Advancing Microfluidic Immunity Testing Systems: New Trends for Microbial Pathogen Detection
Abstract
1. Introduction
2. Fabrication Methods for Microfluidic Chip in Microbial Pathogen Detection
3. The Application of Microfluidic Chip in Microbial Pathogen Detection
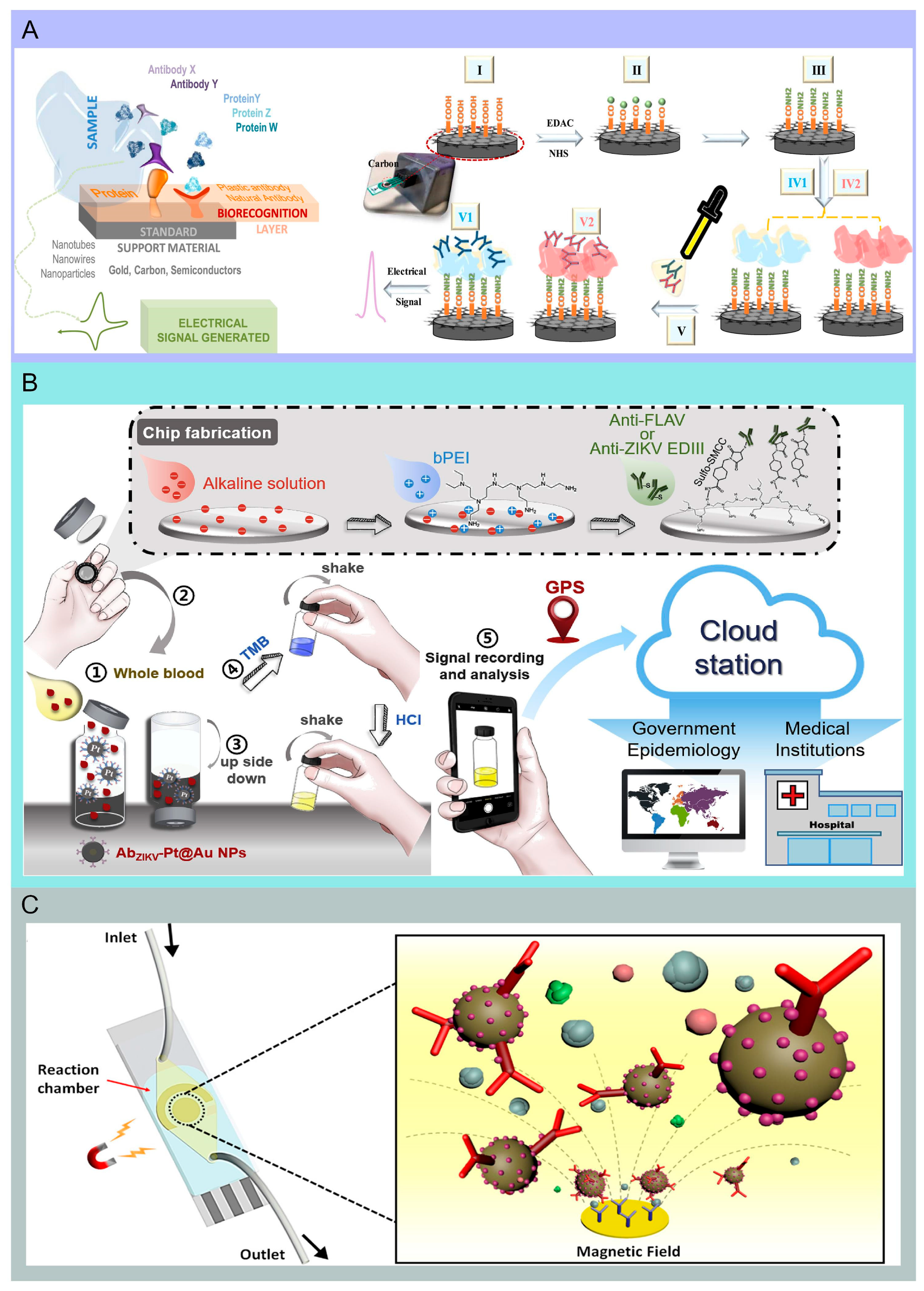
4. Immunosensor-Based Microfluidic Chip
5. Single Molecule Arrays
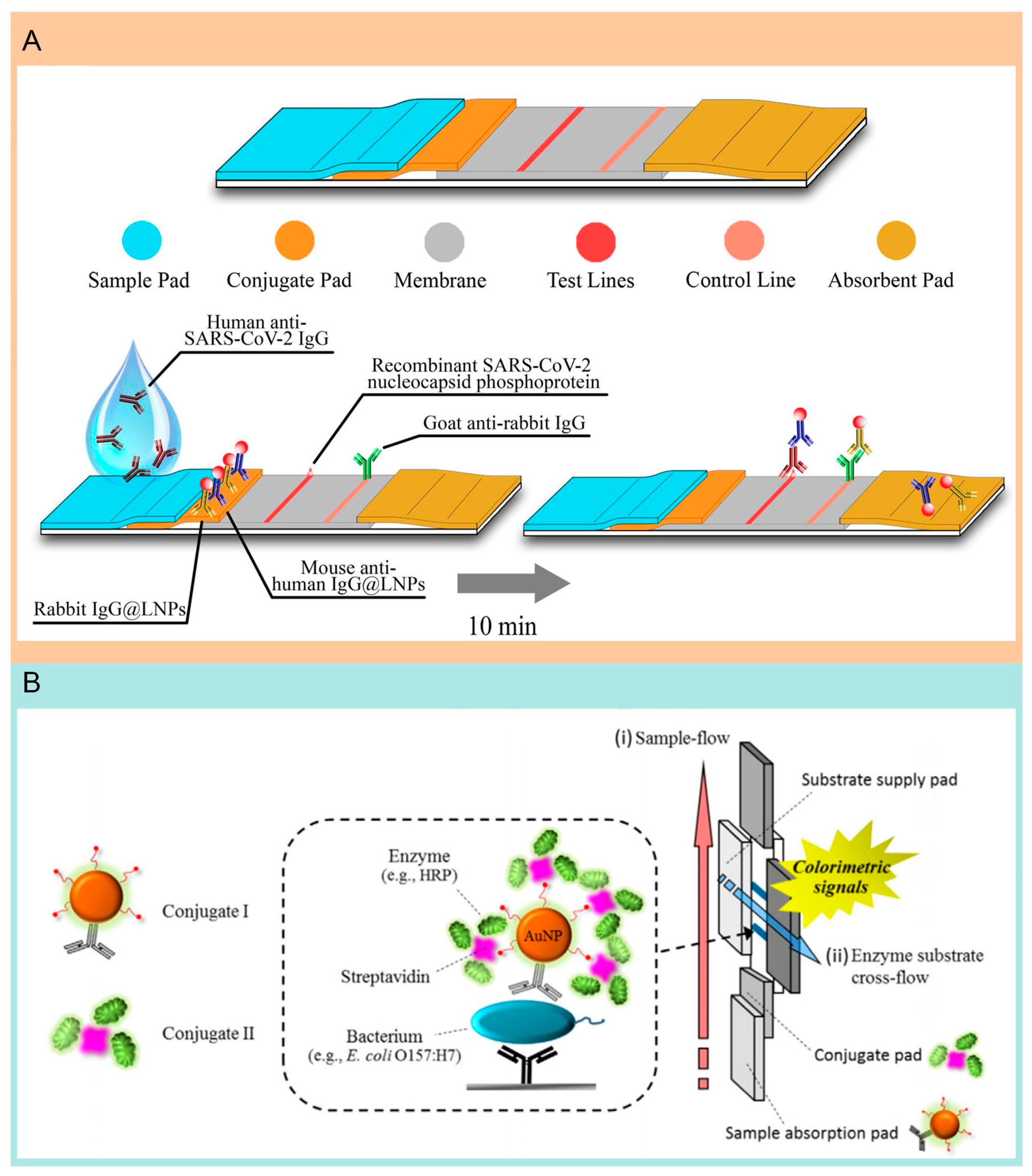
6. Lateral Flow Assay
7. Smartphones Integrated with Microfluidic System
8. Challenges in the Design and Optimization of Immunoassays in Microfluidic System
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Songca, S.P. Applications of Nanozymology in the Detection and Identification of Viral, Bacterial and Fungal Pathogens. Int. J. Mol. Sci. 2022, 23, 4638. [Google Scholar] [CrossRef] [PubMed]
- Hanif, K.; Riaz, M.; Zahoor, I. Pathogens detection and identification in drinking water. IJCBS 2023, 24, 236–250. [Google Scholar]
- Wang, J.; Davidson, J.L.; Kaur, S.; Dextre, A.A.; Ranjbaran, M.; Kamel, M.S.; Athalye, S.M.; Verma, M.S. Paper-Based Biosensors for the Detection of Nucleic Acids from Pathogens. Biosensors 2022, 12, 1094. [Google Scholar] [CrossRef]
- Zhang, S.; Duan, M.; Li, S.; Hou, J.; Qin, T.; Teng, Z.; Hu, J.; Zhang, H.; Xia, X. Current status of recombinase polymerase amplification technologies for the detection of pathogenic microorganisms. Diagn. Microbiol. Infect. Dis. 2024, 108, 116097. [Google Scholar] [CrossRef]
- Bedford, T.; Riley, S.; Barr, I.G.; Broor, S.; Chadha, M.; Cox, N.J.; Daniels, R.S.; Gunasekaran, C.P.; Hurt, A.C.; Kelso, A.; et al. Global circulation patterns of seasonal influenza viruses vary with antigenic drift. Nature 2015, 523, 217–220. [Google Scholar] [CrossRef] [PubMed]
- Vemula, S.V.; Zhao, J.; Liu, J.; Xue, X.W.; Biswas, S.; Hewlett, I. Current approaches for diagnosis of influenza virus infections in humans. Viruses 2016, 8, 96. [Google Scholar] [CrossRef] [PubMed]
- Gong, F.; Wei, H.X.; Li, Q.; Liu, L.; Li, B. Evaluation and Comparison of Serological Methods for COVID-19 Diagnosis. Front. Mol. Biosci. 2021, 8, 682405. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.; Huang, D.; Wang, Y.; Cai, D.; Luo, Y.; Zhong, Z.; Liu, D. Active droplet-array microfluidics-based chemiluminescence immunoassay for point-of-care detection of procalcitonin. Biosens Bioelectron. 2022, 195, 113684. [Google Scholar] [CrossRef] [PubMed]
- Poltronieri, P.; Mezzolla, V.; Primiceri, E.; Maruccio, G. Biosensors for the detection of food pathogens. Foods 2014, 3, 511–526. [Google Scholar] [CrossRef]
- Foudeh, A.M.; Fatanat Didar, T.; Veres, T.; Tabrizian, M. Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab Chip 2012, 12, 3249–3266. [Google Scholar] [CrossRef]
- Rodriguez-Moncayo, R.; Cedillo-Alcantar, D.F.; Guevara-Pantoja, P.E.; Chavez-Pineda, O.G.; Hernandez-Ortiz, J.A.; Amador-Hernandez, J.U.; Rojas-Velasco, G.; Sanchez-Muñoz, F.; Manzur-Sandoval, D.; Patino-Lopez, L.D.; et al. A high-throughput multiplexed microfluidic device for COVID-19 serology assays. Lab Chip 2021, 21, 93–104. [Google Scholar] [CrossRef]
- Swank, Z.; Michielin, G.; Yip, H.M.; Cohen, P.; Andrey, D.O.; Vuilleumier, N.; Kaiser, L.; Eckerle, I.; Meyer, B.; Maerkl, S.J. A high-throughput microfluidic nanoimmunoassay for detecting anti–SARS-CoV-2 antibodies in serum or ultralow-volume blood samples. Proc. Natl. Acad. Sci. USA 2021, 118, e2025289118. [Google Scholar] [CrossRef]
- Heggestad, J.T.; Kinnamon, D.S.; Olson, L.B.; Liu, J.; Kelly, G.; Wall, S.A.; Oshabaheebwa, S.; Quinn, Z.; Fontes, C.M.; Joh, D.Y.; et al. Multiplexed, quantitative serological profiling of COVID-19 from blood by a point-of-care test. Sci. Adv. 2021, 7, eabg4901. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wu, S.; Wang, G.; Liu, W.; Chu, L.T.; Jiang, T.; Kwong, H.K.; Chow, H.L.; Li, I.W.S.; Chen, T.H. Microfluidic particle dam for direct visualization of SARS-CoV-2 antibody levels in COVID-19 vaccinees. Sci. Adv. 2022, 8, eabn6064. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Hu, C.; Jahan, S.; Yuan, B.; Saleh, M.S.; Ju, E.; Gao, S.J.; Panat, R. Sensing of COVID-19 Antibodies in Seconds via Aerosol Jet Nanoprinted Reduced-Graphene-Oxide-Coated 3D Electrodes. Adv. Mater. 2021, 33, 2006647. [Google Scholar] [CrossRef] [PubMed]
- Samper, I.C.; Sánchez-Cano, A.; Khamcharoen, W.; Jang, I.; Siangproh, W.; Baldrich, E.; Geiss, B.J.; Dandy, D.S.; Henry, C.S. Electrochemical Capillary-Flow Immunoassay for Detecting Anti-SARS-CoV-2 Nucleocapsid Protein Antibodies at the Point of Care. ACS Sens. 2021, 6, 4067–4075. [Google Scholar] [CrossRef]
- Siavash Moakhar, R.; del Real Mata, C.; Jalali, M.; Shafique, H.; Sanati, A.; de Vries, J.; Strauss, J.; AbdElFatah, T.; Ghasemi, F.; McLean, M.; et al. A Versatile Biomimic Nanotemplating Fluidic Assay for Multiplex Quantitative Monitoring of Viral Respiratory Infections and Immune Responses in Saliva and Blood. Adv. Sci. 2022, 9, 2204246. [Google Scholar] [CrossRef]
- Feng, M.; Chen, J.; Xun, J.; Dai, R.; Zhao, W.; Lu, H.; Xu, J.; Chen, L.; Sui, G.; Cheng, X.; et al. Development of a Sensitive Immunochromatographic Method Using Lanthanide Fluorescent Microsphere for Rapid Serodiagnosis of COVID-19. ACS Sens. 2020, 5, 2331–2337. [Google Scholar] [CrossRef]
- Chen, S.; Meng, L.; Wang, L.; Huang, X.; Ali, S.; Chen, X.; Yu, M.; Yi, M.; Li, L.; Chen, X.; et al. SERS-based lateral flow immunoassay for sensitive and simultaneous detection of anti-SARS-CoV-2 IgM and IgG antibodies by using gap-enhanced Raman nanotags. Sens. Actuators B Chem. 2021, 348, 130706. [Google Scholar] [CrossRef]
- Grzelak, L.; Temmam, S.; Planchais, C.; Demeret, C.; Tondeur, L.; Huon, C.; Guivel-Benhassine, F.; Staropoli, I.; Chazal, M.; Dufloo, J.; et al. A comparison of four serological assays for detecting anti–SARS-CoV-2 antibodies in human serum samples from different populations. Sci. Transl. Med. 2020, 12, eabc3103. [Google Scholar] [CrossRef]
- Xu, J.; Suo, W.; Goulev, Y.; Sun, L.; Kerr, L.; Paulsson, J.; Zhang, Y.; Lao, T. Handheld Microfluidic Filtration Platform Enables Rapid, Low-Cost, and Robust Self-Testing of SARS-CoV-2 Virus. Small 2021, 17, 2104009. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lillehoj, P.B. Microfluidic Magneto Immunosensor for Rapid, High Sensitivity Measurements of SARS-CoV-2 Nucleocapsid Protein in Serum. ACS Sens. 2021, 6, 1270–1278. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, W.; Pallás-Tamarit, Y.; Juste-Dolz, A.; Sena-Torralba, A.; Gozalbo-Rovira, R.; Rodríguez-Díaz, J.; Navarro, D.; Carrascosa, J.; Gimenez-Romero, D.; Maquieira, Á.; et al. An all-in-one point-of-care testing device for multiplexed detection of respiratory infections. Biosens. Bioelectron. 2022, 213, 114454. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Krel, M.; Dolgov, E.; Park, S.; Li, X.; Wu, W.; Sun, Y.L.; Zhang, J.; Khaing Oo, M.K.; Perlin, D.S.; et al. Rapid and quantitative detection of SARS-CoV-2 specific IgG for convalescent serum evaluation. Biosens. Bioelectron. 2020, 169, 112572. [Google Scholar] [CrossRef]
- Shokr, A.; Pacheco, L.G.C.; Thirumalaraju, P.; Kanakasabapathy, M.K.; Gandhi, J.; Kartik, D.; Silva, F.S.R.; Erdogmus, E.; Kandula, H.; Luo, S.; et al. Mobile Health (mHealth) Viral Diagnostics Enabled with Adaptive Adversarial Learning. ACS Nano 2021, 15, 665–673. [Google Scholar] [CrossRef]
- Wang, C.; Cheng, X.; Liu, L.; Zhang, X.; Yang, X.; Zheng, S.; Rong, Z.; Wang, S. Ultrasensitive and Simultaneous Detection of Two Specific SARS-CoV-2 Antigens in Human Specimens Using Direct/Enrichment Dual-Mode Fluorescence Lateral Flow Immunoassay. ACS Appl. Mater. Interfaces 2021, 13, 40342–40353. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Jiao, X.; Liang, Z.; Zhao, H.; Zhao, Y.; Xie, J.; Jiang, Y.; Yu, X.; Fang, X.; Dai, X. Lateral flow immunoassay coupled with copper enhancement for rapid and sensitive SARS-CoV-2 nucleocapsid protein detection. Biosensors 2022, 12, 13. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Hong, S.; Jang, J. Label-free Detection of Influenza Viruses using a Reduced Graphene Oxide-based Electrochemical Immunosensor Integrated with a Microfluidic Platform. Sci. Rep. 2017, 7, 42771. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.H.; Ma, Y.D.; Fu, C.Y.; Lee, G. Bin A structure-free digital microfluidic platform for detection of influenza a virus by using magnetic beads and electromagnetic forces. Lab Chip 2020, 20, 789–797. [Google Scholar] [CrossRef]
- Han, J.H.; Lee, D.; Chew, C.H.C.; Kim, T.; Pak, J.J. A multi-virus detectable microfluidic electrochemical immunosensor for simultaneous detection of H1N1, H5N1, and H7N9 virus using ZnO nanorods for sensitivity enhancement. Sens. Actuators B Chem. 2016, 228, 36–42. [Google Scholar] [CrossRef]
- Wang, S.; Ai, Z.; Zhang, Z.; Tang, M.; Zhang, N.; Liu, F.; Han, G.; Hong, S.L.; Liu, K. Simultaneous and automated detection of influenza A virus hemagglutinin H7 and H9 based on magnetism and size mediated microfluidic chip. Sens. Actuators B Chem. 2020, 308, 127675. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.L.; Wang, X.; Bao, Z.H.; Zhang, M.F.; Tang, M.; Zhang, N.; Liu, H.; Zhu, Z.Y.; Liu, K.; Chen, Z.L.; et al. Simultaneous detection of multiple influenza virus subtypes based on microbead-encoded microfluidic chip. Anal. Chim. Acta 2023, 1279, 341773. [Google Scholar] [CrossRef] [PubMed]
- Bosch, I.; De Puig, H.; Hiley, M.; Carré-Camps, M.; Perdomo-Celis, F.; Narváez, C.F.; Salgado, D.M.; Senthoor, D.; Grady, M.O.; Phillips, E.; et al. Rapid antigen tests for dengue virus serotypes and zika virus in patient serum. Sci. Transl. Med. 2017, 9, eaan1589. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.P.; Li, N.S.; Chen, Y.T.; Pang, H.H.; Wei, K.C.; Yang, H.W. A serological point-of-care test for Zika virus detection and infection surveillance using an enzyme-free vial immunosensor with a smartphone. Biosens. Bioelectron. 2020, 151, 111960. [Google Scholar] [CrossRef] [PubMed]
- Rong, Z.; Wang, Q.; Sun, N.; Jia, X.; Wang, K.; Xiao, R.; Wang, S. Smartphone-based fluorescent lateral flow immunoassay platform for highly sensitive point-of-care detection of Zika virus nonstructural protein 1. Anal. Chim. Acta 2019, 1055, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Bautista, J.F.; Tarriño, M.; González, A.; Olivares Durán, M.J.; Cobo, F.; Reguera, J.A.; Rodríguez-Granger, J.; Sampedro, A. Comparison of an Enzyme Linked-Immunosorbent Assay and a Chemiluminescent Immunoassay with an Immunofluorescence Assay for Detection of Phase II IgM and IgG Antibodies to Coxiella burnetii. Microorganisms 2024, 12, 552. [Google Scholar] [CrossRef] [PubMed]
- Ghosh Dastider, S.; Barizuddin, S.; Yuksek, N.S.; Dweik, M.; Almasri, M.F. Efficient and rapid detection of salmonella using microfluidic impedance based sensing. J. Sens. 2015, 2015, 293461. [Google Scholar] [CrossRef]
- Park, C.; Lee, J.; Kim, Y.; Kim, J.; Lee, J.; Park, S. 3D-printed microfluidic magnetic preconcentrator for the detection of bacterial pathogen using an ATP luminometer and antibody-conjugated magnetic nanoparticles. J. Microbiol. Methods 2017, 132, 128–133. [Google Scholar] [CrossRef]
- Yang, Y.; Kim, S.; Chae, J. Separating and detecting Escherichia coli in a microfluidic channel for urinary tract infection applications. J. Microelectromech. Syst. 2011, 20, 819–827. [Google Scholar] [CrossRef]
- Park, S.; Min, J.; Kim, Y.K. Chemiluminescent enzyme-linked immunosorbent assay on a strip to detect Escherichia coli O157:H7. Int. J. Environ. Anal. Chem. 2012, 92, 655–664. [Google Scholar] [CrossRef]
- Hossain, S.M.Z.; Ozimok, C.; Sicard, C.; Aguirre, S.D.; Ali, M.M.; Li, Y.; Brennan, J.D. Multiplexed paper test strip for quantitative bacterial detection. Anal. Bioanal. Chem. 2012, 403, 1567–1576. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Q.; Inci, F.; Chaunzwa, T.L.; Ramanujam, A.; Vasudevan, A.; Subramanian, S.; Fai Ip, A.C.; Sridharan, B.; Gurkan, U.A.; Demirci, U. Portable microfluidic chip for detection of Escherichia coli in produce and blood. Int. J. Nanomed. 2012, 7, 2591–2600. [Google Scholar] [CrossRef][Green Version]
- Cho, I.H.; Bhunia, A.; Irudayaraj, J. Rapid pathogen detection by lateral-flow immunochromatographic assay with gold nanoparticle-assisted enzyme signal amplification. Int. J. Food Microbiol. 2015, 206, 60–66. [Google Scholar] [CrossRef]
- Costa, S.P.; Caneira, C.R.F.; Chu, V.; Freitas, P.P.; Conde, J.P.; Carvalho, C.M. A microfluidic platform combined with bacteriophage receptor binding proteins for multiplex detection of Escherichia coli and Pseudomonas aeruginosa in blood. Sens. Actuators B Chem. 2023, 376, 132917. [Google Scholar] [CrossRef]
- Wu, J.; Gu, M. Microfluidic sensing: State of the art fabrication and detection techniques. J. Biomed. Opt. 2011, 16, 080901. [Google Scholar] [CrossRef] [PubMed]
- Sathish, S.; Shen, A.Q. Toward the Development of Rapid, Specific, and Sensitive Microfluidic Sensors: A Comprehensive Device Blueprint. JACS Au 2021, 1, 1815–1833. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Park, S.H.; Chung, K.H.; Pyo, H.B. A disposable plastic-silicon micro PCR chip using flexible printed circuit board protocols and its application to genomic DNA amplification. IEEE Sens. J. 2008, 8, 558–564. [Google Scholar] [CrossRef]
- Annabestani, M.; Shaegh, A.M.; Esmaeili-Dokht, P.; Fardmanesh, M. An Intelligent Machine Learning-Based Sheath-free Microfluidic Impedance Flow cytometer. In Proceedings of the 2020 10th International Conference on Computer and Knowledge Engineering (ICCKE), Mashhad, Iran, 29–30 October 2020; pp. 284–288. [Google Scholar] [CrossRef]
- Felix, F.S.; Baccaro, A.L.B.; Angnes, L. Disposable Voltammetric Immunosensors Integrated with Microfluidic Platforms for Biomedical, Agricultural and Food Analyses: A Review. Sensors 2018, 18, 4124. [Google Scholar] [CrossRef]
- Ali, S.; Hassan, A.; Hassan, G.; Eun, C.H.; Bae, J.; Lee, C.H.; Kim, I.J. Disposable all-printed electronic biosensor for instantaneous detection and classification of pathogens. Sci. Rep. 2018, 8, 5920. [Google Scholar] [CrossRef]
- Ballacchino, G.; Weaver, E.; Mathew, E.; Dorati, R.; Genta, I.; Conti, B.; Lamprou, D.A. Manufacturing of 3d-printed microfluidic devices for the synthesis of drug-loaded liposomal formulations. Int. J. Mol. Sci. 2021, 22, 8064. [Google Scholar] [CrossRef]
- Yafia, M.; Ymbern, O.; Olanrewaju, A.O.; Parandakh, A.; Sohrabi Kashani, A.; Renault, J.; Jin, Z.; Kim, G.; Ng, A.; Juncker, D. Microfluidic chain reaction of structurally programmed capillary flow events. Nature 2022, 605, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Abedini-Nassab, R.; Pouryosef Miandoab, M.; Şaşmaz, M. Microfluidic Synthesis, Control, and Sensing of Magnetic Nanoparticles: A Review. Micromachines 2021, 12, 768. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Zhang, J.; Zou, Z.; Lu, G.; Wu, M.; Niu, L.; Zhang, Y. A Dual-Encoded Bead-Based Immunoassay with Tunable Detection Range for COVID-19 Serum Evaluation. Angew. Chemie—Int. Ed. 2022, 61, e202203706. [Google Scholar] [CrossRef]
- Petrova, V.N.; Russell, C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018, 16, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Tewawong, N.; Vichiwattana, P.; Korkong, S.; Klinfueng, S.; Suntronwong, N.; Thongmee, T.; Theamboonlers, A.; Vongpunsawad, S.; Poovorawan, Y. Evolution of the neuraminidase gene of seasonal influenza A and B viruses in Thailand between 2010 and 2015. PLoS ONE 2017, 12, e0175655. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.S.; Venkatesalu, S.; Dilliyappan, S.; Pasupulla, A.P.; Prathap, L.; Palaniyandi, T.; Baskar, G.; Ravi, M.; Sugumaran, A. Microfluidics as diagnostic tools. Clin. Chim. Acta 2024, 556, 117841. [Google Scholar] [CrossRef] [PubMed]
- Gholivand, A.; Korculanin, O.; Dahlhoff, K.; Babaki, M.; Dickscheid, T.; Lettinga, M.P. Effect of in-plane and out-of-plane bifurcated microfluidic channels on the flow of aggregating red blood cells. Lab Chip 2024, 24, 2317–2326. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Hsieh, T.M.; Lee, D.Y.S.; Ali, E.M.; Xie, H.; Looi, X.L.; Koay, E.S.C.; Li, M.H.; Ying, J.Y. A self-contained all-in-one cartridge for sample preparation and real-time PCR in rapid influenza diagnosis. Lab Chip 2010, 10, 3103–3111. [Google Scholar] [CrossRef]
- Draz, M.S.; Venkataramani, M.; Lakshminarayanan, H.; Saygili, E.; Moazeni, M.; Vasan, A.; Li, Y.; Sun, X.; Hua, S.; Yu, X.G.; et al. Nanoparticle-enhanced electrical detection of Zika virus on paper microchips. Nanoscale 2018, 10, 11841–11849. [Google Scholar] [CrossRef]
- Qiu, J.; Zhou, Y.; Chen, H.; Lin, J.M. Immunomagnetic separation and rapid detection of bacteria using bioluminescence and microfluidics. Talanta 2009, 79, 787–795. [Google Scholar] [CrossRef]
- Chang, M.S.; Yoo, J.H.; Woo, D.H.; Chun, M.S. Efficient detection of Escherichia coli O157:H7 using a reusable microfluidic chip embedded with antimicrobial peptide-labeled beads. Analyst 2015, 140, 7997–8006. [Google Scholar] [CrossRef]
- Chin, C.; Linder, V.; Sia, S. Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip 2012, 12, 2118–2134. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Lv, S.; Zhang, W.; Cui, Y. Microfluidic Point-of-Care (POC) Devices in Early Diagnosis: A Review of Opportunities and Challenges. Sensors 2022, 22, 1620. [Google Scholar] [CrossRef] [PubMed]
- Najjar, D.; Rainbow, J.; Sharma Timilsina, S.; Jolly, P.; de Puig, H.; Yafia, M.; Durr, N.; Sallum, H.; Alter, G.; Li, J.Z.; et al. A lab-on-a-chip for the concurrent electrochemical detection of SARS-CoV-2 RNA and anti-SARS-CoV-2 antibodies in saliva and plasma. Nat. Biomed. Eng. 2022, 6, 968–978. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Zhang, Y.; Li, Y.; Miao, Y.; Gao, S.; Lin, F.; Deng, Y.; Geng, L. Microfluidic chip coupled with optical biosensors for simultaneous detection of multiple analytes: A review. Biosens Bioelectron. 2019, 126, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Qian, C.; Liu, C.; Shen, H.; Wang, Z.; Ping, J.; Wu, J.; Chen, H. Nucleic acid amplification free biosensors for pathogen detection. Biosens. Bioelectron. 2020, 153, 112049. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, L.; Bendali, A.; Pereiro, I.; Azimani, M.; Dumas, S.; Malaquin, L.; Mai, T.D.; Descroix, S. Modular microfluidic system for on-chip extraction, preconcentration and detection of the cytokine biomarker IL-6 in biofluid. Sci. Rep. 2022, 12, 9468. [Google Scholar] [CrossRef] [PubMed]
- Illath, K.; Kar, S.; Gupta, P.; Shinde, A.; Wankhar, S.; Tseng, F.G.; Lim, K.T.; Nagai, M.; Santra, T.S. Microfluidic nanomaterials: From synthesis to biomedical applications. Biomaterials 2022, 280, 121247. [Google Scholar] [CrossRef] [PubMed]
- Mollarasouli, F.; Kurbanoglu, S.; Ozkan, S.A. The role of electrochemical immunosensors in clinical analysis. Biosensors 2019, 9, 86. [Google Scholar] [CrossRef]
- Singh, R.; Mukherjee, M.D.; Sumana, G.; Gupta, R.K.; Sood, S.; Malhotra, B.D. Biosensors for pathogen detection: A smart approach towards clinical diagnosis. Sens. Actuators B Chem. 2014, 197, 385–404. [Google Scholar] [CrossRef]
- Mahato, K.; Kumar, S.; Srivastava, A.; Maurya, P.K.; Singh, R.; Chandra, P. Chapter 14—Electrochemical Immunosensors: Fundamentals and Applications in Clinical Diagnostics. In Handbook of Immunoassay Technologies; Academic Press: Cambridge, MA, USA, 2018; pp. 359–414. [Google Scholar] [CrossRef]
- Zhang, H.; Miller, B.L. Immunosensor-based label-free and multiplex detection of influenza viruses: State of the art. Biosens. Bioelectron. 2019, 141, 111476. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.G.; Lee, T.J.; Jeong, S.W.; Choi, H.W.; Heo, N.S.; Park, J.Y.; Park, T.J.; Lee, S.J. Development of a plastic-based microfluidic immunosensor chip for detection of H1N1 influenza. Sensors 2012, 12, 10810–10819. [Google Scholar] [CrossRef] [PubMed]
- Barro, C.; Zetterberg, H. The blood biomarkers puzzle—A review of protein biomarkers in neurodegenerative diseases. J. Neurosci. Methods 2021, 361, 109281. [Google Scholar] [CrossRef]
- Dong, R.; Yi, N.; Jiang, D. Advances in single molecule arrays (SIMOA) for ultra-sensitive detection of biomolecules. Talanta 2024, 270, 125529. [Google Scholar] [CrossRef] [PubMed]
- Rissin, D.M.; Kan, C.W.; Campbell, T.G.; Howes, S.C.; Fournier, D.R.; Song, L.; Piech, T.; Patel, P.P.; Chang, L.; Rivnak, A.J.; et al. Duffy Single-Molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010, 28, 595–599. [Google Scholar] [CrossRef]
- Cai, G.; Bossé, Y.; Xiao, F.; Kheradmand, F.; Amos, C.I. Tobacco smoking increases the lung gene expression of ACE2, the Receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020, 201, 1557–1559. [Google Scholar] [CrossRef] [PubMed]
- Gilboa, T.; Cohen, L.; Cheng, C.A.; Lazarovits, R.; Uwamanzu-Nna, A.; Han, I.; Griswold, K.; Barry, N.; Thompson, D.B.; Kohman, R.E.; et al. A SARS-CoV-2 Neutralization Assay Using Single Molecule Arrays. Angew. Chem.—Int. Ed. 2021, 60, 25966–25972. [Google Scholar] [CrossRef]
- Thomas, S.N.; Karger, A.B.; Altawallbeh, G.; Nelson, K.M.; Jacobs, D.R., Jr.; Gorlin, J.; Barcelo, H.; Thyagarajan, B. Ultrasensitive detection of salivary SARS-CoV-2 IgG antibodies in individuals with natural and COVID-19 vaccine-induced immunity. Sci. Rep. 2022, 12, 8890. [Google Scholar] [CrossRef]
- Song, L.; Shan, D.; Zhao, M.; Pink, B.A.; Minnehan, K.A.; York, L.; Gardel, M.; Sullivan, S.; Phillips, A.F.; Hayman, R.B.; et al. Direct detection of bacterial genomic DNA at sub-femtomolar concentrations using single molecule arrays. Anal. Chem. 2013, 85, 1932–1939. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.H.; Rissin, D.M.; Kan, C.W.; Fournier, D.R.; Piech, T.; Campbell, T.G.; Meyer, R.E.; Fishburn, M.W.; Cabrera, C.; Patel, P.P.; et al. The Simoa HD-1 Analyzer: A Novel Fully Automated Digital Immunoassay Analyzer with Single-Molecule Sensitivity and Multiplexing. J. Lab. Autom. 2016, 21, 533–547. [Google Scholar] [CrossRef]
- Wu, C.; Garden, P.M.; Walt, D.R. Ultrasensitive Detection of Attomolar Protein Concentrations by Dropcast Single Molecule Assays. J. Am. Chem. Soc. 2020, 142, 12314–12323. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Fu, Y.; Guo, J.; Guo, J. Single-molecule immunoassay technology: Recent advances. Talanta 2023, 265, 124903. [Google Scholar] [CrossRef] [PubMed]
- Duffy, D.C. Digital detection of proteins. Lab Chip. 2023, 23, 818–847. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, Z.; Zhai, X.; Li, Y.; Lin, L.; Zhao, H.; Bian, L.; Li, P.; Yu, L.; Wu, Y.; et al. Rapid and Sensitive Detection of anti-SARS-CoV-2 IgG, Using Lanthanide-Doped Nanoparticles-Based Lateral Flow Immunoassay. Anal. Chem. 2020, 92, 7226–7231. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chen, Y.; Liu, W.; Fang, H.; Li, X.; Hou, L.; Liu, Y.; Lai, W.; Huang, X.; Xiong, Y. Development of a rapid and sensitive quantum dot nanobead-based double-antigen sandwich lateral flow immunoassay and its clinical performance for the detection of SARS-CoV-2 total antibodies. Sens. Actuators B Chem. 2021, 343, 130139. [Google Scholar] [CrossRef]
- Kim, S.; Akarapipad, P.; Nguyen, B.T.; Breshears, L.E.; Sosnowski, K.; Baker, J.; Uhrlaub, J.L.; Nikolich-Žugich, J.; Yoon, J.Y. Direct capture and smartphone quantification of airborne SARS-CoV-2 on a paper microfluidic chip. Biosens. Bioelectron. 2022, 200, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Juncker, D.; Bergeron, S.; Laforte, V.; Li, H. Cross-reactivity in antibody microarrays and multiplexed sandwich assays: Shedding light on the dark side of multiplexing. Curr. Opin. Chem. Biol. 2014, 18, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.A.; Kullo, I.J.; Bailey, K.R.; Klee, G.G. Antibody-based protein multiplex platforms: Technical and operational challenges. Clin. Chem. 2010, 56, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Ng, A.H.C.; Fobel, R.; Chang-Yen, D.A.; Yarnell, L.E.; Pearson, E.L.; Oleksak, C.M.; Fischer, A.T.; Luoma, R.P.; Robinson, J.M.; et al. Automated digital microfluidic platform for magnetic-particle-based immunoassays with optimization by design of experiments. Anal. Chem. 2013, 85, 9638–9646. [Google Scholar] [CrossRef]
- Hartmann, M.; Roeraade, J.; Stoll, D.; Templin, M.F.; Joos, T.O. Protein microarrays for diagnostic assays. Anal. Bioanal. Chem. 2009, 393, 1407–1416. [Google Scholar] [CrossRef]
- Fischer, K.A.F. Brief Communication. Centaurus 1988, 31, 164–167. [Google Scholar] [CrossRef]
- Valentin, M.; Ma, S.; Zhao, A.; Avrameas, A. Validation of immunoassay for protein biomarkers: Bioanalytical study plan implementation to support pre-clinical and clinical studies. J. Pharm. Biomed. Anal. 2011, 55, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.A.; Kullo, I.J.; Bailey, K.R.; Klee, G.G. Measurement and quality control issues in multiplex protein assays: A case study. Clin. Chem. 2009, 55, 1092–1099. [Google Scholar] [CrossRef] [PubMed]
- Eckels, J.; Nathe, C.; Nelson, E.K.; Shoemaker, S.G.; Nostrand, E.V.; Yates, N.L.; Ashley, V.C.; Harris, L.J.; Bollenbeck, M.; Fong, Y.; et al. Quality control, analysis and secure sharing of Luminex® immunoassay data using the open source LabKey Server platform. BMC Bioinform. 2013, 14, 28–30. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Yang, H.; Wang, Y. Inverse design of microfluidic concentration gradient generator using deep learning and physics-based component model. Microfluid. Nanofluidics 2020, 24, 44. [Google Scholar] [CrossRef]
- Boja, E.S.; Jortani, S.A.; Ritchie, J.; Hoofnagle, A.N.; Težak, Ž.; Mansfield, E.; Keller, P.; Rivers, R.C.; Rahbar, A.; Anderson, N.L.; et al. The journey to regulation of protein-based multiplex quantitative assays. Clin. Chem. 2011, 57, 560–567. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bastarache, J.A.; Koyama, T.; Wickersham, N.E.; Mitchell, D.B.; Mernaugh, R.L.; Ware, L.B. Accuracy and reproducibility of a multiplex immunoassay platform: A validation study. J. Immunol. Methods 2011, 367, 33–39. [Google Scholar] [CrossRef]
- Callaghan, D.; Burger, J.; Mishra, A.K. A machine learning approach to radar sea clutter suppression. In Proceedings of the 2017 IEEE Radar Conference (RadarConf), Seattle, WA, USA, 8–12 May 2017; pp. 1222–1227. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury Google, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An imperative style, high-performance deep learning library. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; pp. 8026–8037. [Google Scholar]
- Barupal, D.K.; Fiehn, O. Generating the blood exposome database using a comprehensive text mining and database fusion approach. Environ. Health Perspect. 2019, 127, 2825–2830. [Google Scholar] [CrossRef]
- McIntyre, D.; Lashkaripour, A.; Fordyce, P.; Densmore, D. Machine learning for microfluidic design and control. Lab Chip 2022, 22, 2925–2937. [Google Scholar] [CrossRef] [PubMed]




| Pathogenic Microorganisms | Detection Limit | Sensitivity | Selectivity | Ref. |
|---|---|---|---|---|
| SARS-CoV-2 | 1.6 ng/mL | 95 | 91 | [11] |
| SARS-CoV-2 | 0.12 ng/mL | 100 | 100 | [12] |
| SARS-CoV-2 | 1 nM | 98 | 100 | [13] |
| SARS-CoV-2 | 13.3 ng/mL | - | - | [14] |
| SARS-CoV-2 | 2.8 × 10−15 M | - | - | [15] |
| SARS-CoV-2 | 5 ng/mL | - | - | [16] |
| SARS-CoV-2 | 3.13 ng/mL | 100 | 100 | [17] |
| SARS-CoV-2 | - | 98 | 100 | [18] |
| SARS-CoV-2 | 0.1 ng/mL | - | - | [19] |
| SARS-CoV-2 | - | 99 | 99 | [20] |
| SARS-CoV-2 | - | 95 | 100 | [21] |
| SARS-CoV-2 | 230 pg/mL | - | - | [22] |
| SARS-CoV-2 | 17 ng/mL | 100 | 98 | [23] |
| SARS-CoV-2 | 4 pg/mL | - | - | [24] |
| SARS-CoV-2 | - | 100 | 100 | [25] |
| SARS-CoV-2 | 0.5 pg/mL | - | - | [26] |
| SARS-CoV-2 | 10 pg/mL | - | - | [27] |
| H1N1 | 0.5 PFU/mL | - | - | [28] |
| H1N1 | 0.032 HAU | - | - | [29] |
| H1N1 | 1 pg/mL | - | - | [30] |
| H5N1 | 1 pg/mL | - | - | [30] |
| H7N9 | 1 pg/mL | - | - | [30] |
| H7N9 | 3.4 ng/mL | - | - | [31] |
| H9N2 | 4.5 ng/mL | - | - | [31] |
| H1N1 | 2.2 ng/mL | - | - | [32] |
| H3N2 | 3.4 ng/mL | - | - | [32] |
| H7N3 | 2.9 ng/mL | - | - | [32] |
| Dengue | 1 ng/mL | 76 | 100 | [33] |
| ZIKV | 20 ng/mL | 81 | 86 | [33] |
| ZIKV | 1 pg/mL | - | - | [34] |
| ZIKV | 45 pg/mL | - | - | [35] |
| Coxiella burnetii | - | 92 | 89 | [36] |
| Salmonella typhimurium | 3 × 103 CFU/mL | - | - | [37] |
| Escherichia coli O157:H7 | 10 CFU/mL | - | - | [38] |
| Escherichia coli K-12 | 3.4 × 104 CFU/mL | - | - | [39] |
| Escherichia coli O157:H7 | 1.1 × 103 CFU/mL | - | - | [40] |
| Escherichia coli O157:H7 | 5 CFU/mL | - | - | [41] |
| Escherichia coli BL21 | 20 CFU/mL | - | - | [41] |
| Escherichia coli | 50 CFU/mL | - | - | [42] |
| Escherichia coli O157:H7 | 100 CFU/mL | - | - | [43] |
| Escherichia coli | 103 CFU/mL | - | - | [44] |
| P. aeruginosa | 103 CFU/mL | - | - | [44] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Chen, J.; Zhang, Y.; Yang, Z.; Zhang, K.; Zhang, D.; Zheng, L. Advancing Microfluidic Immunity Testing Systems: New Trends for Microbial Pathogen Detection. Molecules 2024, 29, 3322. https://doi.org/10.3390/molecules29143322
Wang Y, Chen J, Zhang Y, Yang Z, Zhang K, Zhang D, Zheng L. Advancing Microfluidic Immunity Testing Systems: New Trends for Microbial Pathogen Detection. Molecules. 2024; 29(14):3322. https://doi.org/10.3390/molecules29143322
Chicago/Turabian StyleWang, Yiran, Jingwei Chen, Yule Zhang, Zhijin Yang, Kaihuan Zhang, Dawei Zhang, and Lulu Zheng. 2024. "Advancing Microfluidic Immunity Testing Systems: New Trends for Microbial Pathogen Detection" Molecules 29, no. 14: 3322. https://doi.org/10.3390/molecules29143322
APA StyleWang, Y., Chen, J., Zhang, Y., Yang, Z., Zhang, K., Zhang, D., & Zheng, L. (2024). Advancing Microfluidic Immunity Testing Systems: New Trends for Microbial Pathogen Detection. Molecules, 29(14), 3322. https://doi.org/10.3390/molecules29143322






