Cell-Penetrating Peptide-Mediated Biomolecule Transportation in Artificial Lipid Vesicles and Living Cells
Abstract
1. Introduction
2. Membrane-Active Peptides
2.1. Classification of Membrane-Active Peptides
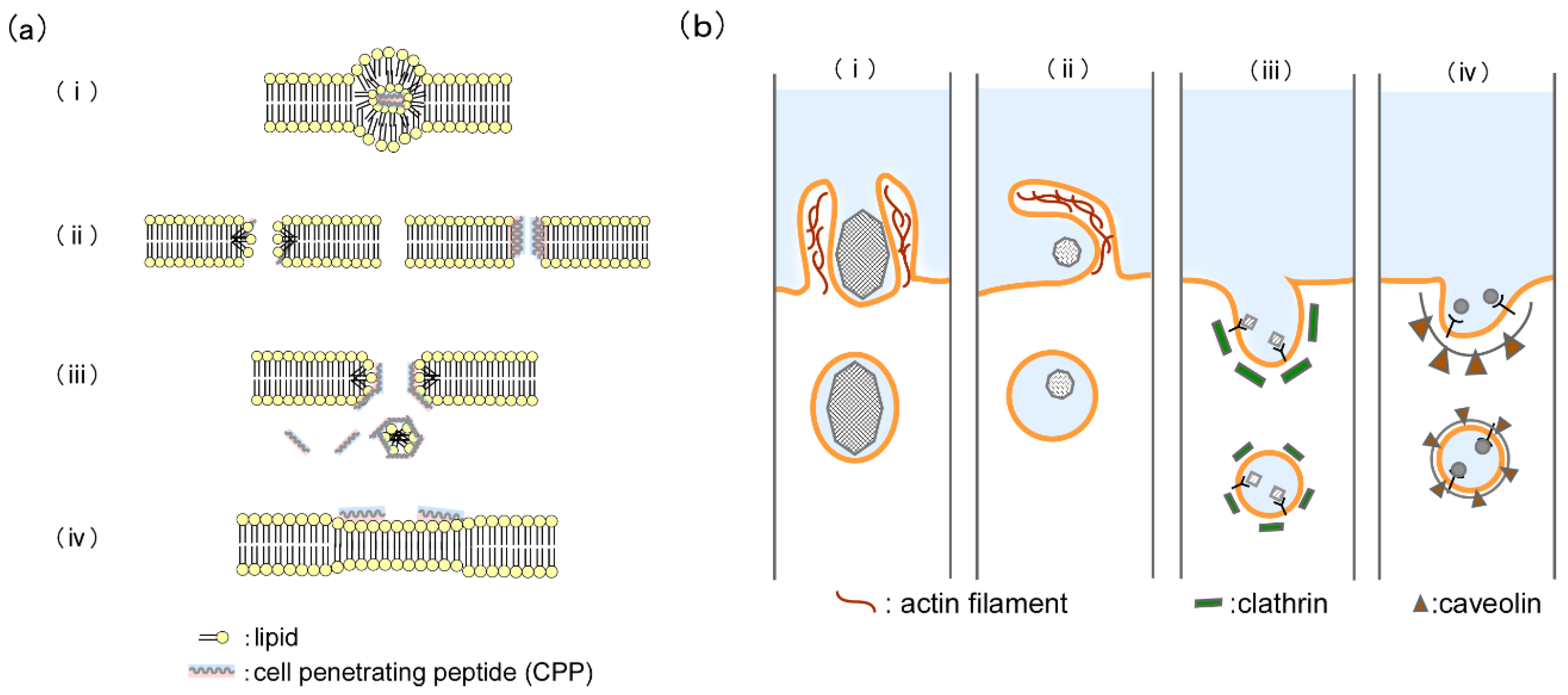
2.2. Internalization Mechanism of Cell-Penetrating Peptides
3. Artificial Membranes
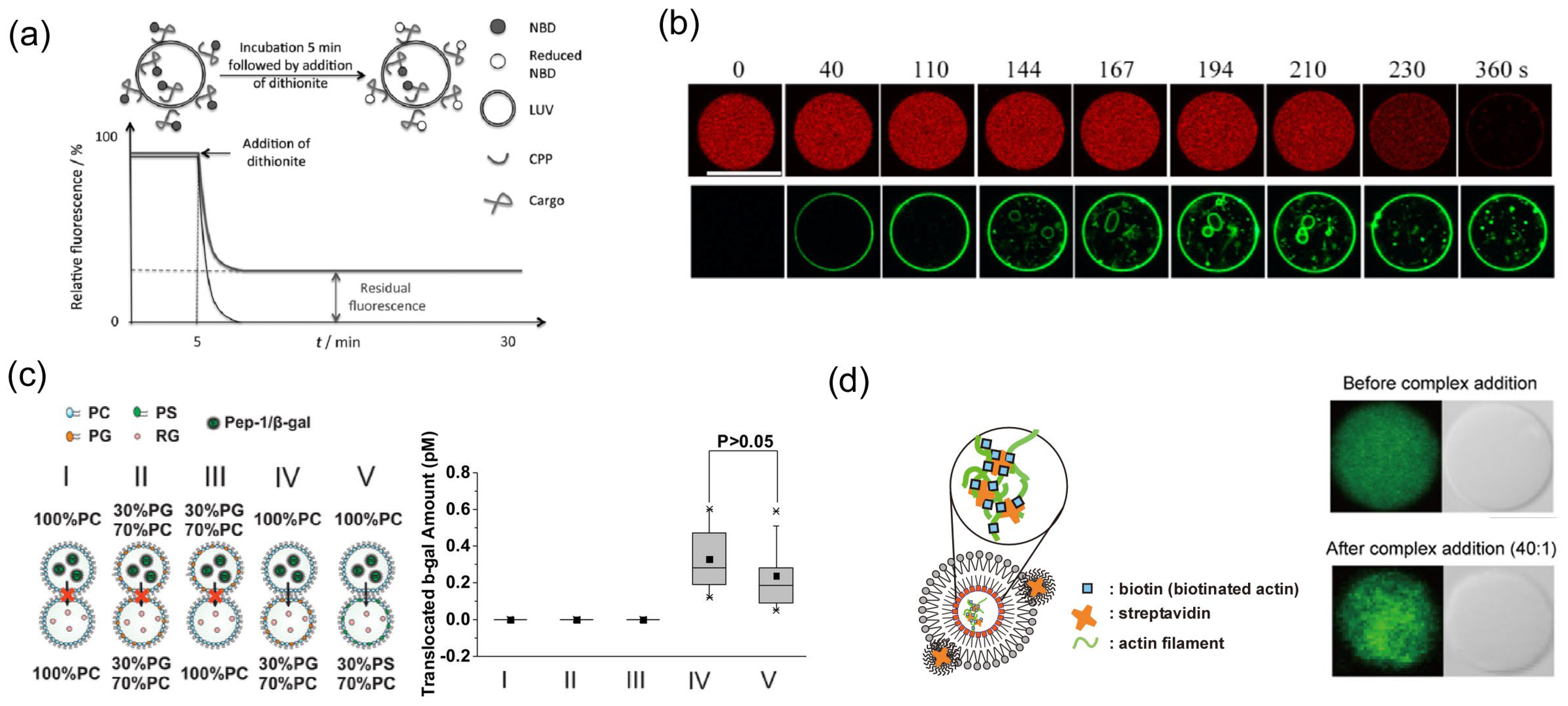
3.1. Observation of Conformation and Interaction Using Small Unilamellar Vesicles (SUVs) and Large Unilamellar Vesicles (LUVs)
3.2. Direct Observation of the Translocation of CPPs Using Giant Unilamellar Vesicles (GUVs)
3.3. Observation of CPP-Mediated Cargo Transportation Using Planar Bilayer Lipid Membrane
3.4. Application of Artificial Cell Models Using GUVs
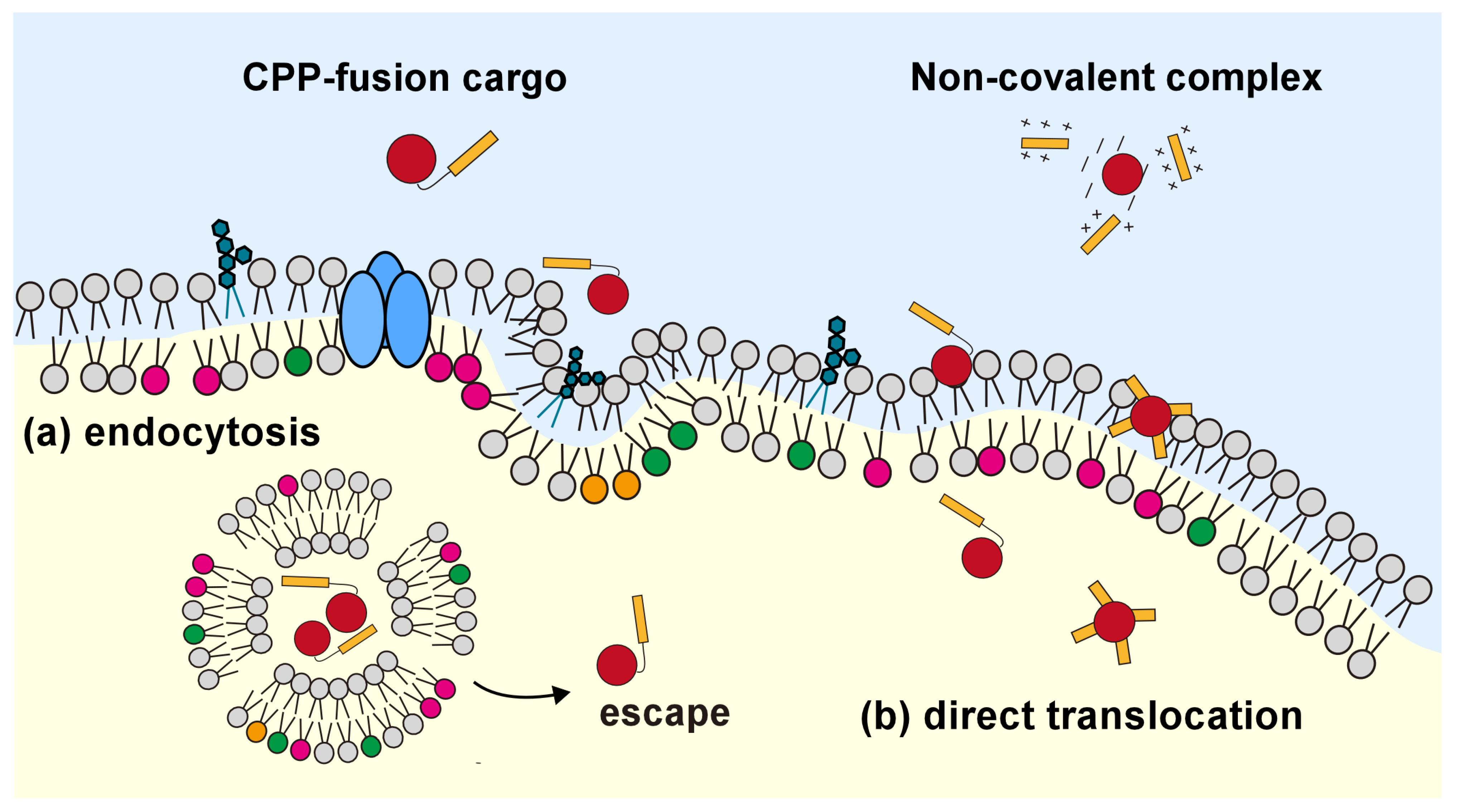
4. Cargo Transportation into the Living Cell for the Control of Cellular Reactions
4.1. CPP-Mediated Biomolecule Transportation into the Living Cell
4.2. Increase in Endosomal Escape Efficiency in the Strategy of Non-Covalent Bonds
4.3. Improving Biomolecules Transportation in the Strategy of Covalent Bonds
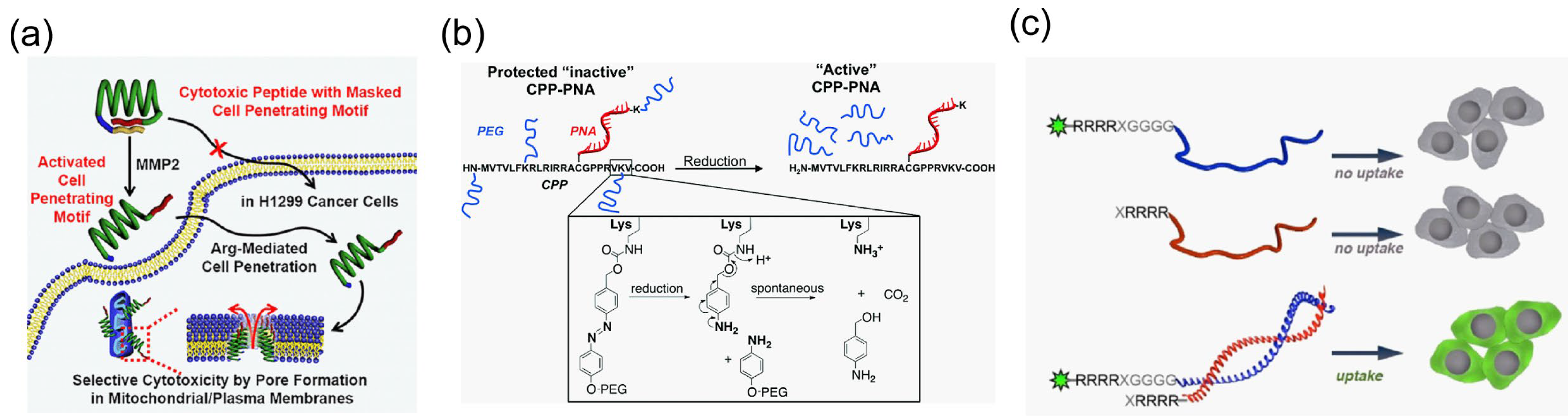
4.3.1. Enhancement of the Endosome Escape Efficiency of Cargo Molecules
4.3.2. Overcoming the Lack of Specificity of Cargo Transportation
5. Application of CPP-Mediated Control of Cellular Reactions: Functional Component Internalization
6. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ye, Z.; Galvanetto, N.; Puppulin, L.; Pifferi, S.; Flechsig, H.; Arndt, M.; Triviño, C.A.S.; Di Palma, M.; Guo, S.; Vogel, H.; et al. Structural Heterogeneity of the Ion and Lipid Channel TMEM16F. Nat. Commun. 2024, 15, 110. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.R.; Salinas, A.M.; Qi, D.; Gupta, S.; Sidote, D.J.; Goldschen-Ohm, M.P. Single-Molecule Imaging with Cell-Derived Nanovesicles Reveals Early Binding Dynamics at a Cyclic Nucleotide-Gated Ion Channel. Nat. Commun. 2021, 12, 6459. [Google Scholar] [CrossRef] [PubMed]
- Blythe, E.E.; von Zastrow, M. β-Arrestin-Independent Endosomal CAMP Signaling by a Polypeptide Hormone GPCR. Nat. Chem. Biol. 2023, 20, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Tan, Q.; Han, S.; Diemar, A.; Löbner, K.; Wang, H.; Schüß, C.; Behr, V.; Mörl, K.; Wang, M.; et al. Receptor-Specific Recognition of NPY Peptides Revealed by Structures of NPY Receptors. Sci. Adv. 2022, 8, eabm1232. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; Yu, X.; Wang, W.; Fan, S.; Li, X.; Wang, J. Crystal Structure of a Phosphorylation-Coupled Vitamin C Transporter. Nat. Struct. Mol. Biol. 2015, 22, 238–241. [Google Scholar] [CrossRef]
- Wu, D.; Chen, Q.; Yu, Z.; Huang, B.; Zhao, J.; Wang, Y.; Su, J.; Zhou, F.; Yan, R.; Li, N.; et al. Transport and Inhibition Mechanisms of Human VMAT2. Nature 2024, 626, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Mita, K.; Sumikama, T.; Iwamoto, M.; Matsuki, Y.; Shigemi, K.; Oiki, S. Conductance Selectivity of Na+ across the K+ Channel via Na+ Trapped in a Tortuous Trajectory. Proc. Natl. Acad. Sci. USA 2021, 118, e2017168118. [Google Scholar] [CrossRef]
- Lee, H.-S.; Min, S.; Jung, Y.-E.; Chae, S.; Heo, J.; Lee, J.-H.; Kim, T.; Kang, H.-C.; Nakanish, M.; Cha, S.-S.; et al. Spatiotemporal Coordination of the RSF1-PLK1-Aurora B Cascade Establishes Mitotic Signaling Platforms. Nat. Commun. 2021, 12, 5931. [Google Scholar] [CrossRef]
- Sebolt-Leopold, J.S.; Herrera, R. Targeting the Mitogen-Activated Protein Kinase Cascade to Treat Cancer. Nat. Rev. Cancer 2004, 4, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Stipanovich, A.; Valjent, E.; Matamales, M.; Nishi, A.; Ahn, J.; Maroteaux, M.; Bertran-Gonzalez, J.; Brami-Cherrier, K.; Enslen, H.; Corbillé, A.; et al. A Phosphatase Cascade by Which Rewarding Stimuli Control Nucleosomal Response. Nature 2008, 453, 879–884. [Google Scholar] [CrossRef]
- Valjent, E.; Pascoli, V.; Svenningsson, P.; Paul, S.; Enslen, H.; Corvol, J.-C.; Stipanovich, A.; Caboche, J.; Lombroso, P.J.; Nairn, A.C.; et al. Regulation of a Protein Phosphatase Cascade Allows Convergent Dopamine and Glutamate Signals to Activate ERK in the Striatum. Proc. Natl. Acad. Sci. USA 2005, 102, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Chahwan, R.; Nepal, R.M.; Frieder, D.; Panier, S.; Roa, S.; Zaheen, A.; Durocher, D.; Scharff, M.D.; Martin, A. The RNF8/RNF168 Ubiquitin Ligase Cascade Facilitates Class Switch Recombination. Proc. Natl. Acad. Sci. USA 2010, 107, 809–814. [Google Scholar] [CrossRef] [PubMed]
- Cervia, L.D.; Shibue, T.; Borah, A.A.; Gaeta, B.; He, L.; Leung, L.; Li, N.; Moyer, S.M.; Shim, B.H.; Dumont, N.; et al. A Ubiquitination Cascade Regulating the Integrated Stress Response and Survival in Carcinomas. Cancer Discov. 2023, 13, 766–795. [Google Scholar] [CrossRef] [PubMed]
- Tew, K.D.; Manevich, Y.; Grek, C.; Xiong, Y.; Uys, J.; Townsend, D.M. The Role of Glutathione S-Transferase P in Signaling Pathways and S-Glutathionylation in Cancer. Free Radic. Biol. Med. 2011, 51, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Đukić, N.; Strømland, Ø.; Elsborg, J.D.; Munnur, D.; Zhu, K.; Schuller, M.; Chatrin, C.; Kar, P.; Duma, L.; Suyari, O.; et al. PARP14 Is a PARP with Both ADP-Ribosyl Transferase and Hydrolase Activities. Sci. Adv. 2023, 9, eadi2687. [Google Scholar] [CrossRef]
- Lee, M.J.; Yaffe, M.B. Protein Regulation in Signal Transduction. Cold Spring Harb. Perspect. Biol. 2016, 8, a005918. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, S.; Tomar, A.; Yen, F.S.; Unlu, G.; Ropek, N.; Weber, R.A.; Wang, Y.; Khan, A.; Gad, M.; et al. Autoregulatory Control of Mitochondrial Glutathione Homeostasis. Science 2023, 382, 820–828. [Google Scholar] [CrossRef]
- Fakhiri, J.; Schneider, M.A.; Puschhof, J.; Stanifer, M.; Schildgen, V.; Holderbach, S.; Voss, Y.; El Andari, J.; Schildgen, O.; Boulant, S.; et al. Novel Chimeric Gene Therapy Vectors Based on Adeno-Associated Virus and Four Different Mammalian Bocaviruses. Mol. Ther. Methods Clin. Dev. 2019, 12, 202–222. [Google Scholar] [CrossRef]
- Kube, S.; Hersch, N.; Naumovska, E.; Gensch, T.; Hendriks, J.; Franzen, A.; Landvogt, L.; Siebrasse, J.-P.; Kubitscheck, U.; Hoffmann, B.; et al. Fusogenic Liposomes as Nanocarriers for the Delivery of Intracellular Proteins. Langmuir 2017, 33, 1051–1059. [Google Scholar] [CrossRef]
- Hörner, S.; Fabritz, S.; Herce, H.D.; Avrutina, O.; Dietz, C.; Stark, R.W.; Cardoso, M.C.; Kolmar, H. Cube-Octameric Silsesquioxane-Mediated Cargo Peptide Delivery into Living Cancer Cells. Org. Biomol. Chem. 2013, 11, 2258–2265. [Google Scholar] [CrossRef]
- Cao, A.; Ye, Z.; Cai, Z.; Dong, E.; Yang, X.; Liu, G.; Deng, X.; Wang, Y.; Yang, S.; Wang, H.; et al. A Facile Method To Encapsulate Proteins in Silica Nanoparticles: Encapsulated Green Fluorescent Protein as a Robust Fluorescence Probe. Angew. Chem. Int. Ed. 2010, 49, 3022–3025. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.J.; Park, K.; Kim, K.S.; Kim, J.; Yang, J.-A.; Kong, J.-H.; Lee, M.Y.; Hoffman, A.S.; Hahn, S.K. Target Specific and Long-Acting Delivery of Protein, Peptide, and Nucleotide Therapeutics Using Hyaluronic Acid Derivatives. J. Control. Release 2010, 141, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Neumann, E.; Schaefer-Ridder, M.; Wang, Y.; Hofschneider, P.H. Gene Transfer into Mouse Lyoma Cells by Electroporation in High Electric Fields. EMBO J. 1982, 1, 841–845. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, E.; Potier, E.; Logeart-Avramoglou, D.; Salomskaite-Davalgiene, S.; Mir, L.M.; Petite, H. Optimization of a Gene Electrotransfer Method for Mesenchymal Stem Cell Transfection. Gene Ther. 2008, 15, 537–544. [Google Scholar] [CrossRef] [PubMed]
- REJMAN, J.; OBERLE, V.; ZUHORN, I.S.; HOEKSTRA, D. Size-Dependent Internalization of Particles via the Pathways of Clathrin- and Caveolae-Mediated Endocytosis. Biochem. J. 2004, 377, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Hedlund, H.; Du Rietz, H.; Johansson, J.M.; Eriksson, H.C.; Zedan, W.; Huang, L.; Wallin, J.; Wittrup, A. Single-Cell Quantification and Dose-Response of Cytosolic SiRNA Delivery. Nat. Commun. 2023, 14, 1075. [Google Scholar] [CrossRef] [PubMed]
- Fawell, S.; Seery, J.; Daikh, Y.; Moore, C.; Chen, L.L.; Pepinsky, B.; Barsoum, J. Tat-Mediated Delivery of Heterologous Proteins into Cells. Proc. Natl. Acad. Sci. USA 1994, 91, 664–668. [Google Scholar] [CrossRef] [PubMed]
- Caron, N.J.; Torrente, Y.; Camirand, G.; Bujold, M.; Chapdelaine, P.; Leriche, K.; Bresolin, N.; Tremblay, J.P. Intracellular Delivery of a Tat-EGFP Fusion Protein into Muscle Cells. Mol. Ther. 2001, 3, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Trabulo, S.; Cardoso, A.L.; Mano, M.; De Lima, M.C.P. Cell-Penetrating Peptides—Mechanisms of Cellular Uptake and Generation of Delivery Systems. Pharmaceuticals 2010, 3, 961–993. [Google Scholar] [CrossRef]
- Khan, M.M.; Filipczak, N.; Torchilin, V.P. Cell Penetrating Peptides: A Versatile Vector for Co-Delivery of Drug and Genes in Cancer. J. Control. Release 2021, 330, 1220–1228. [Google Scholar] [CrossRef]
- Mansur, A.A.P.; Carvalho, S.M.; Lobato, Z.I.P.; Leite, M.D.F.; Cunha, A.D.S.; Mansur, H.S. Design and Development of Polysaccharide-Doxorubicin-Peptide Bioconjugates for Dual Synergistic Effects of Integrin-Targeted and Cell-Penetrating Peptides for Cancer Chemotherapy. Bioconjug. Chem. 2018, 29, 1973–2000. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, K. Development of Artificial Cell Models Using Microfluidic Technology and Synthetic Biology. Micromachines 2020, 11, 559. [Google Scholar] [CrossRef]
- Ohnishi, S.; Kamiya, K. Formation of Giant Lipid Vesicle Containing Dual Functions Facilitates Outer Membrane Phospholipase. ACS Synth. Biol. 2021, 10, 1837–1846. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.H.-L.; Wang, C.-H.; Liao, Y.-T.; Chan, F.-Y.; Kanaoka, Y.; Uchihashi, T.; Kato, K.; Lai, L.; Chang, Y.-W.; Ho, M.-C.; et al. Visualizing the Membrane Disruption Action of Antimicrobial Peptides by Cryo-Electron Tomography. Nat. Commun. 2023, 14, 5464. [Google Scholar] [CrossRef]
- Ghosh, A.; Kar, R.K.; Jana, J.; Saha, A.; Jana, B.; Krishnamoorthy, J.; Kumar, D.; Ghosh, S.; Chatterjee, S.; Bhunia, A. Indolicidin Targets Duplex DNA: Structural and Mechanistic Insight through a Combination of Spectroscopy and Microscopy. ChemMedChem 2014, 9, 2052–2058. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, I.; Breukink, E.; van Kraaij, C.; Kuipers, O.P.; Bierbaum, G.; de Kruijff, B.; Sahl, H.-G. Specific Binding of Nisin to the Peptidoglycan Precursor Lipid II Combines Pore Formation and Inhibition of Cell Wall Biosynthesis for Potent Antibiotic Activity. J. Biol. Chem. 2001, 276, 1772–1779. [Google Scholar] [CrossRef]
- Splith, K.; Neundorf, I. Antimicrobial Peptides with Cell-Penetrating Peptide Properties and Vice Versa. Eur. Biophys. J. 2011, 40, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-T.; Sun, T.-L.; Hung, W.-C.; Huang, H.W. Process of Inducing Pores in Membranes by Melittin. Proc. Natl. Acad. Sci. USA 2013, 110, 14243–14248. [Google Scholar] [CrossRef] [PubMed]
- Lorenzón, E.N.; Nobre, T.M.; Caseli, L.; Cilli, E.M.; da Hora, G.C.A.; Soares, T.A.; Oliveira, O.N. The “Pre-Assembled State” of Magainin 2 Lysine-Linked Dimer Determines Its Enhanced Antimicrobial Activity. Colloids Surf. B Biointerfaces 2018, 167, 432–440. [Google Scholar] [CrossRef]
- Bolintineanu, D.; Hazrati, E.; Davis, H.T.; Lehrer, R.I.; Kaznessis, Y.N. Antimicrobial Mechanism of Pore-Forming Protegrin Peptides: 100 Pores to Kill E. coli. Peptides 2010, 31, 1–8. [Google Scholar] [CrossRef]
- Modugno, C.; Loupiac, C.; Bernard, A.; Jossier, A.; Neiers, F.; Perrier-Cornet, J.-M.; Simonin, H. Effect of High Pressure on the Antimicrobial Activity and Secondary Structure of the Bacteriocin Nisin. Innov. Food Sci. Emerg. Technol. 2018, 47, 9–15. [Google Scholar] [CrossRef]
- Selsted, M.E.; Novotny, M.J.; Morris, W.L.; Tang, Y.Q.; Smith, W.; Cullor, J.S. Indolicidin, a Novel Bactericidal Tridecapeptide Amide from Neutrophils. J. Biol. Chem. 1992, 267, 4292–4295. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.M.; Smart, A.; Bloomberg, G.; Burgess, L.; Millar, M.R. Lactoferricin, a New Antimicrobial Peptide. J. Appl. Bacteriol. 1994, 77, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, Y.; Xue, Z.; Jia, Y.; Li, R.; He, C.; Chen, H. The Structure-Mechanism Relationship and Mode of Actions of Antimicrobial Peptides: A Review. Trends Food Sci. Technol. 2021, 109, 103–115. [Google Scholar] [CrossRef]
- Han, E.; Lee, H. Synergistic Effects of Magainin 2 and PGLa on Their Heterodimer Formation, Aggregation, and Insertion into the Bilayer. RSC Adv. 2015, 5, 2047–2055. [Google Scholar] [CrossRef]
- Lee, H.; Lim, S.I.; Shin, S.-H.; Lim, Y.; Koh, J.W.; Yang, S. Conjugation of Cell-Penetrating Peptides to Antimicrobial Peptides Enhances Antibacterial Activity. ACS Omega 2019, 4, 15694–15701. [Google Scholar] [CrossRef]
- Bucki, R.; Leszczyńska, K.; Namiot, A.; Sokołowski, W. Cathelicidin LL-37: A Multitask Antimicrobial Peptide. Arch. Immunol. Ther. Exp. 2010, 58, 15–25. [Google Scholar] [CrossRef]
- Oren, Z.; Lerman, J.C.; Gudmundsson, G.H.; Agerberth, B.; Shai, Y. Structure and Organization of the Human Antimicrobial Peptide LL-37 in Phospholipid Membranes: Relevance to the Molecular Basis for Its Non-Cell-Selective Activity. Biochem. J. 1999, 341, 501. [Google Scholar] [CrossRef]
- Drab, E.; Sugihara, K. Cooperative Function of LL-37 and HNP1 Protects Mammalian Cell Membranes from Lysis. Biophys. J. 2020, 119, 2440–2450. [Google Scholar] [CrossRef]
- Futaki, S.; Suzuki, T.; Ohashi, W.; Yagami, T.; Tanaka, S.; Ueda, K.; Sugiura, Y. Arginine-Rich Peptides. J. Biol. Chem. 2001, 276, 5836–5840. [Google Scholar] [CrossRef]
- Nakase, I.; Niwa, M.; Takeuchi, T.; Sonomura, K.; Kawabata, N.; Koike, Y.; Takehashi, M.; Tanaka, S.; Ueda, K.; Simpson, J.C.; et al. Cellular Uptake of Arginine-Rich Peptides: Roles for Macropinocytosis and Actin Rearrangement. Mol. Ther. 2004, 10, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Vivès, E.; Brodin, P.; Lebleu, B. A Truncated HIV-1 Tat Protein Basic Domain Rapidly Translocates through the Plasma Membrane and Accumulates in the Cell Nucleus. J. Biol. Chem. 1997, 272, 16010–16017. [Google Scholar] [CrossRef] [PubMed]
- Rizzuti, M.; Nizzardo, M.; Zanetta, C.; Ramirez, A.; Corti, S. Therapeutic Applications of the Cell-Penetrating HIV-1 Tat Peptide. Drug Discov. Today 2015, 20, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Derossi, D.; Joliot, A.H.; Chassaing, G.; Prochiantz, A. The Third Helix of the Antennapedia Homeodomain Translocates through Biological Membranes. J. Biol. Chem. 1994, 269, 10444–10450. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.C.; Deshayes, S.; Heitz, F.; Divita, G. Cell-penetrating Peptides: From Molecular Mechanisms to Therapeutics. Biol. Cell 2008, 100, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Oehlke, J.; Scheller, A.; Wiesner, B.; Krause, E.; Beyermann, M.; Klauschenz, E.; Melzig, M.; Bienert, M. Cellular Uptake of an α-Helical Amphipathic Model Peptide with the Potential to Deliver Polar Compounds into the Cell Interior Non-Endocytically. Biochim. Biophys. Acta Biomembr. 1998, 1414, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Pooga, M.; Hällbrink, M.; Zorko, M.; Langel, Ü. Cell Penetration by Transportan. FASEB J. 1998, 12, 67–77. [Google Scholar] [CrossRef]
- Elmquist, A.; Lindgren, M.; Bartfai, T.; Langel, Ü. VE-Cadherin-Derived Cell-Penetrating Peptide, PVEC, with Carrier Functions. Exp. Cell Res. 2001, 269, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Li, W. GALA: A Designed Synthetic PH-Responsive Amphipathic Peptide with Applications in Drug and Gene Delivery. Adv. Drug Deliv. Rev. 2004, 56, 967–985. [Google Scholar] [CrossRef]
- Gao, C.; Mao, S.; Ditzel, H.J.; Farnaes, L.; Wirsching, P.; Lerner, R.A.; Janda, K.D. A Cell-Penetrating Peptide from a Novel PVII–PIX Phage-Displayed Random Peptide Library. Bioorg. Med. Chem. 2002, 10, 4057–4065. [Google Scholar] [CrossRef]
- Järver, P.; Langel, Ü. Cell-Penetrating Peptides—A Brief Introduction. Biochim. Biophys. Acta Biomembr. 2006, 1758, 260–263. [Google Scholar] [CrossRef] [PubMed]
- Teo, S.L.Y.; Rennick, J.J.; Yuen, D.; Al-Wassiti, H.; Johnston, A.P.R.; Pouton, C.W. Unravelling Cytosolic Delivery of Cell Penetrating Peptides with a Quantitative Endosomal Escape Assay. Nat. Commun. 2021, 12, 3721. [Google Scholar] [CrossRef] [PubMed]
- Niikura, K.; Horisawa, K.; Doi, N. A Fusogenic Peptide from a Sea Urchin Fertilization Protein Promotes Intracellular Delivery of Biomacromolecules by Facilitating Endosomal Escape. J. Control. Release 2015, 212, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Buyanova, M.; Sahni, A.; Yang, R.; Sarkar, A.; Salim, H.; Pei, D. Discovery of a Cyclic Cell-Penetrating Peptide with Improved Endosomal Escape and Cytosolic Delivery Efficiency. Mol. Pharm. 2022, 19, 1378–1388. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Tan, X.; Liu, Z.; Dou, L.; Liu, D.; Pan, Y.; Ma, Y.; Yu, J. Engineered Phage with Cell-Penetrating Peptides for Intracellular Bacterial Infections. mSystems 2023, 8, e0064623. [Google Scholar] [CrossRef] [PubMed]
- Milletti, F. Cell-Penetrating Peptides: Classes, Origin, and Current Landscape. Drug Discov. Today 2012, 17, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Morris, M. A New Peptide Vector for Efficient Delivery of Oligonucleotides into Mammalian Cells. Nucleic Acids Res. 1997, 25, 2730–2736. [Google Scholar] [CrossRef]
- Oehlke, J.; Krause, E.; Wiesner, B.; Beyermann, M.; Bienert, M. Extensive Cellular Uptake into Endothelial Cells of an Amphipathic Β-sheet Forming Peptide. FEBS Lett. 1997, 415, 196–199. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Ikonomova, S.P.; Karlsson, A.J. Secondary Structure of Cell-penetrating Peptides during Interaction with Fungal Cells. Protein Sci. 2018, 27, 702–713. [Google Scholar] [CrossRef]
- Kondo, E.; Saito, K.; Tashiro, Y.; Kamide, K.; Uno, S.; Furuya, T.; Mashita, M.; Nakajima, K.; Tsumuraya, T.; Kobayashi, N.; et al. Tumour Lineage-Homing Cell-Penetrating Peptides as Anticancer Molecular Delivery Systems. Nat. Commun. 2012, 3, 951. [Google Scholar] [CrossRef]
- Numata, K.; Horii, Y.; Oikawa, K.; Miyagi, Y.; Demura, T.; Ohtani, M. Library Screening of Cell-Penetrating Peptide for BY-2 Cells, Leaves of Arabidopsis, Tobacco, Tomato, Poplar, and Rice Callus. Sci. Rep. 2018, 8, 10966. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, K.; Islam, M.M.; Horii, Y.; Yoshizumi, T.; Numata, K. Screening of a Cell-Penetrating Peptide Library in Escherichia Coli: Relationship between Cell Penetration Efficiency and Cytotoxicity. ACS Omega 2018, 3, 16489–16499. [Google Scholar] [CrossRef]
- Marks, J.R.; Placone, J.; Hristova, K.; Wimley, W.C. Spontaneous Membrane-Translocating Peptides by Orthogonal High-Throughput Screening. J. Am. Chem. Soc. 2011, 133, 8995–9004. [Google Scholar] [CrossRef] [PubMed]
- Di Pisa, M.; Chassaing, G.; Swiecicki, J.M. Translocation Mechanism(s) of Cell-Penetrating Peptides: Biophysical Studies Using Artificial Membrane Bilayers. Biochemistry 2015, 54, 194–207. [Google Scholar] [CrossRef]
- Madani, F.; Lindberg, S.; Langel, Ü.; Futaki, S.; Gräslund, A. Mechanisms of Cellular Uptake of Cell-Penetrating Peptides. J. Biophys. 2011, 41429, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Heitz, F.; Morris, M.C.; Divita, G. Twenty Years of Cell-penetrating Peptides: From Molecular Mechanisms to Therapeutics. Br. J. Pharmacol. 2009, 157, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Ruseska, I.; Zimmer, A. Internalization Mechanisms of Cell-Penetrating Peptides. Beilstein J. Nanotechnol. 2020, 11, 101–123. [Google Scholar] [CrossRef] [PubMed]
- Ludtke, S.; He, K.; Huang, H. Membrane Thinning Caused by Magainin 2. Biochemistry 1995, 34, 16764–16769. [Google Scholar] [CrossRef]
- Schmid, S.L.; Conner, S.D. Regulated Portals of Entry into the Cell. Nature 2003, 422, 37–44. [Google Scholar]
- Mercer, J.; Helenius, A. Virus Entry by Macropinocytosis. Nat. Cell Biol. 2009, 11, 510–520. [Google Scholar] [CrossRef]
- Bangham, A.D.; Horne, R.W. Negative Staining of Phospholipids and Their Structural Modification by Surface-Active Agents as Observed in the Electron Microscope. J. Mol. Biol. 1964, 8, 660-IN10. [Google Scholar] [CrossRef] [PubMed]
- Tosaka, T.; Kamiya, K. Function Investigations and Applications of Membrane Proteins on Artificial Lipid Membranes. Int. J. Mol. Sci. 2023, 24, 7231. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, K. Formation and Function of OmpG or OmpA-Incorporated Liposomes Using an in Vitro Translation System. Sci. Rep. 2022, 12, 2376. [Google Scholar] [CrossRef]
- Kawano, R.; Tsuji, Y.; Sato, K.; Osaki, T.; Kamiya, K.; Hirano, M.; Ide, T.; Miki, N.; Takeuchi, S. Automated Parallel Recordings of Topologically Identified Single Ion Channels. Sci. Rep. 2013, 3, 1995. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, K.; Osaki, T.; Nakao, K.; Kawano, R.; Fujii, S.; Misawa, N.; Hayakawa, M.; Takeuchi, S. Electrophysiological Measurement of Ion Channels on Plasma/Organelle Membranes Using an on-Chip Lipid Bilayer System. Sci. Rep. 2018, 8, 17498. [Google Scholar] [CrossRef]
- Guha, A.; McGuire, M.L.; Leriche, G.; Yang, J.; Mayer, M. A Single-Liposome Assay That Enables Temperature-Dependent Measurement of Proton Permeability of Extremophile-Inspired Lipid Membranes. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183567. [Google Scholar] [CrossRef] [PubMed]
- Swiecicki, J.; Bartsch, A.; Tailhades, J.; Di Pisa, M.; Heller, B.; Chassaing, G.; Mansuy, C.; Burlina, F.; Lavielle, S. The Efficacies of Cell-Penetrating Peptides in Accumulating in Large Unilamellar Vesicles Depend on Their Ability To Form Inverted Micelles. ChemBioChem 2014, 15, 884–891. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.Z.; Ariyama, H.; Alam, J.M.; Yamazaki, M. Entry of Cell-Penetrating Peptide Transportan 10 into a Single Vesicle by Translocating Across Lipid Membrane and Its Induced Pores. Biochemistry 2014, 53, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, J.; Holden, M.A.; Chen, M. Peptide-Mediated Membrane Transport of Macromolecular Cargo Driven by Membrane Asymmetry. Anal. Chem. 2017, 89, 12369–12374. [Google Scholar] [CrossRef]
- Miwa, A.; Kamiya, K. Control of Enzyme Reaction Initiation inside Giant Unilamellar Vesicles by the Cell-Penetrating Peptide-Mediated Translocation of Cargo Proteins. ACS Synth. Biol. 2022, 11, 3836–3846. [Google Scholar] [CrossRef]
- Magzoub, M.; Kilk, K.; Eriksson, L.E.G.; Langel, Ü.; Gräslund, A. Interaction and Structure Induction of Cell-Penetrating Peptides in the Presence of Phospholipid Vesicles. Biochim. Biophys. Acta Biomembr. 2001, 1512, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Eiríksdóttir, E.; Konate, K.; Langel, Ü.; Divita, G.; Deshayes, S. Secondary Structure of Cell-Penetrating Peptides Controls Membrane Interaction and Insertion. Biochim. Biophys. Acta Biomembr. 2010, 1798, 1119–1128. [Google Scholar] [CrossRef] [PubMed]
- Magzoub, M.; Eriksson, L.E.G.; Gräslund, A. Conformational States of the Cell-Penetrating Peptide Penetratin When Interacting with Phospholipid Vesicles: Effects of Surface Charge and Peptide Concentration. Biochim. Biophys. Acta Biomembr. 2002, 1563, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, B.; Grooten, J.; Reusens, M.; Joliot, A.; Goethals, M.; Vandekerckhove, J.; Prochiantz, A.; Rosseneu, M. Membrane Interaction and Cellular Internalization of Penetratin Peptides. Eur. J. Biochem. 2004, 271, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, F.; Castro, T.G.; Nogueira, E.; Pires, R.; Silva, C.; Ribeiro, A.; Cavaco-Paulo, A. OBP Fused with Cell-Penetrating Peptides Promotes Liposomal Transduction. Colloids Surfaces B Biointerfaces 2018, 161, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Magzoub, M.; Pramanik, A.; Gräslund, A. Modeling the Endosomal Escape of Cell-Penetrating Peptides: Transmembrane PH Gradient Driven Translocation across Phospholipid Bilayers. Biochemistry 2005, 44, 14890–14897. [Google Scholar] [CrossRef] [PubMed]
- Terrone, D.; Sang, S.L.W.; Roudaia, L.; Silvius, J.R. Penetratin and Related Cell-Penetrating Cationic Peptides Can Translocate Across Lipid Bilayers in the Presence of a Transbilayer Potential. Biochemistry 2003, 42, 13787–13799. [Google Scholar] [CrossRef]
- Henriques, S.T.; Costa, J.; Castanho, M.A.R.B. Translocation of β-Galactosidase Mediated by the Cell-Penetrating Peptide Pep-1 into Lipid Vesicles and Human HeLa Cells Is Driven by Membrane Electrostatic Potential. Biochemistry 2005, 44, 10189–10198. [Google Scholar] [CrossRef] [PubMed]
- Björklund, J.; Biverståhl, H.; Gräslund, A.; Mäler, L.; Brzezinski, P. Real-Time Transmembrane Translocation of Penetratin Driven by Light-Generated Proton Pumping. Biophys. J. 2006, 91, L29–L31. [Google Scholar] [CrossRef]
- Pautot, S.; Frisken, B.J.; Weitz, D.A. Engineering Asymmetric Vesicles. Proc. Natl. Acad. Sci. USA 2003, 100, 10718–10721. [Google Scholar] [CrossRef]
- Kamiya, K.; Osaki, T.; Takeuchi, S. Formation of Vesicles-in-a-Vesicle with Asymmetric Lipid Components Using a Pulsed-Jet Flow Method. RSC Adv. 2019, 9, 30071–30075. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Schertzer, J.W.; Chiarot, P.R. Continuous Microfluidic Fabrication of Synthetic Asymmetric Vesicles. Lab Chip 2015, 15, 3591–3599. [Google Scholar] [CrossRef]
- Bárány-Wallje, E.; Keller, S.; Serowy, S.; Geibel, S.; Pohl, P.; Bienert, M.; Dathe, M. A Critical Reassessment of Penetratin Translocation Across Lipid Membranes. Biophys. J. 2005, 89, 2513–2521. [Google Scholar] [CrossRef] [PubMed]
- Walrant, A.; Matheron, L.; Cribier, S.; Chaignepain, S.; Jobin, M.; Sagan, S.; Alves, I.D. Direct Translocation of Cell-Penetrating Peptides in Liposomes: A Combined Mass Spectrometry Quantification and Fluorescence Detection Study. Anal. Biochem. 2013, 438, 1–10. [Google Scholar] [CrossRef]
- Islam, M.Z.; Sharmin, S.; Moniruzzaman, M.; Yamazaki, M. Elementary Processes for the Entry of Cell-Penetrating Peptides into Lipid Bilayer Vesicles and Bacterial Cells. Appl. Microbiol. Biotechnol. 2018, 102, 3879–3892. [Google Scholar] [CrossRef]
- Shuma, M.L.; Moghal, M.M.R.; Yamazaki, M. Detection of the Entry of Nonlabeled Transportan 10 into Single Vesicles. Biochemistry 2020, 59, 1780–1790. [Google Scholar] [CrossRef]
- Islam, M.Z.; Sharmin, S.; Levadnyy, V.; Alam Shibly, S.U.; Yamazaki, M. Effects of Mechanical Properties of Lipid Bilayers on the Entry of Cell-Penetrating Peptides into Single Vesicles. Langmuir 2017, 33, 2433–2443. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Morishita, T.; Aburai, K.; Ito, D.; Imura, T.; Sakai, K.; Abe, M.; Nakase, I.; Futaki, S.; Sakai, H. Direct Entry of Cell-Penetrating Peptide Can Be Controlled by Maneuvering the Membrane Curvature. Sci. Rep. 2021, 11, 31. [Google Scholar] [CrossRef]
- Billah, M.M.; Or Rashid, M.M.; Ahmed, M.; Yamazaki, M. Antimicrobial Peptide Magainin 2-Induced Rupture of Single Giant Unilamellar Vesicles Comprising E. coli Polar Lipids. Biochim. Biophys. Acta Biomembr. 2023, 1865, 184112. [Google Scholar] [CrossRef]
- Ciobanasu, C.; Siebrasse, J.P.; Kubitscheck, U. Cell-Penetrating HIV1 TAT Peptides Can Generate Pores in Model Membranes. Biophys. J. 2010, 99, 153–162. [Google Scholar] [CrossRef]
- Sun, S.; Xia, Y.; Liu, J.; Dou, Y.; Yang, K.; Yuan, B.; Kang, Z. Real-Time Monitoring the Interfacial Dynamic Processes at Model Cell Membranes: Taking Cell Penetrating Peptide TAT as an Example. J. Colloid Interface Sci. 2022, 609, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, K.; Suzuki, H.; Takeuchi, S. Lipid Bilayer Formation by Contacting Monolayers in a Microfluidic Device for Membrane Protein Analysis. Anal. Chem. 2006, 78, 8169–8174. [Google Scholar] [CrossRef] [PubMed]
- Gotanda, M.; Kamiya, K.; Osaki, T.; Fujii, S.; Misawa, N.; Miki, N.; Takeuchi, S. Sequential Generation of Asymmetric Lipid Vesicles Using a Pulsed-Jetting Method in Rotational Wells. Sens. Actuators B Chem. 2018, 261, 392–397. [Google Scholar] [CrossRef]
- Tsuji, Y.; Kawano, R.; Osaki, T.; Kamiya, K.; Miki, N.; Takeuchi, S. Droplet Split-and-Contact Method for High-Throughput Transmembrane Electrical Recording. Anal. Chem. 2013, 85, 10913–10919. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, K.; Arisaka, C.; Suzuki, M. Investigation of Fusion between Nanosized Lipid Vesicles and a Lipid Monolayer Toward Formation of Giant Lipid Vesicles with Various Kinds of Biomolecules. Micromachines 2021, 12, 133. [Google Scholar] [CrossRef] [PubMed]
- Gehan, P.; Kulifaj, S.; Soule, P.; Bodin, J.B.; Amoura, M.; Walrant, A.; Sagan, S.; Thiam, A.R.; Ngo, K.; Vivier, V.; et al. Penetratin Translocation Mechanism through Asymmetric Droplet Interface Bilayers. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183415. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lein, M.; Gunderson, C.; Holden, M. a Direct Quantitation of Peptide-Mediated Protein Transport across a Droplet–Interface Bilayer. J. Am. Chem. Soc. 2011, 133, 15818–15821. [Google Scholar] [CrossRef] [PubMed]
- Lein, M.; DeRonde, B.M.; Sgolastra, F.; Tew, G.N.; Holden, M.A. Protein Transport across Membranes: Comparison between Lysine and Guanidinium-Rich Carriers. Biochim. Biophys. Acta Biomembr. 2015, 1848, 2980–2984. [Google Scholar] [CrossRef]
- Kamiya, K.; Osaki, T.; Takeuchi, S. Formation of Nano-Sized Lipid Vesicles with Asymmetric Lipid Components Using a Pulsed-Jet Flow Method. Sens. Actuators B Chem. 2021, 327, 128917. [Google Scholar] [CrossRef]
- Kamiya, K.; Takeuchi, S. Giant Liposome Formation toward the Synthesis of Well-Defined Artificial Cells. J. Mater. Chem. B 2017, 5, 5911–5923. [Google Scholar] [CrossRef]
- Mishra, A.; Lai, G.H.; Schmidt, N.W.; Sun, V.Z.; Rodriguez, A.R.; Tong, R.; Tang, L.; Cheng, J.; Deming, T.J.; Kamei, D.T.; et al. Translocation of HIV TAT Peptide and Analogues Induced by Multiplexed Membrane and Cytoskeletal Interactions. Proc. Natl. Acad. Sci. USA 2011, 108, 16883–16888. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.; Depollier, J.; Heitz, F.; Divita, G. A Peptide Carrier for the Delivery of Biologically Active Proteins into Mammalian CellsApplication to the Delivery of Antibodies and Therapeutic Proteins. In Cell Biology; Academic Press: Cambridge, MA, USA, 2006; Volume 4, pp. 13–18. ISBN 9780121647308. [Google Scholar]
- Crombez, L.; Aldrian-Herrada, G.; Konate, K.; Nguyen, Q.N.; McMaster, G.K.; Brasseur, R.; Heitz, F.; Divita, G. A New Potent Secondary Amphipathic Cell-Penetrating Peptide for SiRNA Delivery into Mammalian Cells. Mol. Ther. 2009, 17, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Kurzawa, L.; Pellerano, M.; Morris, M.C. PEP and CADY-Mediated Delivery of Fluorescent Peptides and Proteins into Living Cells. Biochim. Biophys. Acta Biomembr. 2010, 1798, 2274–2285. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.F.L.; Wallabregue, A.L.D.; Franz, L.; Hackenberger, C.P.R. Targeted Subcellular Protein Delivery Using Cleavable Cyclic Cell-Penetrating Peptides. Bioconjug. Chem. 2019, 30, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Angeles-Boza, A.M.; Erazo-Oliveras, A.; Lee, Y.-J.; Pellois, J.-P. Generation of Endosomolytic Reagents by Branching of Cell-Penetrating Peptides: Tools for the Delivery of Bioactive Compounds to Live Cells in Cis or Trans. Bioconjug. Chem. 2010, 21, 2164–2167. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.H.; Chong, S.; Nam, S.; Hyun, S.; Choi, S.; Gye, H.; Jang, S.; Jang, J.; Hwang, S.W.; Yu, J.; et al. Multimeric Amphipathic A-Helical Sequences for Rapid and Efficient Intracellular Protein Transport at Nanomolar Concentrations. Adv. Sci. 2018, 5, 1800240. [Google Scholar] [CrossRef] [PubMed]
- Akita, T.; Kimura, R.; Akaguma, S.; Nagai, M.; Nakao, Y.; Tsugane, M.; Suzuki, H.; Oka, J.; Yamashita, C. Usefulness of Cell-Penetrating Peptides and Penetration Accelerating Sequence for Nose-to-Brain Delivery of Glucagon-like Peptide-2. J. Control. Release 2021, 335, 575–583. [Google Scholar] [CrossRef]
- Schneider, A.F.L.; Kithil, M.; Cardoso, M.C.; Lehmann, M.; Hackenberger, C.P.R. Cellular Uptake of Large Biomolecules Enabled by Cell-Surface-Reactive Cell-Penetrating Peptide Additives. Nat. Chem. 2021, 13, 530–539. [Google Scholar] [CrossRef]
- Lee, S.H.; Moroz, E.; Castagner, B.; Leroux, J.-C. Activatable Cell Penetrating Peptide–Peptide Nucleic Acid Conjugate via Reduction of Azobenzene PEG Chains. J. Am. Chem. Soc. 2014, 136, 12868–12871. [Google Scholar] [CrossRef]
- Bode, S.A.; Kruis, I.C.; Adams, H.P.J.H.M.; Boelens, W.C.; Pruijn, G.J.M.; van Hest, J.C.M.; Löwik, D.W.P.M. Coiled-Coil-Mediated Activation of Oligoarginine Cell-Penetrating Peptides. ChemBioChem 2017, 18, 185–188. [Google Scholar] [CrossRef]
- Morris, M.C.; Depollier, J.; Mery, J.; Heitz, F.; Divita, G. A Peptide Carrier for the Delivery of Biologically Active Proteins into Mammalian Cells. Nat. Biotechnol. 2001, 19, 1173–1176. [Google Scholar] [CrossRef]
- Imesch, P.; Scheiner, D.; Szabo, E.; Fink, D.; Fedier, A. Conjugates of Cytochrome c and Antennapedia Peptide Activate Apoptosis and Inhibit Proliferation of HeLa Cancer Cells. Exp. Ther. Med. 2013, 6, 786–790. [Google Scholar] [CrossRef]
- Zamaleeva, A.I.; Collot, M.; Bahembera, E.; Tisseyre, C.; Rostaing, P.; Yakovlev, A.V.; Oheim, M.; de Waard, M.; Mallet, J.-M.; Feltz, A. Cell-Penetrating Nanobiosensors for Pointillistic Intracellular Ca2+ -Transient Detection. Nano Lett. 2014, 14, 2994–3001. [Google Scholar] [CrossRef]
- Qian, Z.; Xu, X.; Amacher, J.F.; Madden, D.R.; Cormet-Boyaka, E.; Pei, D. Intracellular Delivery of Peptidyl Ligands by Reversible Cyclization: Discovery of a PDZ Domain Inhibitor That Rescues CFTR Activity. Angew. Chem. Int. Ed. 2015, 54, 5874–5878. [Google Scholar] [CrossRef]
- Ferrari, A.; Pellegrini, V.; Arcangeli, C.; Fittipaldi, A.; Giacca, M.; Beltram, F. Caveolae-Mediated Internalization of Extracellular HIV-1 Tat Fusion Proteins Visualized in Real Time. Mol. Ther. 2003, 8, 284–294. [Google Scholar] [CrossRef]
- Liu, Y.; Song, Z.; Zheng, N.; Nagasaka, K.; Yin, L.; Cheng, J. Systemic SiRNA Delivery to Tumors by Cell-Penetrating α-Helical Polypeptide-Based Metastable Nanoparticles. Nanoscale 2018, 10, 15339–15349. [Google Scholar] [CrossRef]
- van Asbeck, A.H.; Beyerle, A.; McNeill, H.; Bovee-Geurts, P.H.M.; Lindberg, S.; Verdurmen, W.P.R.; Hällbrink, M.; Langel, Ü.; Heidenreich, O.; Brock, R. Molecular Parameters of SiRNA–Cell Penetrating Peptide Nanocomplexes for Efficient Cellular Delivery. ACS Nano 2013, 7, 3797–3807. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Kawamura, Y.; Hirose, H.; Kiyokawa, M.; Hirate, M.; Hirata, T.; Higuchi, Y.; Futaki, S. E3MPH16: An Efficient Endosomolytic Peptide for Intracellular Protein Delivery. J. Control. Release 2024, 367, 877–891. [Google Scholar] [CrossRef]
- Liu, B.R.; Huang, Y.W.; Winiarz, J.G.; Chiang, H.J.; Lee, H.J. Intracellular Delivery of Quantum Dots Mediated by a Histidine- and Arginine-Rich HR9 Cell-Penetrating Peptide through the Direct Membrane Translocation Mechanism. Biomaterials 2011, 32, 3520–3537. [Google Scholar] [CrossRef]
- Del’Guidice, T.; Lepetit-Stoffaes, J.-P.; Bordeleau, L.-J.; Roberge, J.; Théberge, V.; Lauvaux, C.; Barbeau, X.; Trottier, J.; Dave, V.; Roy, D.-C.; et al. Membrane Permeabilizing Amphiphilic Peptide Delivers Recombinant Transcription Factor and CRISPR-Cas9/Cpf1 Ribonucleoproteins in Hard-to-Modify Cells. PLoS ONE 2018, 13, e0195558. [Google Scholar] [CrossRef]
- de Oliveira, E.C.L.; Santana, K.; Josino, L.; Lima e Lima, A.H.; de Souza de Sales Júnior, C. Predicting Cell-Penetrating Peptides Using Machine Learning Algorithms and Navigating in Their Chemical Space. Sci. Rep. 2021, 11, 7628. [Google Scholar] [CrossRef]
- Akishiba, M.; Takeuchi, T.; Kawaguchi, Y.; Sakamoto, K.; Yu, H.H.; Nakase, I.; Takatani-Nakase, T.; Madani, F.; Gräslund, A.; Futaki, S. Cytosolic Antibody Delivery by Lipid-Sensitive Endosomolytic Peptide. Nat. Chem. 2017, 9, 751–761. [Google Scholar] [CrossRef]
- Allen, J.K.; Brock, D.J.; Kondow-McConaghy, H.M.; Pellois, J.-P. Efficient Delivery of Macromolecules into Human Cells by Improving the Endosomal Escape Activity of Cell-Penetrating Peptides: Lessons Learned from DfTAT and Its Analogs. Biomolecules 2018, 8, 50. [Google Scholar] [CrossRef]
- Yu, S.; Yang, H.; Li, T.; Pan, H.; Ren, S.; Luo, G.; Jiang, J.; Yu, L.; Chen, B.; Zhang, Y.; et al. Efficient Intracellular Delivery of Proteins by a Multifunctional Chimaeric Peptide in Vitro and in Vivo. Nat. Commun. 2021, 12, 5131. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Johnson, G.; Peltier, G.C.; Pellois, J.-P. A HA2-Fusion Tag Limits the Endosomal Release of Its Protein Cargo despite Causing Endosomal Lysis. Biochim. Biophys. Acta Gen. Subj. 2011, 1810, 752–758. [Google Scholar] [CrossRef]
- Erazo-Oliveras, A.; Muthukrishnan, N.; Baker, R.; Wang, T.-Y.; Pellois, J.-P. Improving the Endosomal Escape of Cell-Penetrating Peptides and Their Cargos: Strategies and Challenges. Pharmaceuticals 2012, 5, 1177–1209. [Google Scholar] [CrossRef]
- Nischan, N.; Herce, H.D.; Natale, F.; Bohlke, N.; Budisa, N.; Cardoso, M.C.; Hackenberger, C.P.R. Covalent Attachment of Cyclic TAT Peptides to GFP Results in Protein Delivery into Live Cells with Immediate Bioavailability. Angew. Chem. Int. Ed. 2015, 54, 1950–1953. [Google Scholar] [CrossRef]
- Takayama, K.; Nakase, I.; Michiue, H.; Takeuchi, T.; Tomizawa, K.; Matsui, H.; Futaki, S. Enhanced Intracellular Delivery Using Arginine-Rich Peptides by the Addition of Penetration Accelerating Sequences (Pas). J. Control. Release 2009, 138, 128–133. [Google Scholar] [CrossRef]
- Jiang, T.; Olson, E.S.; Nguyen, Q.T.; Roy, M.; Jennings, P.A.; Tsien, R.Y. Tumor Imaging by Means of Proteolytic Activation of Cell-Penetrating Peptides. Proc. Natl. Acad. Sci. USA 2004, 101, 17867–17872. [Google Scholar] [CrossRef]
- Olson, E.S.; Jiang, T.; Aguilera, T.A.; Nguyen, Q.T.; Ellies, L.G.; Scadeng, M.; Tsien, R.Y. Activatable Cell Penetrating Peptides Linked to Nanoparticles as Dual Probes for in Vivo Fluorescence and MR Imaging of Proteases. Proc. Natl. Acad. Sci. USA 2010, 107, 4311–4316. [Google Scholar] [CrossRef]
- Li, J.; Liu, F.; Shao, Q.; Min, Y.; Costa, M.; Yeow, E.K.L.; Xing, B. Enzyme-Responsive Cell-Penetrating Peptide Conjugated Mesoporous Silica Quantum Dot Nanocarriers for Controlled Release of Nucleus-Targeted Drug Molecules and Real-Time Intracellular Fluorescence Imaging of Tumor Cells. Adv. Healthc. Mater. 2014, 3, 1230–1239. [Google Scholar] [CrossRef]
- Zhao, N.; Bardine, C.; Lourenço, A.L.; Wang, Y.; Huang, Y.; Cleary, S.J.; Wilson, D.M.; Oh, D.Y.; Fong, L.; Looney, M.R.; et al. In Vivo Measurement of Granzyme Proteolysis from Activated Immune Cells with PET. ACS Cent. Sci. 2021, 7, 1638–1649. [Google Scholar] [CrossRef]
- Hingorani, D.V.; Camargo, M.F.; Quraishi, M.A.; Adams, S.R.; Advani, S.J. Tumor Activated Cell Penetrating Peptides to Selectively Deliver Immune Modulatory Drugs. Pharmaceutics 2021, 13, 365. [Google Scholar] [CrossRef]
- van Duijnhoven, S.M.J.; Robillard, M.S.; Nicolay, K.; Grüll, H. Tumor Targeting of MMP-2/9 Activatable Cell-Penetrating Imaging Probes Is Caused by Tumor-Independent Activation. J. Nucl. Med. 2011, 52, 279–286. [Google Scholar] [CrossRef]
- Lee, J.; Oh, E.-T.; Lee, H.-J.; Lee, E.; Kim, H.G.; Park, H.J.; Kim, C. Tuning of Peptide Cytotoxicity with Cell Penetrating Motif Activatable by Matrix Metalloproteinase-2. ACS Omega 2022, 7, 29684–29691. [Google Scholar] [CrossRef]
- Weinstain, R.; Savariar, E.N.; Felsen, C.N.; Tsien, R.Y. In Vivo Targeting of Hydrogen Peroxide by Activatable Cell-Penetrating Peptides. J. Am. Chem. Soc. 2014, 136, 874–877. [Google Scholar] [CrossRef]
- Perez-Lopez, A.M.; Valero, E.; Bradley, M. Synthesis and Optimization of a Reactive Oxygen Species Responsive Cellular Delivery System. New J. Chem. 2017, 41, 2392–2400. [Google Scholar] [CrossRef]
- Hansen, M.B.; Van Gaal, E.; Minten, I.; Storm, G.; Van Hest, J.C.M.; Löwik, D.W.P.M. Constrained and UV-Activatable Cell-Penetrating Peptides for Intracellular Delivery of Liposomes. J. Control. Release 2012, 164, 87–94. [Google Scholar] [CrossRef]
- Lin, Y.; Mazo, M.M.; Skaalure, S.C.; Thomas, M.R.; Simon, R. Activatable Cell-Biomaterial Interfacing with Photo-Caged Peptides. Chem. Sci. 2018, 10, 1158–1167. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, F.; Chen, X. Integrin Targeted Delivery of Chemotherapeutics. Theranostics 2011, 1, 201–210. [Google Scholar] [CrossRef]
- Myrberg, H.; Zhang, L.; Mäe, M.; Langel, Ü. Design of a Tumor-Homing Cell-Penetrating Peptide. Bioconjug. Chem. 2008, 19, 70–75. [Google Scholar] [CrossRef]
- Allred, C.A.; Gormley, C.; Venugopal, I.; Li, S.; McGuire, M.J.; Brown, K.C. Tumor-Specific Intracellular Delivery: Peptide-Guided Transport of a Catalytic Toxin. Commun. Biol. 2023, 6, 60. [Google Scholar] [CrossRef]
- Zahid, M.; Phillips, B.E.; Albers, S.M.; Giannoukakis, N.; Watkins, S.C.; Robbins, P.D. Identification of a Cardiac Specific Protein Transduction Domain by In Vivo Biopanning Using a M13 Phage Peptide Display Library in Mice. PLoS ONE 2010, 5, e12252. [Google Scholar] [CrossRef]
- Higa, M.; Katagiri, C.; Shimizu-Okabe, C.; Tsumuraya, T.; Sunagawa, M.; Nakamura, M.; Ishiuchi, S.; Takayama, C.; Kondo, E.; Matsushita, M. Identification of a Novel Cell-Penetrating Peptide Targeting Human Glioblastoma Cell Lines as a Cancer-Homing Transporter. Biochem. Biophys. Res. Commun. 2015, 457, 206–212. [Google Scholar] [CrossRef]
- Youn, P.; Chen, Y.; Furgeson, D.Y. A Myristoylated Cell-Penetrating Peptide Bearing a Transferrin Receptor-Targeting Sequence for Neuro-Targeted SiRNA Delivery. Mol. Pharm. 2014, 11, 486–495. [Google Scholar] [CrossRef]
- Laroui, N.; Cubedo, N.; Rossel, M.; Bettache, N. Improvement of Cell Penetrating Peptide for Efficient SiRNA Targeting of Tumor Xenografts in Zebrafish Embryos. Adv. Ther. 2020, 3, 1–9. [Google Scholar] [CrossRef]
- Tan, M.; Lan, K.-H.; Yao, J.; Lu, C.-H.; Sun, M.; Neal, C.L.; Lu, J.; Yu, D. Selective Inhibition of ErbB2-Overexpressing Breast Cancer In Vivo by a Novel TAT-Based ErbB2-Targeting Signal Transducers and Activators of Transcription 3–Blocking Peptide. Cancer Res. 2006, 66, 3764–3772. [Google Scholar] [CrossRef]
- Park, H.; Otte, A.; Park, K. Evolution of Drug Delivery Systems: From 1950 to 2020 and Beyond. J. Control. Release 2022, 342, 53–65. [Google Scholar] [CrossRef]
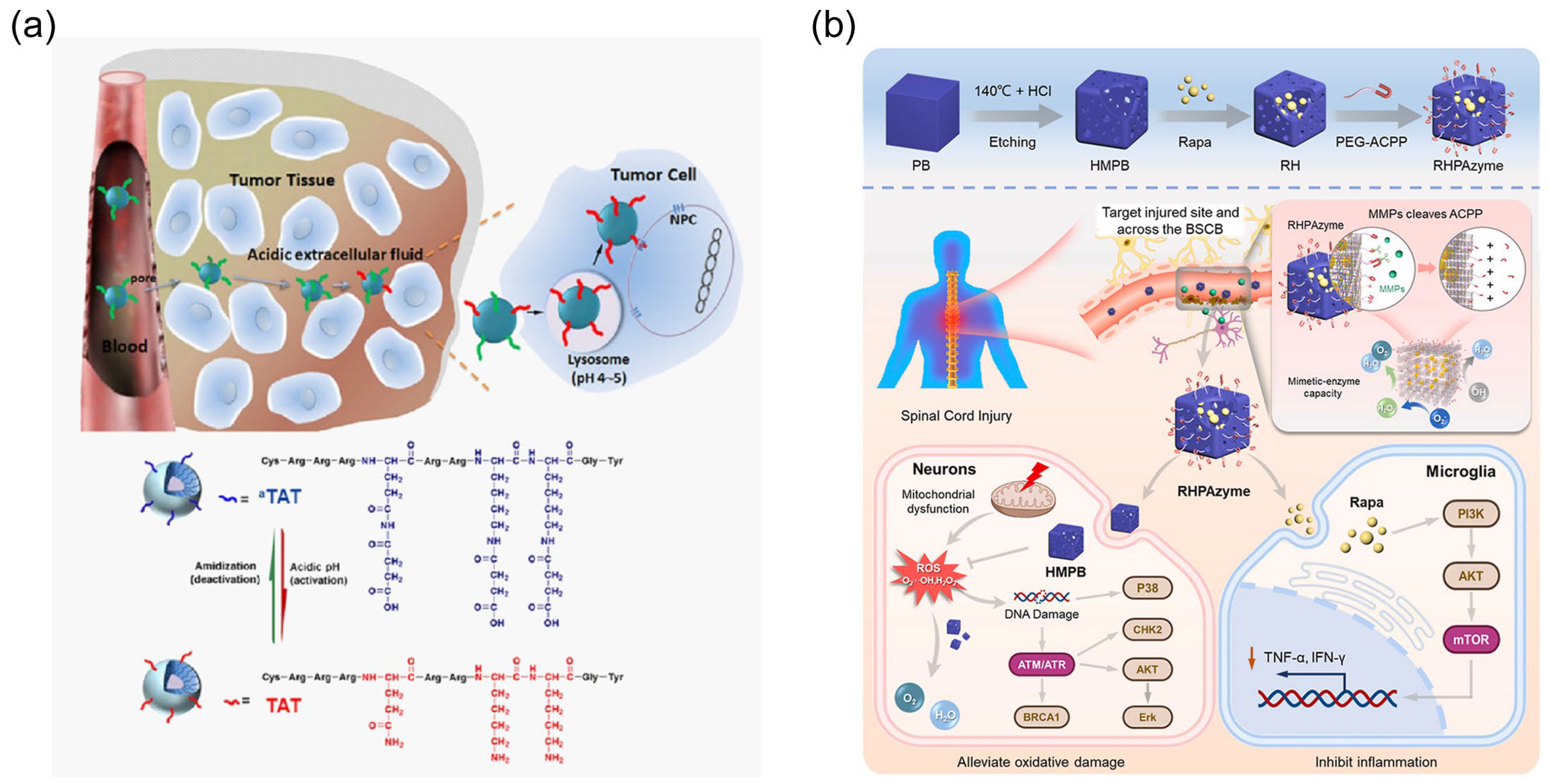
- Jin, E.; Zhang, B.; Sun, X.; Zhou, Z.; Ma, X.; Sun, Q.; Tang, J.; Shen, Y.; Van Kirk, E.; Murdoch, W.J.; et al. Acid-Active Cell-Penetrating Peptides for in Vivo Tumor-Targeted Drug Delivery. J. Am. Chem. Soc. 2013, 135, 933–940. [Google Scholar] [CrossRef]
- Shen, K.; Li, X.; Huang, G.; Yuan, Z.; Xie, B.; Chen, T.; He, L. High Rapamycin-Loaded Hollow Mesoporous Prussian Blue Nanozyme Targets Lesion Area of Spinal Cord Injury to Recover Locomotor Function. Biomaterials 2023, 303, 122358. [Google Scholar] [CrossRef]
- Zhang, Z.; Baxter, A.E.; Ren, D.; Qin, K.; Chen, Z.; Collins, S.M.; Huang, H.; Komar, C.A.; Bailer, P.F.; Parker, J.B.; et al. Efficient Engineering of Human and Mouse Primary Cells Using Peptide-Assisted Genome Editing. Nat. Biotechnol. 2024, 42, 305–315. [Google Scholar] [CrossRef]
- Khafagy, E.S.; Morishita, M. Oral Biodrug Delivery Using Cell-Penetrating Peptide. Adv. Drug Deliv. Rev. 2012, 64, 531–539. [Google Scholar] [CrossRef]
- Zhu, Y.; Jiang, Y.; Meng, F.; Deng, C.; Cheng, R.; Zhang, J.; Feijen, J.; Zhong, Z. Highly Efficacious and Specific Anti-Glioma Chemotherapy by Tandem Nanomicelles Co-Functionalized with Brain Tumor-Targeting and Cell-Penetrating Peptides. J. Control. Release 2018, 278, 1–8. [Google Scholar] [CrossRef]
- Yin, H.; Lu, H.; Xiong, Y.; Ye, L.; Teng, C.; Cao, X.; Li, S.; Sun, S.; Liu, W.; Lv, W.; et al. Tumor-Associated Neutrophil Extracellular Traps Regulating Nanocarrier-Enhanced Inhibition of Malignant Tumor Growth and Distant Metastasis. ACS Appl. Mater. Interfaces 2021, 13, 59683–59694. [Google Scholar] [CrossRef]
- Xiang, B.; Jia, X.-L.; Qi, J.-L.; Yang, L.-P.; Sun, W.-H.; Yan, X.; Yang, S.-K.; Cao, D.-Y.; Du, Q.; Qi, X.-R. Enhancing SiRNA-Based Cancer Therapy Using a New PH-Responsive Activatable Cell-Penetrating Peptide-Modified Liposomal System. Int. J. Nanomed. 2017, 12, 2385–2405. [Google Scholar] [CrossRef]
- Wang, H.; Jiao, Y.; Ma, S.; Li, Z.; Gong, J.; Jiang, Q.; Shang, Y.; Li, H.; Li, J.; Li, N.; et al. Nebulized Inhalation of Peptide-Modified DNA Origami To Alleviate Acute Lung Injury. Nano Lett. 2024, 24, 6102–6111. [Google Scholar] [CrossRef]
- Kosuge, M.; Takeuchi, T.; Nakase, I.; Jones, A.T.; Futaki, S. Cellular Internalization and Distribution of Arginine-Rich Peptides as a Function of Extracellular Peptide Concentration, Serum, and Plasma Membrane Associated Proteoglycans. Bioconjug. Chem. 2008, 19, 656–664. [Google Scholar] [CrossRef]

| Primary | Name | Origin | Secondary | Activity (Concentration) | Target Membrane or Cell | pH, Temprature | Ref. | |
|---|---|---|---|---|---|---|---|---|
| AMP | cationic | melitin | Apis mellifera | amphipathic α-helix | pore formation (≥8 nM) | DOPC or DOPG liposome | pH 7 | [38] |
| magainin | Xenopus laevis | α-helix | toroidal pore (≥10 nM) | monolayer of E. coli lipid extract and LUV | pH 7.4 | [39] | ||
| protegrin | porcine neutrophils | anti parallel β-sheet | octomer pore (25 mg/mL) | E. coli ML-35p cells | pH 7.4 | [40] | ||
| nisin | Lactococcus lactis | Loop | pore formation/inhibition of cell wall synthesis (−) | bacterial membrane | pH 2.8, 6.8 (pressure treatment) | [41] | ||
| ndolicidin | Bovine neutrophils | α-helix | membrane dissolution/inhibition of DNA synthesis (10 µg/mL) | E. coli ML-35, S. sureus | pH 7.4 | [42] | ||
| Lactferricin | human lactoferrin | βturn/loop | direct transrocation/pore formation (≥7.5 mg/L) | E. coli, S. aureus 8532 and 8530 and so on. | pH 5–8 | [43] | ||
| LL-37 | Human | basic/amphiphathic α-helix | pore formation/carpet model (7.5 µM) | PC/chol or PC/PS SUV and E. coli D21 | pH 7.4, pH 8.1 | [47,48] | ||
| CPP | cationic | R8 | Chemic | random coil | direct transrocation/endocytosis (10 µM) | HeLa cell | pH 7 (α-MEM), 37 °C or 4 °C | [50,51] |
| TAT | HIV-1-TAT protein | random coil/PpII helix | direct transrocation (500 nM)/pore formation (100 µM) | HeLa cell | pH 7 (Opt-MEM), 37 °C or 4 °C | [52,53] | ||
| penetratin | Antennapedia homeodomain | amphipathic α-helix, β-sheet (under PG lipid) | direct translocation/endocytosis (25 µM) | E15 striatal cell | pH 7.4 (DMEM/F12), 37 °C or 4 °C | [54] | ||
| amphipathic | Pep-1 | Chimera (Trp-rich motif-SV40 NLS) | α-helix | direct translocation/water pore (0.1 µM) | HS68 fibroblasts | pH 7 (DMEM), 37 °C | [55] | |
| MAP | Chimeric | α-helix | Multiroute (1.8–5 µM) | Calf aortic endothelial cells (AEC) | pH 7 (DMEM), 37 °C or 0 °C | [56] | ||
| transportan | Galanin-mastoparan | α-helix | endocytosis/direct translocation (5–500 nM) | Bowes’ melanoma cells | pH 7 (MEM), 37 °C or 0 °C | [57] | ||
| pVEC | murine VE-cadherin | β-sheet | direct translocation/transporter mediated (10 µM) | AEC, HBCEC, bEND, Bowes melanoma cells | pH 7(DMEM or MEM), 37 °C or 4 °C | [58] | ||
| anionic | GALA | Chemic (EALA repeat) | α-helix | pore formation/ membrane distavilization (2 µM) | PC LUV, POPC SUV | pH 4.5–8 | [59] | |
| hydrophobic | Pep-7 | Random Library | α-helix/homodimer | endocytosis (1 µM) | B-lymphocyte WI–L2 cells | pH 7 (RPMI 1640 medium), 37 °C | [60] |
| CPP | Cargo | Combining Strategy | Treatment Concentration (of CPP or CPP Conjugated Cargo) | Cell | Efficiency | Ref. |
|---|---|---|---|---|---|---|
| Pep-1 | β-gal, GFP, FITC-labeled peptide | noncovalent complex | 0.5 µM< [Pep-1] < 50 µM | HS-68, Cos-7 cell | >80%(protein), >90% (peptide) | [122] |
| CADY | short peptide, siRNA | noncovalent complex | 40 or 60 µM(peptide), 1.6 µM(siRNA) | HeLa cell | unable to deliver(peptide), 97%(knockdown) | [123,124] |
| Cyclic R10 | mcherry | disulfide bond, maleimide bond | 30 µM, 50 µM | HeLa cell | ー | [125] |
| Branch TAT | fluorescein(FI) | carbonyl bond | 1 µM, 3 µM | HeLa cell | 40 %(1 µM), 80%(3 µM) | [126] |
| tetrameric LK-1 | eGFP, PPAR | peptide bond | 50 nM, 100 nM | HeLa, HEK293T cells | 50%, almost 100% | [127] |
| PAS-CPP | Glucagon-like Peptide-2 | peptide bond | 6.75 µM | A549 cells | >90% | [128] |
| R8 | TAMRA, GBP1, mcherry | disulfide bond | 1 µM, 10 µM, 30 µM | HeLa Kyoto cells | 5%, 90%(under free linear CPP) | [129] |
| activatable M918 | PNA | maleimide bond | 8 µM | HT-29-luc cell | 60% (luciferase expression) | [130] |
| R4 + R4 | sfGFP | peptide bond (Zipper peptide) | 10 µM | HeLa cell | ー | [131] |
| CPP | Composition | Cargo | Conbining Strategy | Target, Effects | Ref. |
|---|---|---|---|---|---|
| TAT | TAT-PGFK-E5 | QD nanoparticles | makeimide linker | cancer (doxorubicin) | [152] |
| aTAT | amine masked TAT | PEG-PCL micelles | makeimide linker | tumor | [170] |
| R9 | E8-PLGLAG-R9-Cys | PB nanoparticles | protease cleaving linker | spinal cord injury | [171] |
| TAT | TAT-4 × NLS-Cas9-2 × NLS, TAT-HA2 | Cas9 protein | expression, mixing | genome editing | [172] |
| TAT | TAT (YGRKKRRQRRRC) | tandem nanomicelles | PEG linler | anti-glioma chemotherapy | [173] |
| R8, TAT, Penetratin | RRRRRRRR, GRKKRRQRRRPPQ, RQIKIWFQNRRMKWKK | insuline | noncovalent | Brain Delivery | [174] |
| TAT | GRKKRRQRRRPQPLGLAGGC | paclitaxel (PTX) prodrug nanoparticle | protease cleaving linker | Inhibition of tumor growth | [175] |
| R8 | RRRRRRRR-hydrazone linker-ehGehGehGehG | liposome containing siRNA | hydrazone bond | gene silencing | [176] |
| R9 | RRRRRRRR | DNA origami nanostructure | azide-alkyne cycroaddition | ros scavenger | [177] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miwa, A.; Kamiya, K. Cell-Penetrating Peptide-Mediated Biomolecule Transportation in Artificial Lipid Vesicles and Living Cells. Molecules 2024, 29, 3339. https://doi.org/10.3390/molecules29143339
Miwa A, Kamiya K. Cell-Penetrating Peptide-Mediated Biomolecule Transportation in Artificial Lipid Vesicles and Living Cells. Molecules. 2024; 29(14):3339. https://doi.org/10.3390/molecules29143339
Chicago/Turabian StyleMiwa, Akari, and Koki Kamiya. 2024. "Cell-Penetrating Peptide-Mediated Biomolecule Transportation in Artificial Lipid Vesicles and Living Cells" Molecules 29, no. 14: 3339. https://doi.org/10.3390/molecules29143339
APA StyleMiwa, A., & Kamiya, K. (2024). Cell-Penetrating Peptide-Mediated Biomolecule Transportation in Artificial Lipid Vesicles and Living Cells. Molecules, 29(14), 3339. https://doi.org/10.3390/molecules29143339







