Constituents from Dolichos lablab L. Flowers and Their Anti-Inflammatory Effects via Inhibition of IL-1β Release
Abstract
1. Introduction
2. Results
3. Discussion
4. Experiment
4.1. Plant Material
4.2. General Experimental Procedures
4.3. Extraction and Isolation
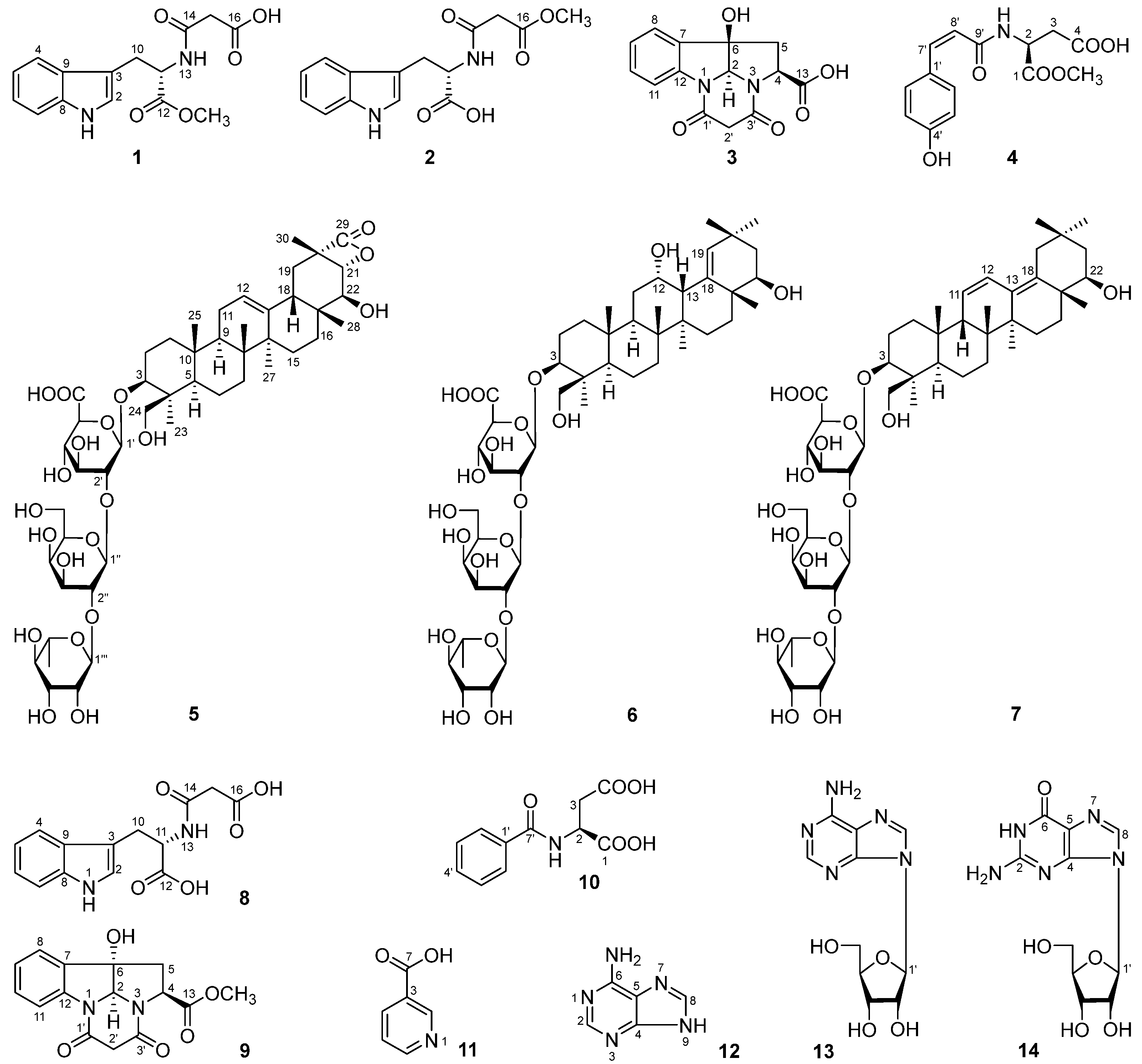
4.4. Spectral Data of 1–7
4.4.1. Flosdolilabnitrogenousol A (1)
4.4.2. Flosdolilabnitrogenousol B (2)
4.4.3. Flosdolilabnitrogenousol C (3)
4.4.4. Flosdolilabnitrogenousol D (4)
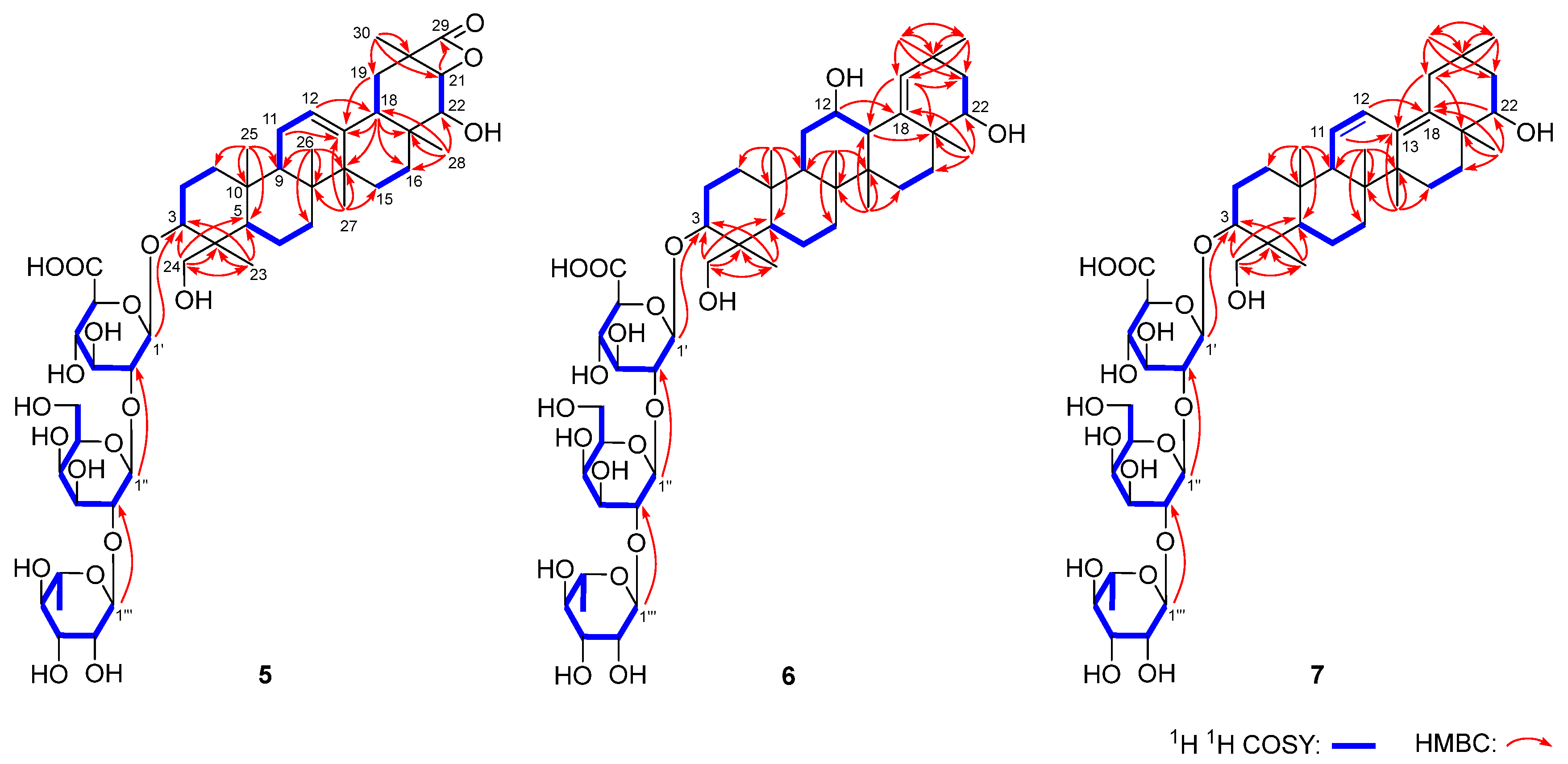
4.4.5. Flosdolilabsaponin A (5)
4.4.6. Flosdolilabsaponin B (6)
4.4.7. Flosdolilabsaponin C (7)
4.5. Determination of the Absolute Configuration of Sugars in Compound 5
4.6. ECD Calculation
4.7. BMDM Preparation
4.8. MTT Assay
4.9. Inhibitory Effect of the Tested Compounds on LPS/Nigericin-Induced IL-1β Release on BMDMs Measured by Mouse IL-1β Elisa Kit
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leuti, A.; Fazio, D.; Fava, M.; Piccoli, A.; Oddi, S.; Maccarrone, M. Bioactive lipids, inflammation and chronic diseases. Adv. Drug Deliv. Rev. 2020, 159, 133–169. [Google Scholar] [CrossRef] [PubMed]
- Vande Walle, L.; Lamkanfi, M. Drugging the NLRP3 inflammasome: From signalling mechanisms to therapeutic targets. Nat. Rev. Drug Discov. 2024, 23, 43–66. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Zhan, X.; Ding, K.; Xu, G.; Shi, W.; Ren, L.; Fang, Z.; Liu, T.; Hou, X.; Zhao, J.; et al. Dihydrotanshinone I specifically inhibits NLRP3 inflammasome activation and protects against septic shock in vivo. Front. Pharmacol. 2021, 12, 750815. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wu, W.; Li, W.; Huang, S.; Li, Z.; Liu, R.; Shan, Z.; Zhang, C.; Li, W.; Wang, S. Activation of NLRP3 inflammasome promotes foam cell formation in vascular smooth muscle cells and atherogenesis via HMGB1. J. Am. Heart Assoc. 2018, 7, e008596. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Liu, B.; Pei, S.; Zheng, D.; Wang, Z.; Ji, T.; Pan, S.; Shen, H.Y.; Wang, H. Higher CSF levels of NLRP3 inflammasome is associated with poor prognosis of anti-N-methyl-D-aspartate receptor encephalitis. Front. Immunol. 2019, 10, 905. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.N.; Guo, X.Y.; Tang, J.; Ding, S.Q.; Shen, L.; Wang, R.; Ma, S.F.; Hu, J.G.; Lü, H.Z. Expression and localization of absent in melanoma 2 in the injured spinal cord. Neural. Regen. Res. 2019, 14, 542–552. [Google Scholar] [PubMed]
- Liu, H.; Zhan, X.; Xu, G.; Wang, Z.; Li, R.; Wang, Y.; Qin, Q.; Shi, W.; Hou, X.; Yang, R.; et al. Cryptotanshinone specifically suppresses NLRP3 inflammasome activation and protects against inflammasome-mediated diseases. Pharmacol. Res. 2021, 164, 105384. [Google Scholar] [CrossRef] [PubMed]
- The National Health and Family Planning Commission releases the latest list of dual-purpose Chinese medicines. Zhongyi Linchuang Yanjiu 2017, 9, 11–14.
- Gao, Y.; Huang, R.; Qiu, Y.; Liu, Y.; Chen, L. Characterization of the chemical composition of different parts of Dolichos lablab L. and revelation of its anti-ulcerative colitis effects by modulating the gut microbiota and host metabolism. J. Ethnopharmacol. 2024, 322, 117629. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, X.; Peng, X.; Wang, Y.; Zhou, L.; Liu, X.; Guo, Q. Analysis of volatile components in different parts of Dolichos lablab by GC-MS. Zhongguo Yaoye 2020, 29, 12–14. [Google Scholar]
- Yue, D. Study on Chemical Constituents, Antioxidant and Hypoglycemic Activities of Flavonoids from Flos Dolichoris lablab L.; Anhui Ploytechnic University: Wuhu, China, 2021. [Google Scholar]
- Shi, Z.W.; Zhang, W.; Liu, W.; Ruan, J.Y.; Wang, T.; Zhang, Y. Isolation and identification of phenolic constituents from the flower bud of Dolichos lablab L. Zhongguo Yaowuhuaxue Zazhi 2023, 33, 857–863. [Google Scholar]
- Song, K.S.; Ishikawa, Y.; Kobayashi, S.; Sankawa, U.; Ebizuka, Y. N-Acylamino acids from Ephedra distachya cultures. Phytochemistry 1992, 31, 823–826. [Google Scholar] [CrossRef]
- Lu, M.; Ruan, J.; Yu, R.; Zhang, Y.; Zhao, W.; Yang, D.; Wang, W.; Zhang, Y.; Wang, T. Neolignan derivatives from Penthorum chinense with antitumor activity in human colorectal cancer cells by regulating Wnt/β-catenin signaling pathway. Phytochemistry 2023, 214, 113827. [Google Scholar] [CrossRef]
- Zhu, D.H.; Zhang, J.K.; Jia, J.F.; Liu, J.J.; Wei, J.J.; Yang, M.; Yang, Y.; Li, M.; Hao, Z.Y.; Zheng, X.K.; et al. Alkaloids from the stem of Ephedra equisetina. J. Asian Nat. Prod. Res. 2023, 25, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cao, H.; Ruan, J.; Wu, Y.; Yang, D.; Gao, Q.; Wang, D.; Chen, Q.; Zhang, Y.; Wang, T. Saponins from Aesculus wilsonii seeds exert anti-inflammatory activity through the suppression of NF-κB and NLRP3 pathway. Arab. J. Chem. 2023, 16, 105077. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Q.; Chen, H.; Wang, B.; An, D.; Zhao, Y. Structural determination of saponins from Hedysarum polybotrys. Magn. Reason. Chem. 2006, 44, 1128–1130. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Ruan, J.; Wang, T.; Dong, Y.; Hao, J.; Liu, E.; Han, L.; Gao, X.; Wang, T. Oleanane type saponins from the stems of Astragalus membranaceus (Fisch.) Bge. var. mongholicus (Bge.) Hsiao. Fitoterapia 2016, 109, 99–105. [Google Scholar] [CrossRef]
- Uchida, K.; Kawakishi, S. Ascorbate-mediated specific oxidation of the imidazole ring in a histidine derivative. Bioorg. Chem. 1989, 17, 330–343. [Google Scholar] [CrossRef]
- Tang, L.C.; Wang, N.; Yao, H.P.; Yang, X.H.; Deng, S.M.; Zhang, C.Y. Chemical components of the Hevea brasiliensis skim. Linye Huaxue Yu Gongye 2013, 33, 125–129. [Google Scholar]
- Lim, S.S.; Jung, Y.J.; Hyun, S.K.; Lee, Y.S.; Choi, J.S. Rat lens aldose reductase inhibitory constituents of Nelumbo nucifera stamens. Phytother. Res. 2006, 20, 825–830. [Google Scholar] [CrossRef]
- Domondon, D.L.; He, W.; De Kimpe, N.; Hofte, M.; Poppe, J. β-Adenosine, a bioactive compound in grass chaff stimulating mushroom production. Phytochemistry 2004, 65, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, C.; Wang, F.; Dong, H.; Guo, S.; Yang, J.; Xiao, P. Chemical constituents of Dendrobium candidum. Zhongguo Zhongyao Zazhi 2010, 35, 1715–1719. [Google Scholar] [PubMed]
- Diaz-Del-Olmo, I.; Worboys, J.; Martin-Sanchez, F.; Gritsenko, A.; Ambrose, A.R.; Tannahill, G.M.; Nichols, E.M.; Lopez-Castejon, G.; Davis, D.M. Internalization of the membrane attack complex triggers NLRP3 inflammasome activation and IL-1β secretion in human macrophages. Front. Immunol. 2021, 12, 720655. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, G.; Yang, X.; Wang, W.; Zhang, J.; He, Y.; Zhang, W.; Jing, T.; Lin, R. Nicotinic acid inhibits NLRP3 inflammasome activation via SIRT1 in vascular endothelial cells. Int. Immunopharmacol. 2016, 40, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Cronstein, B.N. Adenosine, an endogenous anti-inflammatory agent. J. Appl. Physiol. 1994, 76, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Hansel, G.; Tonon, A.C.; Guella, F.L.; Pettenuzzo, L.F.; Duarte, T.; Duarte, M.M.M.F.; Oses, J.P.; Achaval, M.; Souza, D.O. Guanosine protects against cortical focal ischemia. Involvement of inflammatory response. Mol. Neurobiol. 2015, 52, 1791–1803. [Google Scholar] [CrossRef]
- Scott, S.A.; Fu, J.; Chang, P.V. Dopamine receptor D2 confers colonization resistance via microbial metabolites. Nature 2024, 628, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Cervenka, I.; Agudelo, L.Z.; Ruas, J.L. Kynurenines: Tryptophan’s metabolites in exercise, inflammation, and mental health. Science 2017, 357, eaaf9794. [Google Scholar] [CrossRef] [PubMed]
- Farooq, A.; Hoque, R.; Ouyang, X.; Farooq, A.; Ghani, A.; Ahsan, K.; Guerra, M.; Mehal, W.Z. Activation of N-methyl-D-aspartate receptor downregulates inflammasome activity and liver inflammation via a β-arrestin-2 pathway. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, 732–740. [Google Scholar] [CrossRef]
- Sun, S.; Li, Z.; Huang, C.; Liu, J.; Yu, Q.; Jiang, X.; Yue, K.; Zhao, J.; Xu, T.; Liu, Y.; et al. Discovery of novel 2,3-dihydro-1H-indene-5-sulfonamide NLRP3 inflammasome inhibitors targeting colon as a potential therapy for colitis. J. Med. Chem. 2023, 66, 16141–16167. [Google Scholar] [CrossRef]




| No. | 1 | 2 | ||
|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | – | 10.89 (d, 2.5, 1H) | – | 10.88 (d, 2.0, 1H) |
| 2 | 123.7 | 7.16 (d, 2.5, 1H) | 123.5 | 7.14 (d, 2.0, 1H) |
| 3 | 109.0 | – | 109.5 | – |
| 4 | 117.9 | 7.48 (br. d, ca. 8, 1H) | 118.1 | 7.53 (dd, 1.0, 8.0, 1H) |
| 5 | 118.3 | 6.99 (t like, ca. 8, 1H) | 118.3 | 6.98 (dt like, ca. 1, 8, 1H) |
| 6 | 120.9 | 7.07 (t like, ca. 8, 1H) | 120.8 | 7.08 (dt like, ca. 1, 8, 1H) |
| 7 | 111.3 | 7.34 (br. d, ca. 8, 1H) | 111.2 | 7.34 (br. d, ca. 8, 1H) |
| 8 | 136.0 | – | 135.9 | – |
| 9 | 127.0 | – | 127.2 | – |
| 10 | 27.1 | 3.07 (dd, 7.5, 14.5, 1H) | 27.0 | 3.05 (dd, 7.5, 15.0, 1H) |
| 3.14 (dd, 5.5, 14.5, 1H) | 3.18 (dd, 5.5, 15.0, 1H) | |||
| 11 | 53.1 | 4.55 (ddd, 5.5, 7.5, 7.5, 1H) | 53.1 | 4.50 (ddd, 5.5, 7.5, 8.0, 1H) |
| 12 | 172.0 | – | 172.9 | – |
| 13 | – | 8.57 (d, 7.5, 1H) | – | 8.43 (d, 8.0, 1H) |
| 14 | 165.8 | – | 164.9 | – |
| 15 | 42.1 | 3.15 (s, 2H) | 41.8 | 3.30 (s, 2H) |
| 16 | 169.2 | – | 168.1 | – |
| 12-COOCH3 | 51.7 | 3.57 (s, 1H) | ||
| 16-COOCH3 | 51.6 | 3.58 (s, 3H) | ||
| No. | δC | δH (J in Hz) | No. | δC | δH (J in Hz) |
|---|---|---|---|---|---|
| 2 | 83.2 | 5.74 (s, 1H) | 11 | 113.9 | 7.68 (br. d, ca. 8, 1H) |
| 4 | 60.0 | 4.58 (dd, 2.5, 10.0, 1H) | 12 | 142.1 | – |
| 5 | 42.7 | 2.58 (dd, 2.5, 14.0, 1H) | 13 | 170.6 | – |
| 2.94 (dd, 10.0, 14.0, 1H) | 1′ | 165.8 | – | ||
| 6 | 84.4 | – | 2′ | 46.9 | 2.93 (d, 16.0, 1H) |
| 7 | 133.9 | – | 4.35 (d, 16.0, 1H) | ||
| 8 | 124.4 | 7.45 (br. d, ca. 8, 1H) | 3′ | 167.3 | – |
| 9 | 124.2 | 7.10 (t like, ca. 8, 1H) | 6-OH | – | 6.47 (br. s, 1H) |
| 10 | 129.6 | 7.31 (t like, ca. 8, 1H) | 13-COOH | 12.72 (br. s, 1H) |
| No. | δC | δH (J in Hz) | No. | δC | δH (J in Hz) |
|---|---|---|---|---|---|
| 1 | 172.9 | – | 3′,5′ | 116.0 | 6.72 (d, 8.4, 2H) |
| 2 | 50.4 | 4.83 (dd, 5.4, 7.2, 1H) | 4′ | 159.4 | – |
| 3 | 37.0 | 2.79 (dd, 7.2, 17.4, 1H) | 7′ | 139.4 | 6.67 (d, 12.6, 1H) |
| 2.84 (dd, 5.4, 17.4, 1H) | 8′ | 120.5 | 5.84 (d, 12.6, 1H) | ||
| 4 | 174.2 | – | 9′ | 169.9 | – |
| 1′ | 127.8 | – | 1-COOCH3 | 53.0 | 3.73 (s, 3H) |
| 2′,6′ | 132.6 | 7.45 (d, 8.4, 2H) |
| No. | 5 | 6 | 7 | |||
|---|---|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | 38.7 | 0.83 (m, 1H) | 38.7 | 0.80 (m, 1H) | 37.9 | 0.85 (m, o, 1H) |
| 1.39 (m, o, 1H) | 1.41 (m, o, 1H) | 1.62 (m, o, 1H) | ||||
| 2 | 26.7 | 1.88 (m, 1H) | 26.7 | 1.86 (m, o, 1H) | 26.6 | 1.94 (m, o, 1H) |
| 2.23 (m, 1H) | 2.11 (m, 1H) | 2.26 (m, o, 1H) | ||||
| 3 | 91.2 | 3.43 (dd, 3.0, 10.8) | 91.3 | 3.27 (dd, 4.2, 11.4, 1H) | 91.2 | 3.42 (dd, 4.2, 11.4, 1H) |
| 4 | 43.9 | – | 44.0 | – | 43.9 | – |
| 5 | 56.2 | 0.86 (br. d, ca. 11, 1H) | 56.5 | 0.87 (br. d, ca. 11, 1H) | 55.4 | 0.87 (m, o, 1H) |
| 6 | 18.5 | 1.25 (m, 1H) | 18.5 | 1.25 (m, 1H) | 18.6 | 1.29 (m, o, 1H) |
| 1.55 (m, o, 1H) | 1.55 (m, o, 1H) | 1.62 (m, o, 1H) | ||||
| 7 | 33.7 | 1.40 (m, o, 1H) | 35.2 | 1.52 (m, 1H) | 32.8 | 1.32 (m, 1H) |
| 1.44 (m, 1H) | ||||||
| 8 | 39.6 | – | 41.1 | – | 40.5 | – |
| 9 | 47.8 | 1.51 (dd, 7.2, 10.2, 1H) | 45.1 | 1.99 (dd, 7.2, 12.0, 1H) | 54.3 | 1.94 (br. s, 1H) |
| 10 | 36.5 | – | 36.4 | – | 36.2 | – |
| 11 | 23.9 | 1.79 (m, o, 1H) | 31.7 | 1.42 (m, 1H) | 126.2 | 5.56 (br. d, ca. 10, 1H) |
| 1.64 (m, 1H) | ||||||
| 12 | 124.6 | 5.32 (t like, ca. 3, 1H) | 67.9 | 4.43 (m, o, 1H) | 126.5 | 6.49 (d, 10.2, 1H) |
| 13 | 141.8 | – | 43.1 | 2.40 (br. s, 1H) | 135.2 | – |
| 14 | 42.3 | – | 43.1 | – | 42.3 | – |
| 15 | 25.8 | 1.12 (m, o, 1H) | 28.9 | 1.20 (m, 1H) | 24.6 | 1.13 (m, 1H) |
| 1.70 (m, 1H) | 1.87 (m, 1H) | 1.68 (m, 1H) | ||||
| 16 | 31.9 | 1.84 (m, 2H) | 34.3 | 1.70 (m, 1H) | 33.7 | 1.85 (m, o, 1H) |
| 2.24 (m, 1H) | 2.26 (m, o, 1H) | |||||
| 17 | 37.8 | – | 40.4 | – | 41.2 | – |
| 18 | 47.8 | 2.10 (br. d, ca. 13, 1H) | 140.3 | – | 137.9 | – |
| 19 | 35.7 | 1.13 (m, o, 1H) | 130.8 | 5.97 (s, 1H) | 37.8 | 1.88 (d, 14.4, 1H) |
| 2.60 (dd, 13.2, 13.2, 1H) | 2.49 (d, 14.4, 1H) | |||||
| 20 | 55.4 | – | 33.9 | – | 32.3 | – |
| 21 | 83.1 | 4.57 (d, 7.8, 1H) | 42.3 | 1.87 (m, o, 1H) | 44.8 | 1.76 (dd, 3.6, 11.4, 1H) |
| 1.97 (dd, 12.6, 12.6, 1H) | 1.84 (m, o, 1H) | |||||
| 22 | 79.4 | 4.15 (d, 7.8, 1H) | 75.3 | 4.08 (m, 1H) | 76.6 | 3.80 (dd, 3.6, 11.4, 1H) |
| 23 | 23.1 | 1.46 (s, 3H) | 22.9 | 1.41 (s, 3H) | 22.9 | 1.46 (s, 3H) |
| 24 | 63.7 | 3.27 (d, 11.4, 1H) | 63.6 | 3.25 (d, 11.4, 1H) | 63.3 | 3.27 (d, 11.4, 1H) |
| 4.28 (d, 11.4, 1H) | 4.26 (d, 11.4, 1H) | 4.24 (d, 11.4, 1H) | ||||
| 25 | 16.1 | 0.72 (s, 3H) | 17.4 | 0.70 (s, 3H) | 18.2 | 0.71 (s, 3H) |
| 26 | 17.3 | 0.87 (s, 3H) | 16.4 | 1.05 (s, 3H) | 16.6 | 0.76 (s, 3H) |
| 27 | 24.3 | 1.13 (s, 3H) | 20.0 | 1.35 (s, 3H) | 20.3 | 1.10 (s, 3H) |
| 28 | 22.8 | 1.16 (s, 3H) | 18.9 | 1.35 (s, 3H) | 18.7 | 1.35 (s, 3H) |
| 29 | 175.7 | – | 30.8 | 1.17 (s, 3H) | 32.6 | 1.03 (s, 3H) |
| 30 | 19.4 | 1.57 (s, 3H) | 32.2 | 1.06 (s, 3H) | 25.5 | 0.90 (s, 3H) |
| 1′ | 105.5 | 5.00 (d, 7.2, 1H) | 105.5 | 4.97 (d, 6.6, 1H) | 105.6 | 5.03 (d, 7.2, 1H) |
| 2′ | 76.8 | 4.60 (m, o, 1H) | 76.7 | 4.58 (dd, 6.6, 9.0, 1H) | 76.8 | 4.46 (dd, 7.2, 8.4, 1H) |
| 3′ | 78.6 | 4.62 (m, o, 1H) | 78.5 | 4.61 (dd, 9.0, 9.6, 1H) | 78.6 | 4.65 (dd, 8.4, 8.4, 1H) |
| 4′ | 73.9 | 4.46 (dd, 9.6, 9.6, 1H) | 74.0 | 4.43 (m, o, 1H) | 73.9 | 4.48 (dd, 8.4, 9.6, 1H) |
| 5′ | 77.6 | 4.62 (m, o, 1H) | 77.6 | 4.69 (d, 8.4, 1H) | 77.6 | 4.66 (d, 9.6, 1H) |
| 6′ | 172.7 | – | 172.8 | – | 172.6 | – |
| 1″ | 101.8 | 5.80 (d, 7.2, 1H) | 101.8 | 5.77 (d, 6.6, 1H) | 101.8 | 5.81 (d, 6.6, 1H) |
| 2″ | 77.8 | 4.57 (m, o, 1H) | 77.9 | 4.55 (dd, 6.6, 9.0, 1H) | 77.9 | 4.58 (dd, 6.6, 8.4, 1H) |
| 3″ | 76.7 | 4.11 (dd, 3.0, 9.0, 1H) | 76.7 | 4.09 (br. d, ca. 9, 1H) | 76.7 | 4.12 (br. d, ca. 8, 1H) |
| 4″ | 71.2 | 4.40 (br. d, ca. 3, 1H) | 71.2 | 4.39 (br. s, 1H) | 71.2 | 4.41 (br. s, 1H) |
| 5″ | 76.5 | 3.94 (t like, ca. 6, 1H) | 76.5 | 3.93 (t like, ca. 5, 1H) | 76.5 | 3.94 (t like, ca. 5, 1H) |
| 6″ | 61.6 | 4.31 (dd, 6.0, 10.8, 1H) | 61.6 | 4.30 (dd, 4.8, 10.2, 1H) | 61.6 | 4.32 (dd, 5.4, 10.8, 1H) |
| 4.43 (dd, 6.0, 10.8, 1H) | 4.41 (dd, 6.0, 10.2, 1H) | 4.42 (dd, 5.4, 10.8, 1H) | ||||
| 1‴ | 102.5 | 6.30 (br. s, 1H) | 102.5 | 6.28 (br. s, 1H) | 102.6 | 6.31 (br. s, 1H) |
| 2‴ | 72.5 | 4.81 (br. s, 1H) | 72.2 | 4.81 (br. s, 1H) | 72.5 | 4.83 (br. s, 1H) |
| 3‴ | 72.8 | 4.69 (br. d, ca. 8, 1H) | 72.5 | 4.68 (br. d, ca. 8, 1H) | 72.9 | 4.70 (br. d, ca. 9, 1H) |
| 4‴ | 74.4 | 4.35 (dd, 9.0, 9.6, 1H) | 74.4 | 4.32 (dd, 8.4, 9.0, 1H) | 74.4 | 4.36 (dd, 9.0, 10.8, 1H) |
| 5‴ | 69.5 | 5.00 (m, 1H) | 69.4 | 4.99 (m, 1H) | 69.5 | 5.02 (m, 1H) |
| 6‴ | 19.0 | 1.79 (d, 6.0, 3H) | 19.0 | 1.78 (d, 6.0, 3H) | 19.0 | 1.82 (d, 6.0, 3H) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Z.; Li, H.; Cheng, J.; Zhang, W.; Ruan, J.; Zhang, Q.; Dang, Z.; Zhang, Y.; Wang, T. Constituents from Dolichos lablab L. Flowers and Their Anti-Inflammatory Effects via Inhibition of IL-1β Release. Molecules 2024, 29, 3751. https://doi.org/10.3390/molecules29163751
Shi Z, Li H, Cheng J, Zhang W, Ruan J, Zhang Q, Dang Z, Zhang Y, Wang T. Constituents from Dolichos lablab L. Flowers and Their Anti-Inflammatory Effects via Inhibition of IL-1β Release. Molecules. 2024; 29(16):3751. https://doi.org/10.3390/molecules29163751
Chicago/Turabian StyleShi, Zhongwei, Huimin Li, Jiaming Cheng, Wei Zhang, Jingya Ruan, Qianqian Zhang, Zhunan Dang, Yi Zhang, and Tao Wang. 2024. "Constituents from Dolichos lablab L. Flowers and Their Anti-Inflammatory Effects via Inhibition of IL-1β Release" Molecules 29, no. 16: 3751. https://doi.org/10.3390/molecules29163751
APA StyleShi, Z., Li, H., Cheng, J., Zhang, W., Ruan, J., Zhang, Q., Dang, Z., Zhang, Y., & Wang, T. (2024). Constituents from Dolichos lablab L. Flowers and Their Anti-Inflammatory Effects via Inhibition of IL-1β Release. Molecules, 29(16), 3751. https://doi.org/10.3390/molecules29163751






