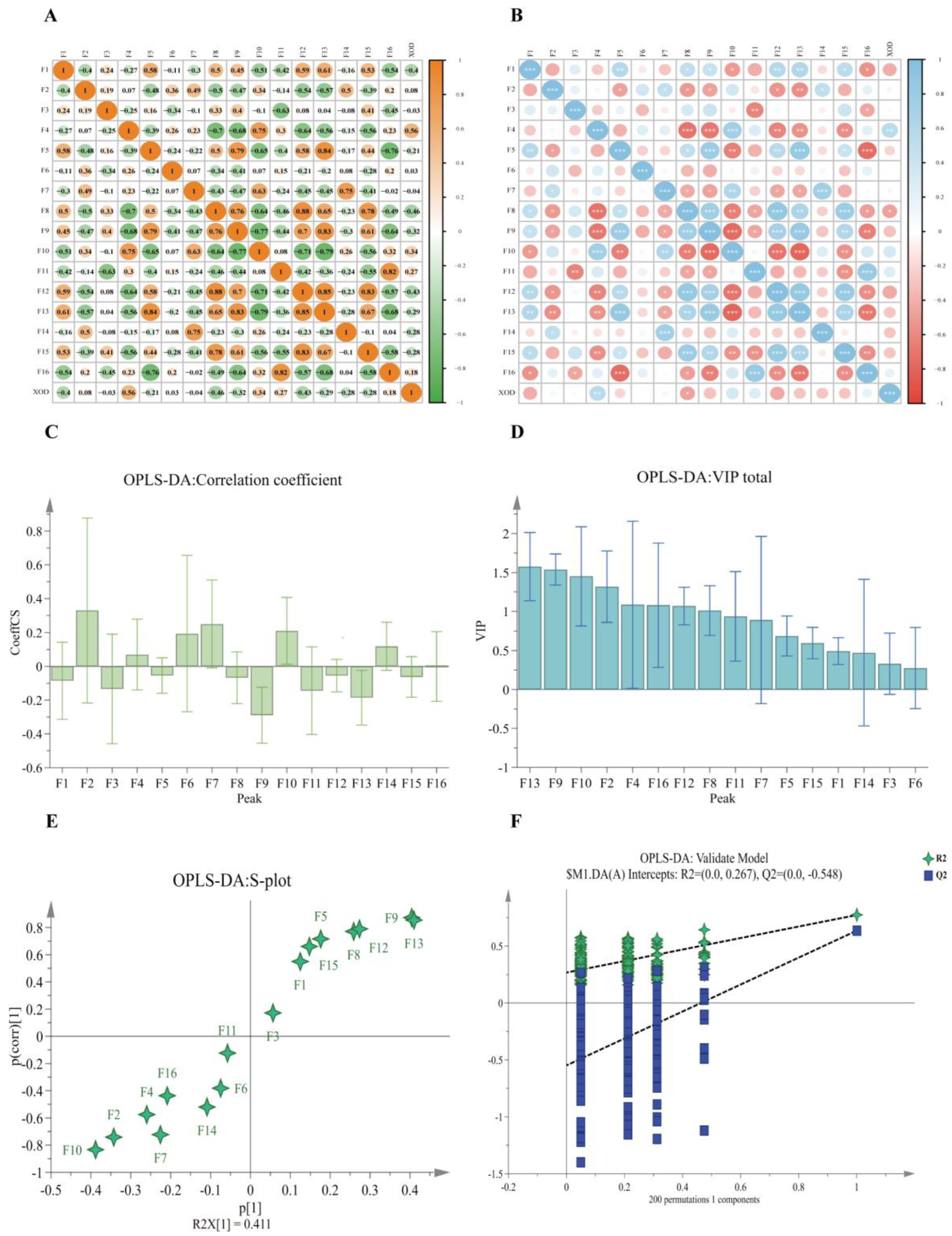
2.6. Chemometric Analyses
2.6.1. Grey Relevance Analysis (GRA)
2.6.2. Bivariate Correlation Analysis (BCA)
2.6.3. Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA)
2.7. Molecular Docking of Constituent Groups to Proteins
3. Discussion
3.1. Determination of Total Polyphenol Content and XOD Activity of the Batch
3.2. Batch UHPLC Fingerprints
3.3. Spectral Effect Correlation Analysis (SECA)
3.4. Molecular Docking Analysis
4. Materials and Methods
4.1. Reagents and Materials
4.2. Solutions Preparation
4.3. Colour Development Method
4.4. Plotting of Standard Curve and Determination of TZ–TPC Content
4.5. Optimisation of Total Polyphenol Extraction Conditions and Determination of TPC in 21 TZ Batches
4.6. Screening of Optimal Reaction Conditions for XOD
4.7. A Study of the Type of Inhibition of XOD
4.8. Determination of XOD Inhibitory Activity of TZ-TPC Batch
4.9. UHPLC Fingerprinting of 21 Batches of TZ-TPC Extracts
4.9.1. Chromatographic Conditions
4.9.2. Preparation of Standard Solution and Sample Solutions
4.9.3. Fingerprinting and Methodological Validation
4.9.4. Batch Fingerprint Analysis
4.10. Spectrum–Effect Relationship
4.11. Molecular Docking
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peng, A.; Lin, L.; Zhao, M. Screening of key flavonoids and monoterpenoids for xanthine oxidase inhibitory activity-oriented quality control of Chrysanthemum morifolium Ramat. ‘Boju’ based on spectrum-effect relationship coupled with UPLC-TOF-MS and HS-SPME-GC/MS. Food Res. Int. 2020, 137, 109448. [Google Scholar] [CrossRef] [PubMed]
- FitzGerald, J.D.; Dalbeth, N.; Mikuls, T.; Brignardello-Petersen, R.; Guyatt, G.; Abeles, A.M.; Gelber, A.C.; Harrold, L.R.; Khanna, D.; King, C.; et al. 2020 American College of Rheumatology Guideline for the Management of Gout. Arthritis Care Res. 2020, 72, 744–760. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Pope, J.E. Cardiovascular effects of urate-lowering therapies in patients with chronic gout: A systematic review and meta-analysis. Rheumatology 2017, 56, 1144–1153. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-W.; Dao, R.-L.; Chung, W.-H. Immunopathogenesis and risk factors for allopurinol severe cutaneous adverse reactions. Curr. Opin. Allergy Clin. Immunol. 2016, 16, 339–345. [Google Scholar] [CrossRef]
- Müller-Wieland, D.; Nitschmann, S. Kardiovaskuläres Risiko bei Patienten mit Gicht: Cardiovascular Safety of Febuxostat or Allopurinol in Participants with Gout and Cardiovascular Comorbidities (CARES). Internist 2018, 59, 1224–1228. [Google Scholar] [CrossRef]
- National Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China 2020 Edition; China Pharmaceutical Science and Technology Press: Beijing, China, 2020; p. 291. [Google Scholar]
- Guizhou Provincial Drug Administration. Quality Standards for Chinese herbal medicines and ethnic herbs in Guizhou Province; Guizhou Science and Technology Press: Guiyang, China, 2003; p. 296. [Google Scholar]
- Hao, J.; Wang, X.; Li, Y.; Li, X.; Wu, L.; Wei, D. Study on Chemical Components of Essential Oil in Peach Juvenile Branchlet by Gas Chromatography-Mass Spectrometry. Chin. J. Exp. Tradit. Med. Formulae 2010, 16, 45–48. [Google Scholar]
- Guo, F.; Li, C.; Dou, J.; Liang, J.; Chen, Z.; Xu, Z.; Wang, T. Research progress on pharmacological properties and application of probiotics in the fermentation of Scutellaria baicalensis Georgi. Front. Nutr. 2024, 11, 1407182. [Google Scholar] [CrossRef]
- Yang, L.; Li, X.; Law, K.; Long, J.; Wu, H.; Wang, X. Qualitative study of the chemical composition of peach branches. J. Guiyang Coll. Tradit. Chin. Med. 2014, 36, 9–11. [Google Scholar]
- Ma, B.; Ning, J.; Wang, F.; Zheng, H.; Han, L. Molecular Mechanism of the Therapeutic Effect of Peach Blossom against Constipation: An Exploratory Study Based on Network Pharmacology Analysis and Molecular Docking Verification. Evid. Based Complement. Altern. Med. 2023, 2023, 8577485. [Google Scholar] [CrossRef]
- Palumbo, M.; Cefola, M.; Pace, B.; Ricci, I.; Siano, F.; Amato, G.; Stocchero, M.; Cozzolino, R. Volatile Metabolites to Assess the Onset of Chilling Injury in Fresh-Cut Nectarines. Foods 2024, 13, 1047. [Google Scholar] [CrossRef]
- Sun, X.; Yuan, S.; Shen, S.; Geng, H.; Liu, J.; Cui, Z.; Wang, F.; Wei, G.; Liu, D.; Wang, H. First Report of Rust Caused by Tranzschelia discolor on Peach Leaves in Shenzhou, China. Plant Dis. 2022, 107, 1235. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhu, Y.; Liu, C.; Cheng, J.; He, F. Taohong Siwu decoction reduces acute myocardial ischemia–reperfusion injury by promoting autophagy to inhibit pyroptosis. J. Ethnopharmacol. 2024, 321, 117515. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, B.T.; Calle, A.; Saski, C.A.; Melgar, J.C. Differential Gene Expression Patterns in Peach Roots under Non-Uniform Soil Conditions in Response to Organic Matter. Genes 2024, 15, 70. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Liu, P.; Yao, X.; Cao, H.; Zhu, H.; Wang, Q.; Liu, Y.; Fang, M.; Wu, Y.; Gong, Z. Hepatoprotective effects of peach gum polysaccharides against alcoholic liver injury: Moderation of oxidative stress and promotion of lipid metabolism. Front. Nutr. 2024, 10, 1325450. [Google Scholar] [CrossRef]
- Ji, Z.; Dai, H.; Jin, W.; Song, H.; Tong, Y.; Xu, J. Study on the occurrence pattern of peach twig blight. Chin. Fruit Tree 2016, 2, 13–17. [Google Scholar]
- Sun, Z.G.; WKUllah, I.; Zhu, H.L. Recent Advances in Xanthine Oxidase Inhibitors. Mini-Rev. Med. Chem. 2024, 24, 1177–1186. [Google Scholar] [CrossRef]
- Shizhen, M.-L. The Compendium of Materia Medica (More Points Book, First and Second Books), 2nd ed.; People’s Health Publishing House: Beijing, China, 2004; pp. 1742–1751. [Google Scholar]
- Coletro, H.N.; Diniz, A.P.; Guimaraes, N.S.; Carraro, J.C.C.; Mendonca, R.D.; Meireles, A.L. Polyphenols for improvement of inflammation and symptoms in rheumatic diseases: Systematic review. Sao Paulo Med. J. 2021, 139, 615–623. [Google Scholar] [CrossRef]
- Li, S.; Huang, X.; Li, Y.; Ding, R.; Wu, X.; Li, L.; Li, C.; Gu, R. Spectrum-Effect Relationship in Chinese Herbal Medicine: Current Status and Future Perspectives. Crit. Rev. Anal. Chem. 2023, 1–22. [Google Scholar] [CrossRef]
- Ma, J.; Li, K.; Shi, S.; Li, J.; Tang, S.; Liu, L. The Application of UHPLC-HRMS for Quality Control of Traditional Chinese Medicine. Front. Pharmacol. 2022, 13, 922488. [Google Scholar] [CrossRef]
- Yang, M.; Li, M.; Huang, Z.; He, Q.; Sun, Q.; Yang, Y. UHPLC Fingerprinting and Spectroscopic Relationship of Antioxidant Activity of the Miao Medicine Ficus Tikoua Bur. Chin. J. Mod. Appl. Pharm. 2024, 41, 787–796. [Google Scholar]
- Cao, X.; Hu, C.; Shang, F.; Lv, Y.; Bian, Z.; Yuan, Q.; Zhang, H.; Wang, Y.; Li, N.; Wang, L.; et al. Identification and quantitation of NF-κB inhibitory components in weichang’an pill based on UHPLC-QE-MS and spectrum-effect relationship. Arab. J. Chem. 2024, 17, 105328. [Google Scholar] [CrossRef]
- Nijat, D.; Lu, C.F.; Lu, J.J.; Abdulla, R.; Hasan, A.; Aidarhan, N.; Aisa, H.A. Spectrum-effect relationship between UPLC fingerprints and antidiabetic and antioxidant activities of Rosa rugosa. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2021, 1179, 122843. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, G.; Pan, J.; Gong, D. Galangin competitively inhibits xanthine oxidase by a ping-pong mechanism. Food Res. Int. 2016, 89, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yu, X.; Yang, R.; Zheng, B.; Zhang, Y.; Zhang, F. Quality evaluation of Lonicerae Japonicae Flos from different origins based on high-performance liquid chromatography (HPLC) fingerprinting and multicomponent quantitative analysis combined with chemical pattern recognition. Phytochem. Anal. 2024, 35, 647–663. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sun, Y.; Gao, Y.; Yu, X.; Zhao, C.; Song, X.; Han, F.; Yu, J. Spectrum-effect relationship analysis based on HPLC-FT-ICR-MS and multivariate statistical analysis to reveal the pharmacodynamic substances of Ling-Gui-Zhu-Gan decoction on Alzheimer’s disease. J. Pharm. Biomed. Anal. 2024, 237, 115765. [Google Scholar] [CrossRef]
- Yu, Y.; Xiong, Y.; Tong, S.; Li, Y.; Cai, R.; Zhang, X.; Gao, F. Inhibitory Activity of Quercetin, Rutin, and Hyperoside against Xanthine Oxidase: Kinetics, Fluorescence, and Molecular Docking. Curr. Pharm. Biotechnol. 2024, 3222. [Google Scholar] [CrossRef]
- Wu, H.; Wang, Y.; Huang, J.; Li, Y.; Lin, Z.; Zhang, B. Rutin ameliorates gout via reducing XOD activity, inhibiting ROS production and NLRP3 inflammasome activation in quail. Biomed. Pharmacother. 2023, 158, 114175. [Google Scholar] [CrossRef]
- Chen, M.; Ji, H.; Song, W.; Zhang, D.; Su, W.; Liu, S. Anserine beneficial effects in hyperuricemic rats by inhibiting XOD, regulating uric acid transporter and repairing hepatorenal injury. Food Funct. 2022, 13, 9434–9442. [Google Scholar] [CrossRef]
- Ganzon, J.G.; Chen, L.G.; Wang, C.C. 4-O-Caffeoylquinic acid as an antioxidant marker for mulberry leaves rich in phenolic compounds. J. Food Drug Anal. 2018, 26, 985–993. [Google Scholar] [CrossRef]
- Zhang, M.; Lai, J.; Wu, Q.; Lai, J.; Su, J.; Zhu, B.; Li, Y. Naringenin Induces HepG2 Cell Apoptosis via ROS-Mediated JAK-2/STAT-3 Signaling Pathways. Molecules 2023, 28, 4506. [Google Scholar] [CrossRef]
- Deepika; Dakal, T.C.; Sharma, N.K.; Ranga, V.; Maurya, P.K. Naringenin Orchestrates and Regulates the Reactive Oxygen Species-Mediated Pathways and Proinflammatory Signaling: Targeting Hallmarks of Aging-Associated Disorders. Rejuvenation Res. 2024, 27, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Musci, M.; Yao, S. Optimization and validation of Folin–Ciocalteu method for the determination of total polyphenol content of Pu-erh tea. Int. J. Food Sci. Nutr. 2017, 68, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Yong, T.; Liang, D.; Chen, S.; Xiao, C.; Gao, X.; Wu, Q.; Xie, Y.; Huang, L.; Hu, H.; Li, X.; et al. Caffeic acid phenethyl ester alleviated hypouricemia in hyperuricemic mice through inhibiting XOD and up-regulating OAT3. Phytomedicine 2022, 103, 154256. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.Y.; Baek, B.S.; Song, S.H.; Kim, M.S.; Huh, J.I.; Shim, K.H.; Kim, K.W.; Lee, K.H. Xanthine dehydrogenase/xanthine oxidase and oxidative stress. AGE 1997, 20, 127–140. [Google Scholar] [CrossRef]
- Noviana, E.; Indrayanto, G.; Rohman, A. Advances in Fingerprint Analysis for Standardization and Quality Control of Herbal Medicines. Front. Pharmacol. 2022, 13, 853023. [Google Scholar] [CrossRef]
- Pervez, H.; Mozumder, M.S.; Mourad, A.-H.I. Optimization of Injection Molding Parameters for HDPE/TiO2 Nanocomposites Fabrication with Multiple Performance Characteristics Using the Taguchi Method and Grey Relational Analysis. Materials 2016, 9, 710. [Google Scholar] [CrossRef]
- Aucar, M.G.; Cavasotto, C.N. Molecular Docking Using Quantum Mechanical-Based Methods. In Quantum Mechanics in Drug Discovery; Heifetz, A., Ed.; Springer: New York, NY, USA, 2020; pp. 269–284. [Google Scholar]
- Stanzione, F.; Giangreco, I.; Cole, J.C. Chapter Four—Use of molecular docking computational tools in drug discovery. In Progress in Medicinal Chemistry; Witty, D.R., Cox, B., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; Volume 60, pp. 273–343. [Google Scholar]


| Order of Inspection | Variant | Prerequisite | Total Polyphenol Yield (mg/g) | Option |
|---|---|---|---|---|
| 1 | Withdrawal method | ultrasound extraction | 4.348 ± 0.008 | √ |
| reflux extraction | 4.234 ± 0.013 | |||
| 2 | Extraction solvents | methanol | 3.887 ± 0.018 | |
| ethanol | 3.948 ± 0.008 | √ | ||
| 3 | Material–liquid ratio (g/mL) | 1:30 | 2.673 ± 0.046 | |
| 1:35 | 2.951 ± 0.026 | |||
| 1:40 | 3.199 ± 0.018 | √ | ||
| 1:45 | 2.343 ± 0.017 | |||
| 1:50 | 2.100 ± 0.010 | |||
| 4 | Ethanol volume fraction (%) | 40 | 2.806 ± 0.10 | |
| 50 | 3.060 ± 0.01 | |||
| 60 | 3.263 ± 0.06 | √ | ||
| 70 | 3.205 ± 0.01 | |||
| 80 | 3.118 ± 0.036 | |||
| 5 | Withdrawal time (min) | 50 | 2.012 ± 0.046 | |
| 60 | 2.136 ± 0.035 | |||
| 70 | 2.352 ± 0.035 | |||
| 80 | 2.468 ± 0.013 | √ | ||
| 90 | 2.174 ± 0.023 |
| NO. | TZ Batches | Total Polyphenol Yield (mg/g) | NO. | TZ Batches | Total Polyphenol Yield (mg/g) |
|---|---|---|---|---|---|
| 1 | TZ-14 | 7.454 ± 0.040 | 12 | TZ-8 | 5.509 ± 0.023 |
| 2 | TZ-11 | 7.245 ± 0.023 | 13 | TZ-19 | 5.462 ± 0.061 |
| 3 | TZ-17 | 6.790 ± 0.154 | 14 | TZ-20 | 5.431 ± 0.074 |
| 4 | TZ-6 | 6.689 ± 0.023 | 15 | TZ-9 | 5.123 ± 0.027 |
| 5 | TZ-13 | 6.635 ± 0.048 | 16 | TZ-5 | 4.852 ± 0.088 |
| 6 | TZ-4 | 6.597 ± 0.061 | 17 | TZ-2 | 4.775 ± 0.053 |
| 7 | TZ-12 | 6.381 ± 0.071 | 18 | TZ-18 | 4.575 ± 0.081 |
| 8 | TZ-10 | 6.373 ± 0.058 | 19 | TZ-15 | 3.888 ± 0.000 |
| 9 | TZ-3 | 6.249 ± 0.023 | 20 | TZ-7 | 2.838 ± 0.013 |
| 10 | TZ-16 | 5.979 ± 0.136 | 21 | TZ-1 | 2.136 ± 0.013 |
| 11 | TZ-21 | 5.624 ± 0.106 |
| Order of Inspection | Variant | Prerequisite | XOD Oxidation Rate (1 × 10−4) | Option |
|---|---|---|---|---|
| 1 | XOD concentration (U/L) | 25 | 3.333 ± 1.155 | |
| 50 | 4.333 ± 1.155 | |||
| 100 | 9.333 ± 1.155 | |||
| 200 | 11.667 ± 1.155 | √ | ||
| 400 | 3.667 ± 0.577 | |||
| 2 | Xanthine concentration (mg/L) | 100 | 112.000 ± 5.292 | |
| 120 | 175.667 ± 10.066 | |||
| 140 | 358.667 ± 3.215 | |||
| 160 | 412.667 ± 2.517 | √ | ||
| 180 | 372.333 ± 2.517 | |||
| 200 | 283.000 ± 1.000 | |||
| 220 | 229.000 ± 3.000 | |||
| 3 | Reaction temperature (°C) | 15 | 165.000 ± 1.732 | |
| 20 | 203.333 ± 8.083 | |||
| 25 | 264.333 ± 1.155 | |||
| 30 | 281.667 ± 7.024 | √ | ||
| 35 | 230.667 ± 9.292 | |||
| 4 | pH (PBS) | 6 | 189.333 ± 2.517 | |
| 6.5 | 218.667 ± 4.041 | |||
| 7 | 226.667 ± 5.859 | |||
| 7.5 | 229.667 ± 0.577 | √ | ||
| 8 | 217.333 ± 5.508 | |||
| 8.5 | 196.000 ± 3.606 |
| Rank | Batch | 1/IC50 (μg/mL) | Rank | Batch | 1/IC50 (μg/mL) |
|---|---|---|---|---|---|
| Positive drug | Allopurinol | 1.166 ± 0.059 | 11 | TZ–20 | 0.582 ± 0.077 |
| 1 | TZ–15 | 1.067 ± 0.091 | 12 | TZ–13 | 0.579 ± 0.011 |
| 2 | TZ–17 | 0.938 ± 0.201 | 13 | TZ–14 | 0.576 ± 0.127 |
| 3 | TZ–1 | 0.855 ± 0.052 | 14 | TZ–7 | 0.548 ± 0.026 |
| 4 | TZ–4 | 0.791 ± 0.027 | 15 | TZ–12 | 0.494 ± 0.016 |
| 5 | TZ–3 | 0.781 ± 0.126 | 16 | TZ–16 | 0.454 ± 0.053 |
| 6 | TZ–11 | 0.747 ± 0.116 | 17 | TZ–10 | 0.449 ± 0.248 |
| 7 | TZ–2 | 0.715 ± 0.407 | 18 | TZ–21 | 0.427 ± 0.027 |
| 8 | TZ–6 | 0.686 ± 0.183 | 19 | TZ–8 | 0.422 ± 0.072 |
| 9 | TZ–18 | 0.661 ± 0.059 | 20 | TZ–19 | 0.412 ± 0.020 |
| 10 | TZ–9 | 0.649 ± 0.022 | 21 | TZ–5 | 0.381 ± 0.083 |
| Rank | Peak | Relatedness Value | Rank | Peak | Relatedness Value |
|---|---|---|---|---|---|
| 1 | F14 | 2.571 | 9 | F15 | 2.309 |
| 2 | F7 | 2.475 | 10 | F9 | 2.226 |
| 3 | F3 | 2.473 | 11 | F2 | 2.218 |
| 4 | F10 | 2.464 | 12 | F4 | 2.211 |
| 5 | F5 | 2.376 | 13 | F13 | 2.203 |
| 6 | F1 | 2.349 | 14 | F16 | 2.127 |
| 7 | F8 | 2.338 | 15 | F11 | 2.125 |
| 8 | F6 | 2.323 | 16 | F12 | 2.106 |
| Peak | XOD | Peak | XOD | ||
|---|---|---|---|---|---|
| Pearson Correlation | Significance (Two–Tailed) | Pearson Correlation | Significance (Two–Tailed) | ||
| F1 | −0.404 | 0.069 | F9 | −0.316 | 0.163 |
| F2 | 0.081 | 0.727 | F10 | 0.338 | 0.134 |
| F3 | −0.031 | 0.893 | F11 | 0.270 | 0.236 |
| F4 | 0.556 ** | 0.009 * | F12 | −0.428 | 0.053 |
| F5 | −0.214 | 0.352 | F13 | −0.288 | 0.205 |
| F6 | 0.032 | 0.890 | F14 | −0.280 | 0.219 |
| F7 | −0.043 | 0.853 | F15 | −0.281 | 0.217 |
| F8 | −0.463 * | 0.034 * | F16 | 0.184 | 0.425 |
| Protein PDB Number | Ingredient | Type of Force | Amino Acid Residue | Residue Binding Site | Active Force |
|---|---|---|---|---|---|
| XOD-1FIQ | F2 4–O–Caffeoylquinic acid | Hydrophobic Interactions | TRP | 336 | 3.93 |
| TRP | 336 | 3.70 | |||
| Hydrogen Bonds | TRP | 336 | 2.54 | ||
| LYS | 422 | 3.23 | |||
| SER | 425 | 2.65 | |||
| Salt Bridges | ARG | 426 | 4.10 | ||
| LYS | 433 | 4.8 | |||
| LYS | 1228 | 5.45 | |||
| pi-Stacking | TRP | 336 | 3.7 | ||
| F10 Naringenin | Hydrophobic Interactions | ILE | 266 | 3.9 | |
| THR | 354 | 3.7 | |||
| ILE | 358 | 3.6 | |||
| Hydrogen Bonds | GLU | 45 | 2.8 | ||
| ASN | 351 | 3.0 | |||
| ASN | 351 | 4.0 | |||
| ARG | 426 | 3.4 | |||
| SER | 1225 | 4.1 |
| NO. | PBS/mL | TZ Extracts/mL | XOD/mL | Thymine/mL |
|---|---|---|---|---|
| A1 | 0.390 | 0.010 | 0.050 | 0.350 |
| A2 | 0.440 | 0.010 | 0.000 | 0.350 |
| A3 | 0.400 | 0.000 | 0.050 | 0.350 |
| A4 | 0.450 | 0.000 | 0.000 | 0.350 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Xu, Y.; Yu, Q.; Li, M.; Yang, L.; Yang, Y. Spectroscopic Relationship between XOD and TAOZHI Total Polyphenols Based on Chemometrics and Molecular Docking Techniques. Molecules 2024, 29, 4288. https://doi.org/10.3390/molecules29184288
Yang M, Xu Y, Yu Q, Li M, Yang L, Yang Y. Spectroscopic Relationship between XOD and TAOZHI Total Polyphenols Based on Chemometrics and Molecular Docking Techniques. Molecules. 2024; 29(18):4288. https://doi.org/10.3390/molecules29184288
Chicago/Turabian StyleYang, Mingyu, Yitang Xu, Qihua Yu, Mengyu Li, Liyong Yang, and Ye Yang. 2024. "Spectroscopic Relationship between XOD and TAOZHI Total Polyphenols Based on Chemometrics and Molecular Docking Techniques" Molecules 29, no. 18: 4288. https://doi.org/10.3390/molecules29184288
APA StyleYang, M., Xu, Y., Yu, Q., Li, M., Yang, L., & Yang, Y. (2024). Spectroscopic Relationship between XOD and TAOZHI Total Polyphenols Based on Chemometrics and Molecular Docking Techniques. Molecules, 29(18), 4288. https://doi.org/10.3390/molecules29184288







