The Discovery, Molecular Cloning, and Characterization of Dextransucrase LmDexA and Its Active Truncated Mutant from Leuconostoc mesenteroides NN710
Abstract
1. Introduction
2. Materials and Methods
2.1. Medium and Plasmid
2.2. Isolation and Identification of Strain
2.3. Cloning of Dextransucrase and Its Truncated Variants Genes
2.4. Protein Expression and Purification of the Recombinant Dextransucrases
2.5. Enzyme Activity Assays
2.6. Biochemical Characterization of Recombinant Dextransucrase
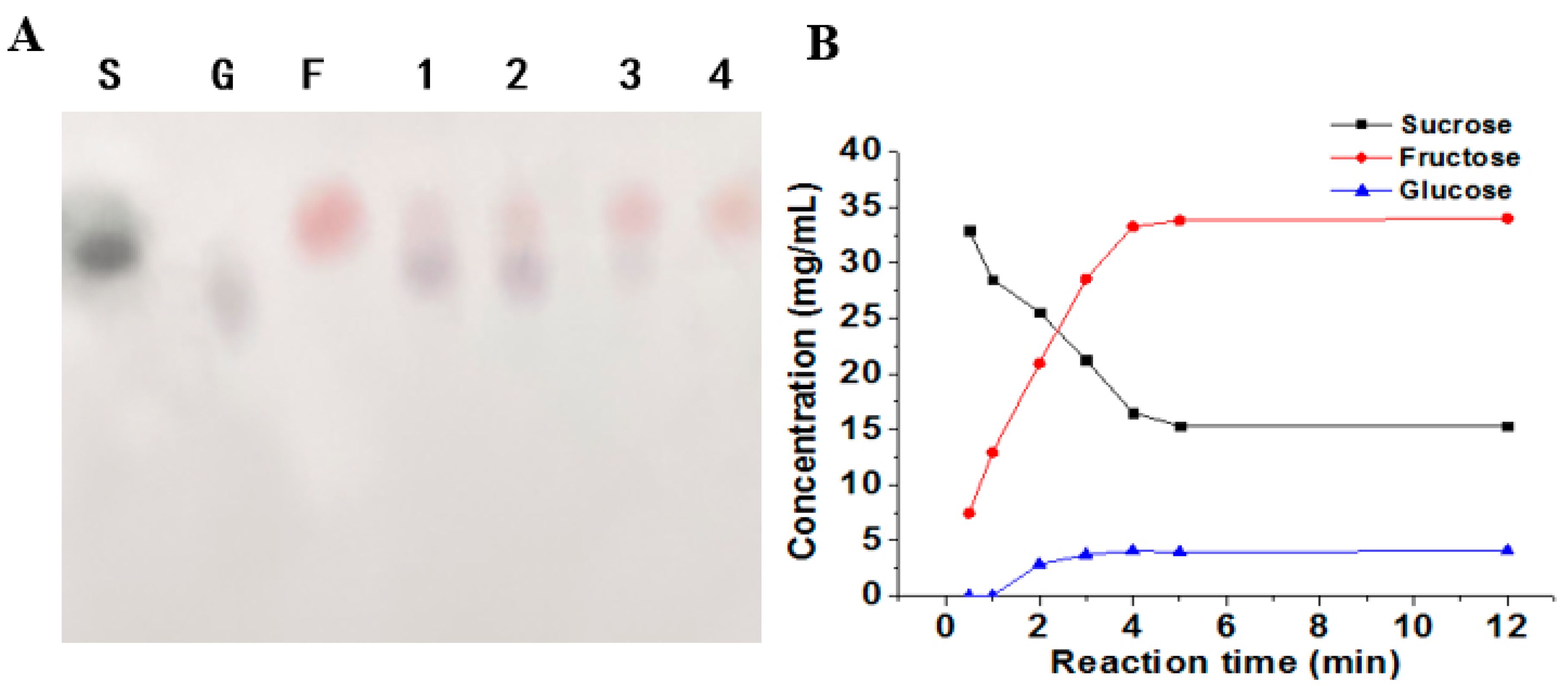
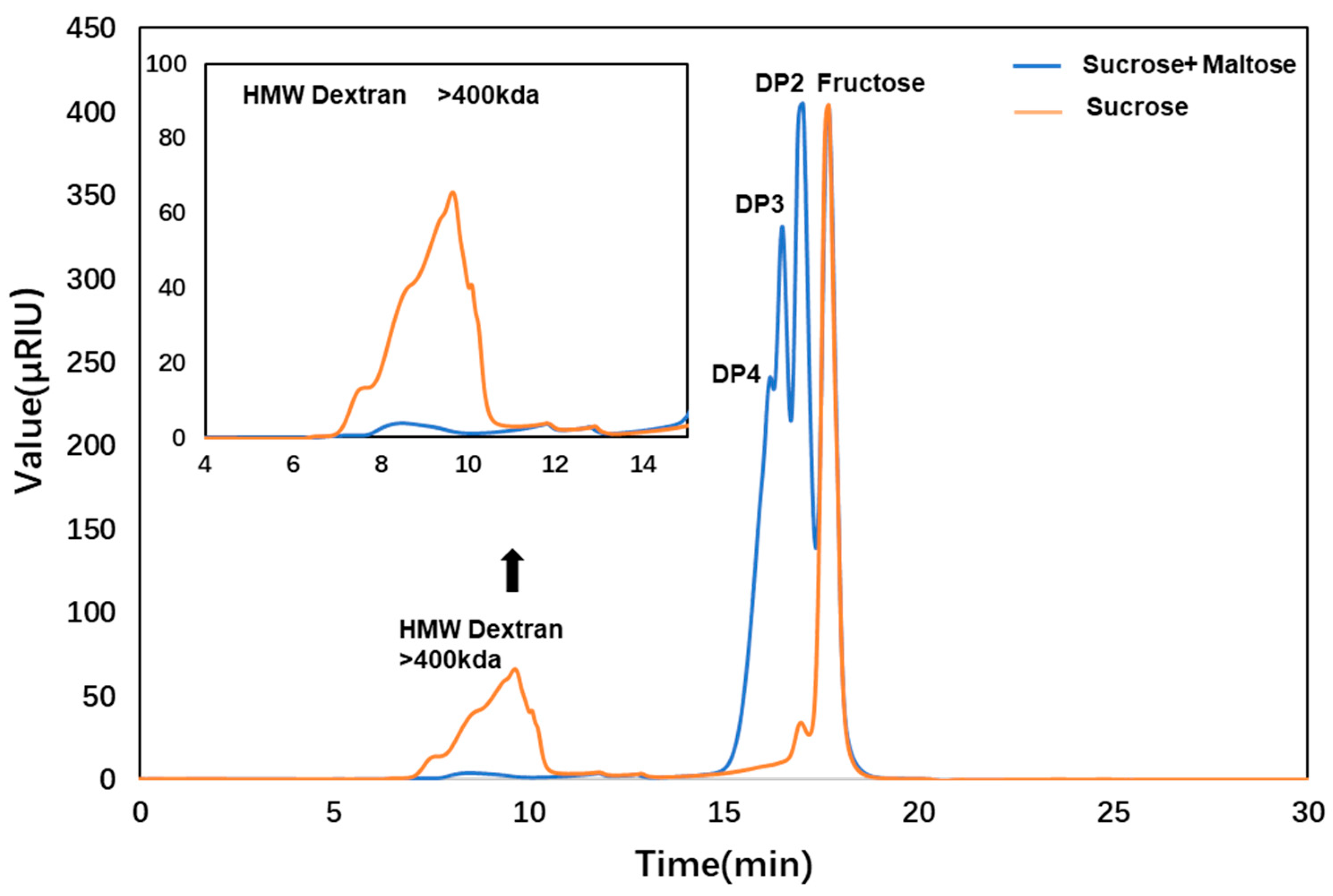
2.7. Product Analysis
3. Results and Discussion
3.1. Screening of the Strains Manifesting Dextran Synthesis Ability
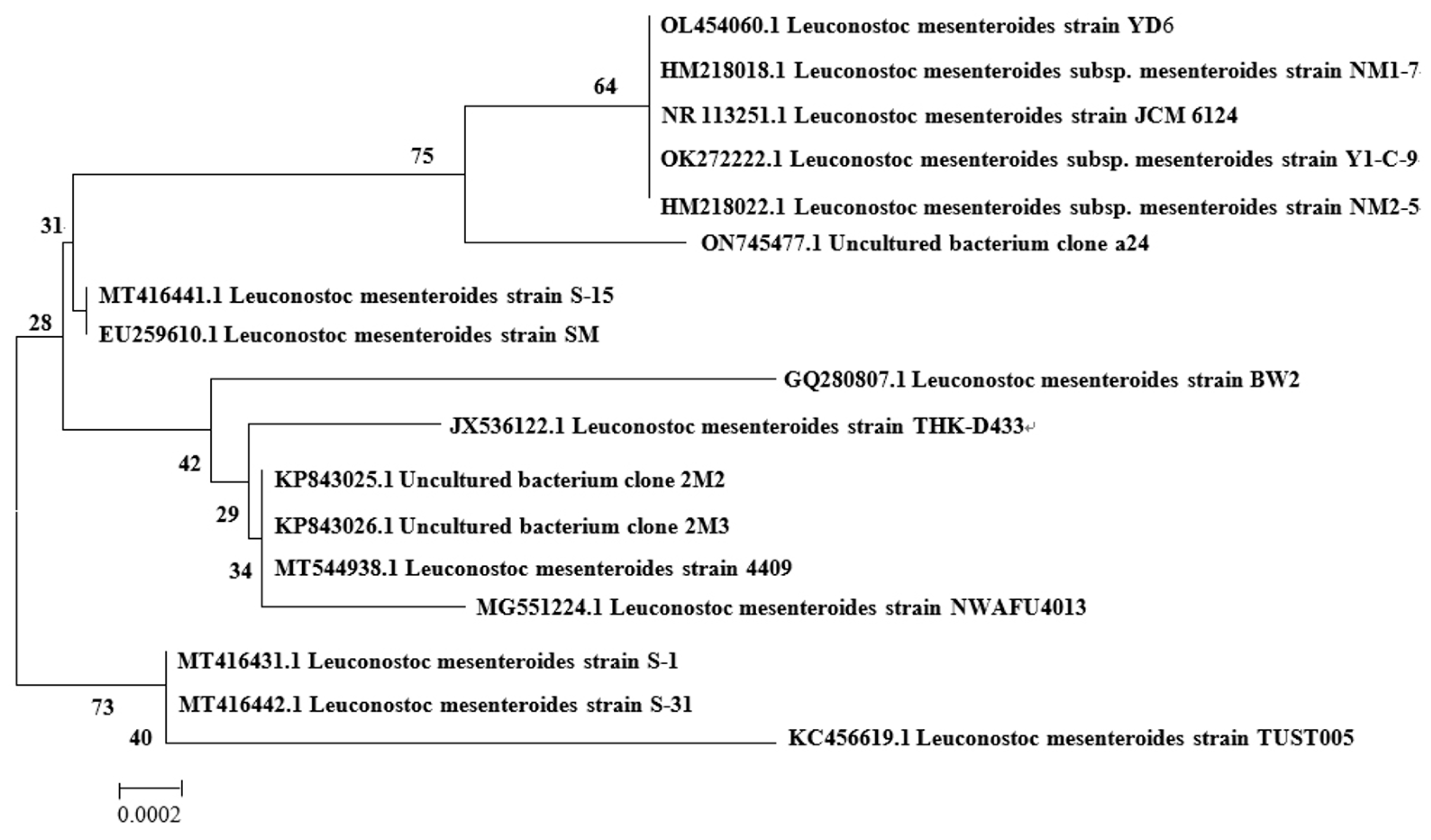
3.2. The Identification of the Strain NN710
3.3. Sequence Analysis of LmDexA
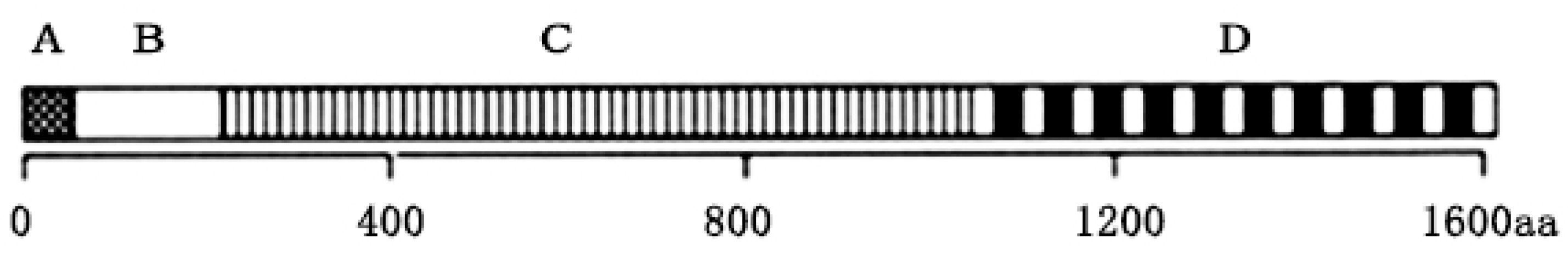
3.4. N-Terminal Truncation Construction Strategy
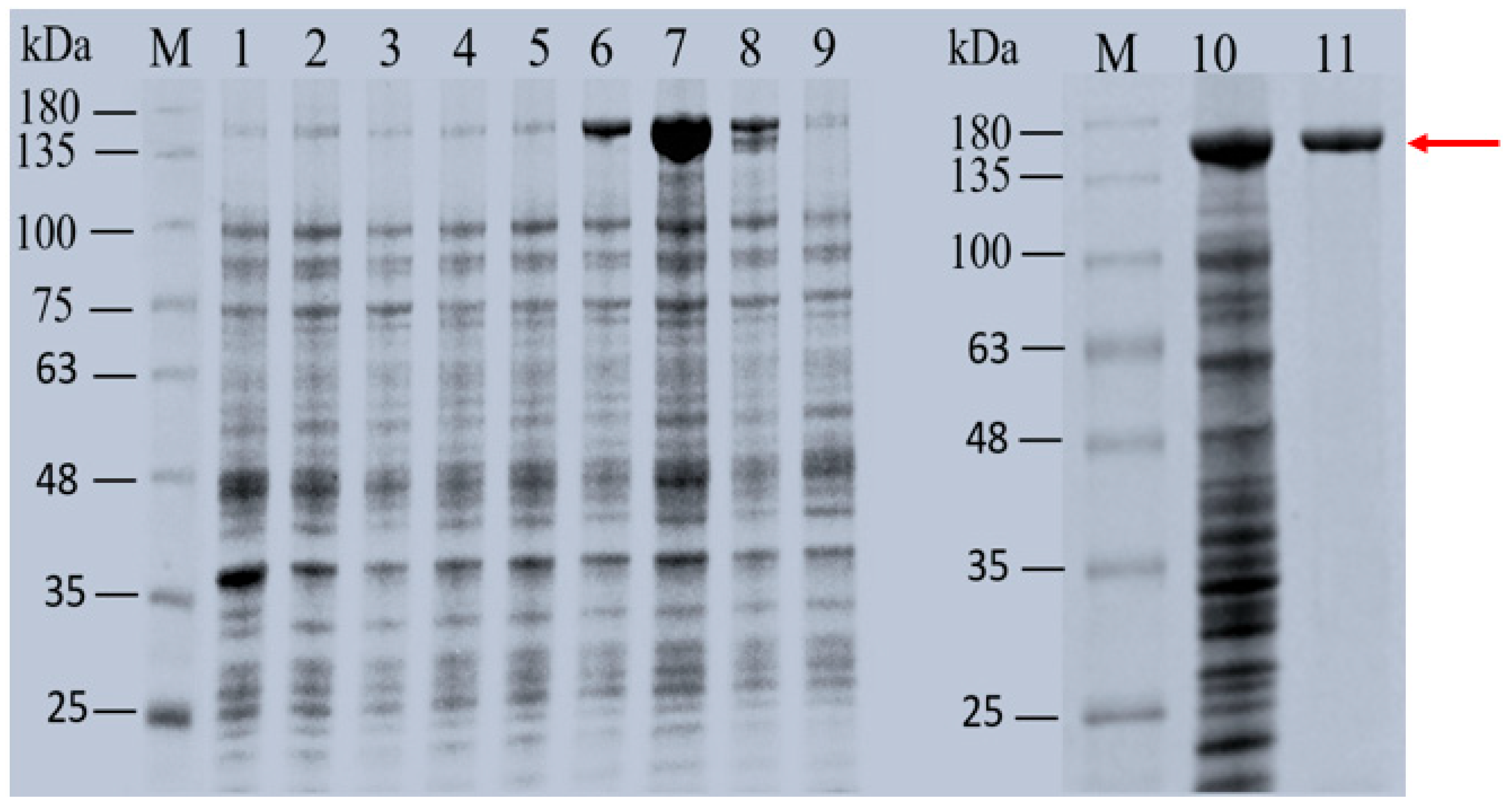
3.5. Expression and Purification of N-Terminal Truncation Mutants
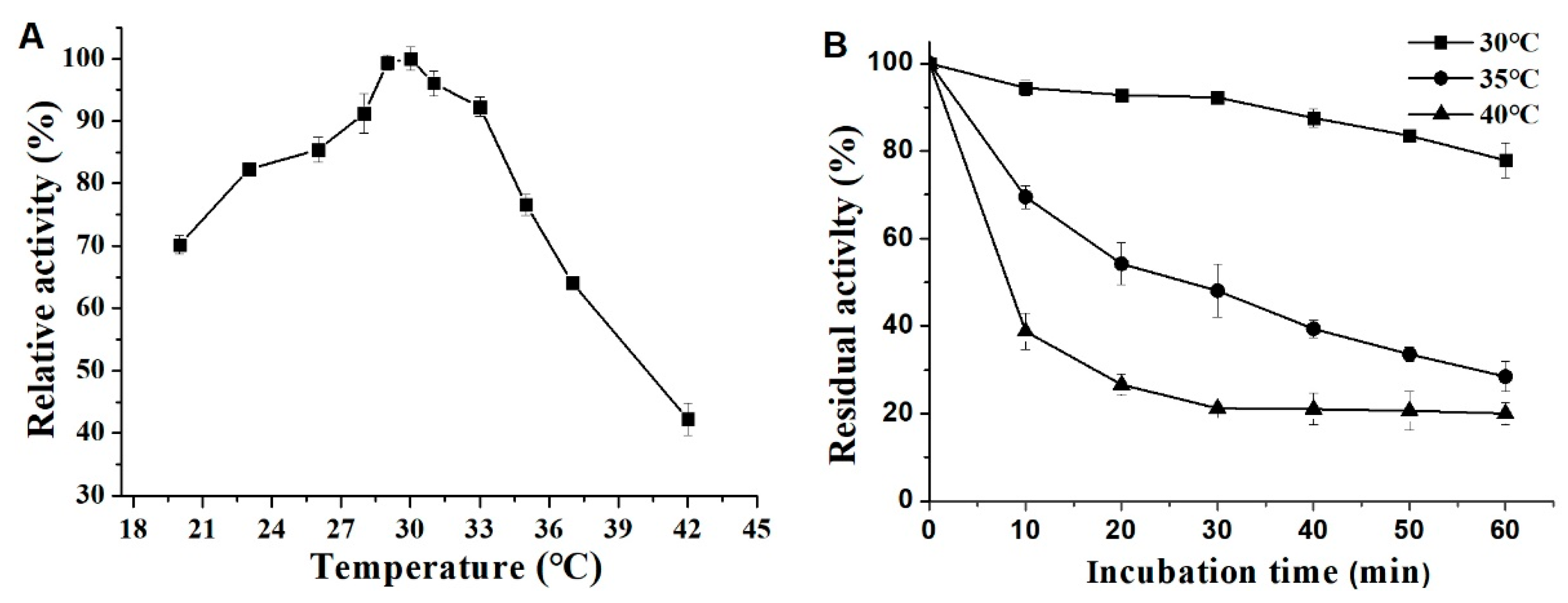
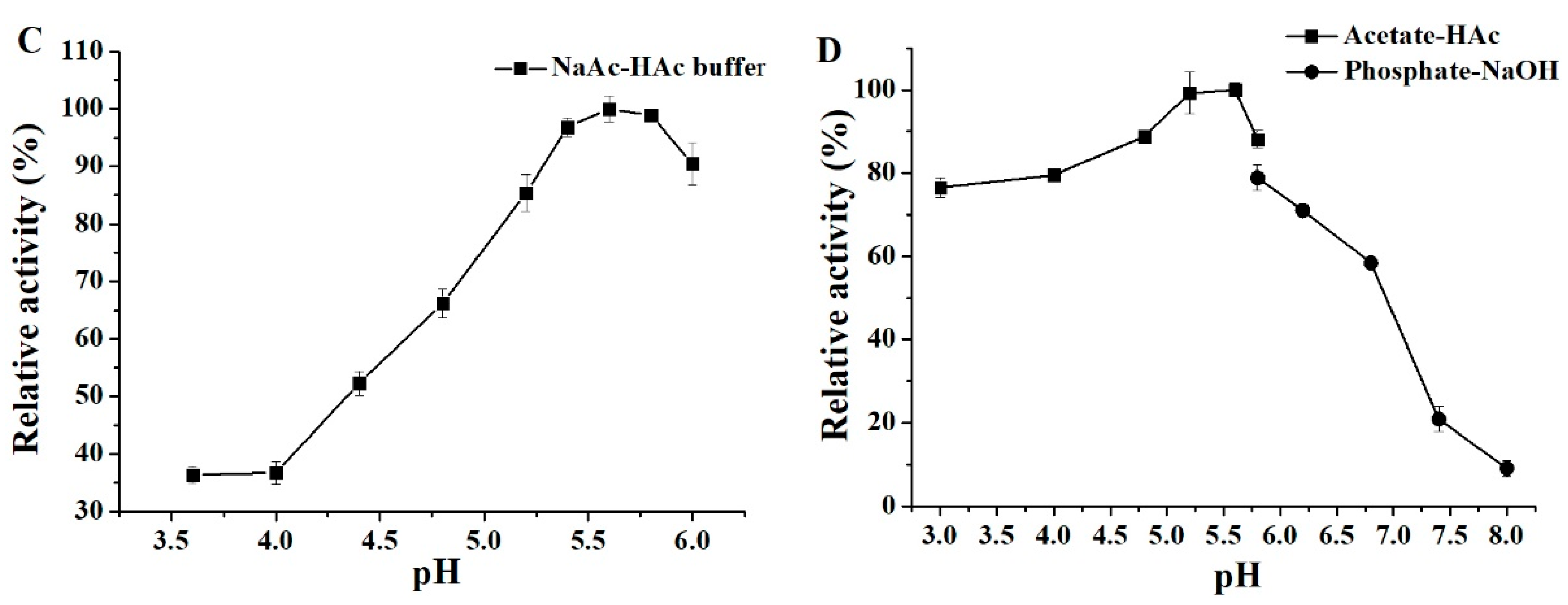
3.6. Effects of pH and Temperature on Activity and Stability of ΔN190LmDexA
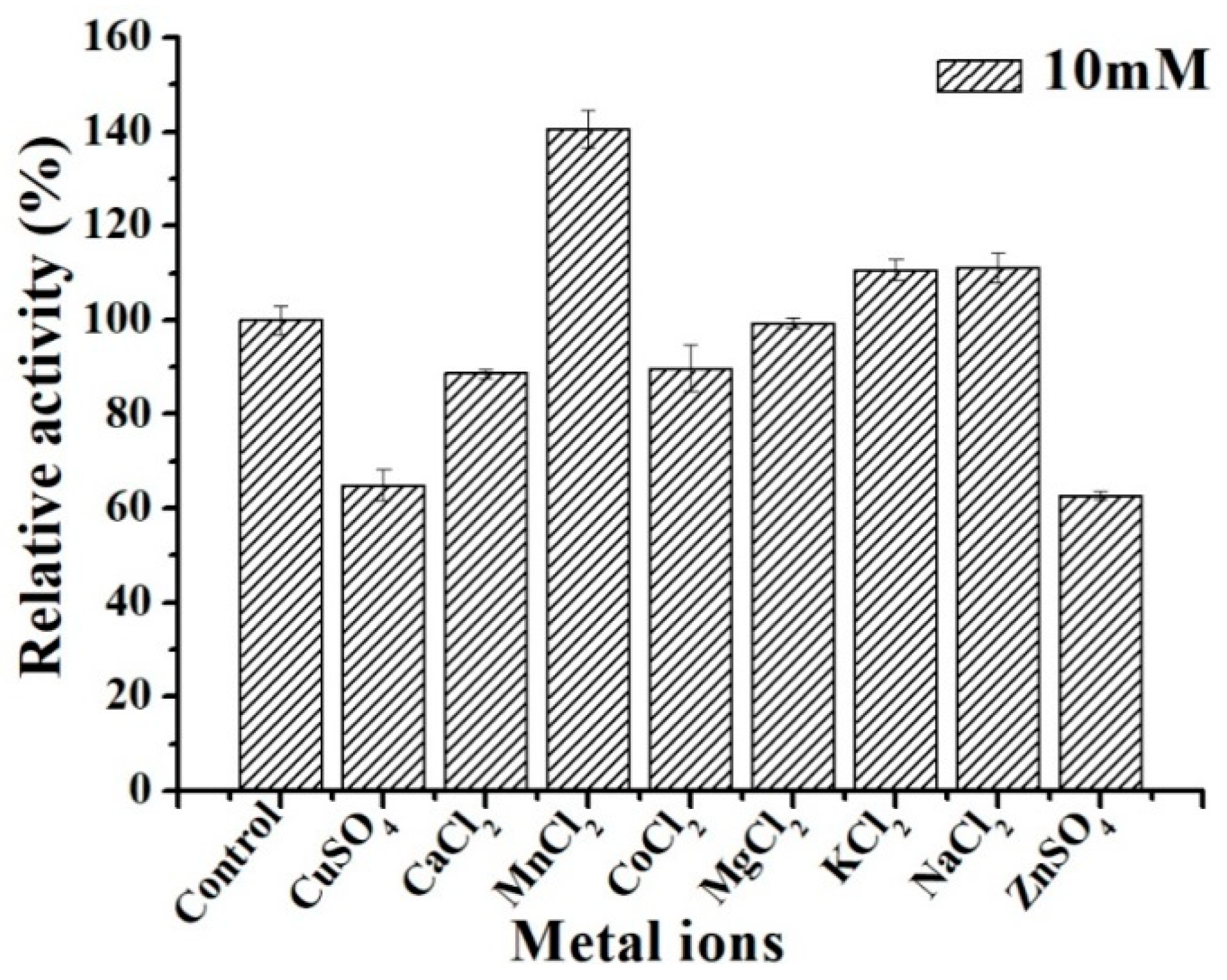
3.7. Effect of Metallic Cations on ΔN190LmDexA Activity
3.8. Effect of Substrate Concentration and Kinetic Parameters of ΔN190LmDexA
3.9. Properties of the ΔN190LmDexA
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Passerini, D.; Vuillemin, M.; Ufarte, L.; Morel, S.; Loux, V.; Fontagne-Faucher, C.; Monsan, P.; Remaud-Simeon, M.; Moulis, C. Inventory of the GH70 enzymes encoded by Leuconostoc citreum NRRL B-1299—Identification of three novel alpha-transglucosylases. FEBS J. 2015, 282, 2115–2130. [Google Scholar] [CrossRef] [PubMed]
- Wangpaiboon, K.; Padungros, P.; Nakapong, S.; Charoenwongpaiboon, T.; Rejzek, M.; Field, R.A.; Pichyangkura, R. An alpha-1,6-and alpha-1,3-linked glucan produced by Leuconostoc citreum ABK-1 alternansucrase with nanoparticle and film-forming properties. Sci. Rep. 2018, 8, 8340. [Google Scholar] [CrossRef]
- Robyt, J.F. Mechanisms in the glucansucrase synthesis of polysaccharides and oligosaccharides from sucrose. Adv. Carbohydr. Chem. Biochem. 1995, 51, 133–168. [Google Scholar]
- Kenney, A.C.; Cole, J.A. Identification of a 1,3-α glucosyltransferase involved in insoluble glucan synthesis by a serotype c strain of Streptococcus mutans. FEMS Microbiol. Lett. 1983, 16, 159–162. [Google Scholar] [CrossRef]
- Kralj, S.; van Geel-Schutten, G.H.; Dondorff, M.M.G.; Kirsanovs, S.; van der Maarel, M.; Dijkhuizen, L. Glucan synthesis in the genus Lactobacillus: Isolation and characterization of glucansucrase genes, enzymes and glucan products from six different strains. Microbiology 2004, 150, 3681–3690. [Google Scholar] [CrossRef] [PubMed]
- Besrour-Aouam, N.; Fhoula, I.; Hernandez-Alcantara, A.M.; Mohedano, M.L.; Najjari, A.; Prieto, A.; Ruas-Madiedo, P.; Lopez, P.; Ouzari, H.I. The role of dextran production in the metabolic context of Leuconostoc and Weissella Tunisian strains. Carbohydr. Polym. 2021, 253, 117254. [Google Scholar] [CrossRef]
- Bounaix, M.S.; Robert, H.; Gabriel, V.; Morel, S.; Remaud-Simeon, M.; Gabriel, B.; Fontagne-Faucher, C. Characterization of dextran-producing Weissella strains isolated from sourdoughs and evidence of constitutive dextransucrase expression. FEMS Microbiol. Lett. 2010, 311, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Leathers, T.D.; Bischoff, K.M. Biofilm formation by strains of Leuconostoc citreum and L. mesenteroides. Biotechnol. Lett. 2011, 33, 517–523. [Google Scholar] [CrossRef]
- Vettori, M.H.P.B.; Blanco, K.C.; Cortezi, M.; Lima, C.J.B.; Contiero, J. Dextran: Effect of process parameters on production, purification and molecular weight and recent applications. Diálogos Ciência 2012, 2012, 171–186. [Google Scholar] [CrossRef]
- Vujicic-Zagar, A.; Pijning, T.; Kralj, S.; Lopez, C.A.; Eeuwema, W.; Dijkhuizen, L.; Dijkstra, B.W. Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes. Proc. Natl. Acad. Sci. USA 2010, 107, 21406–21411. [Google Scholar] [CrossRef] [PubMed]
- Leemhuis, H.; Pijning, T.; Dobruchowska, J.M.; van Leeuwen, S.S.; Kralj, S.; Dijkstra, B.W.; Dijkhuizen, L. Glucansucrases: Three-dimensional structures, reactions, mechanism, alpha-glucan analysis and their implications in biotechnology and food applications. J. Biotechnol. 2013, 163, 250–272. [Google Scholar] [CrossRef] [PubMed]
- Jeanes, A.; Haynes, W.C.; Wilham, C.A.; Rankin, J.C.; Melvin, E.H.; Austin, M.J.; Cluskey, J.E.; Fisher, B.E.; Tsuchiya, H.M.; Rist, C.E. Characterization and classification of dextrans from ninety-six strains of bacteria. J. Am. Chem. Soc. 1954, 76, 5041–5052. [Google Scholar] [CrossRef]
- Pranckute, R.; Kaunietis, A.; Kuisiene, N.; Citavicius, D.J. Combining prebiotics with probiotic bacteria can enhance bacterial growth and secretion of bacteriocins. Int. J. Biol. Macromol. 2016, 89, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Ganzle, M.G. Site Directed Mutagenesis of Dextransucrase DsrM from Weissella cibaria: Transformation to a Reuteransucrase. J. Agric. Food Chem. 2016, 64, 6848–6855. [Google Scholar] [CrossRef]
- Zannini, E.; Waters, D.M.; Coffey, A.; Arendt, E.K. Production, properties, and industrial food application of lactic acid bacteria-derived exopolysaccharides. Appl. Microbiol. Biotechnol. 2016, 100, 1121–1135. [Google Scholar] [CrossRef] [PubMed]
- Mohan Rao, T.J.; Goyal, A. Purification, optimization of assay, and stability studies of dextransucrase isolated from Weissella cibaria JAG8. Prep. Biochem. Biotechnol. 2013, 43, 329–341. [Google Scholar] [CrossRef]
- Naessens, M.; Cerdobbel, A.; Soetaert, W.; Vandamme, E.J. Leuconostoc dextransucrase and dextran: Production, properties and applications. J. Chem. Technol. Biotechnol. 2005, 80, 845–860. [Google Scholar] [CrossRef]
- Argüello-Morales, M.A.; Remaud-Simeon, M.; Pizzut, S.; Sarçabal, P.; Willemot, R.-M.; Monsan, P. Sequence analysis of the gene encoding alternansucrase, a sucrose glucosyltransferase from Leuconostoc mesenteroides NRRL B-1355. FEMS Microbiol. Lett. 2000, 182, 81–85. [Google Scholar] [CrossRef]
- Fabre, E.; Bozonnet, S.; Arcache, A.; Willemot, R.M.; Vignon, M.; Monsan, P.; Remaud-Simeon, M. Role of the two catalytic domains of DSR-E dextransucrase and their involvement in the formation of highly alpha-1,2 branched dextran. J. Bacteriol. 2005, 187, 296–303. [Google Scholar] [CrossRef]
- Molina, M.; Cioci, G.; Moulis, C.; Severac, E.; Remaud-Simeon, M. Bacterial alpha-Glucan and Branching Sucrases from GH70 Family: Discovery, Structure-Function Relationship Studies and Engineering. Microorganisms 2021, 9, 1607. [Google Scholar] [CrossRef]
- Meng, X.; Gangoiti, J.; Bai, Y.; Pijning, T.; Van Leeuwen, S.S.; Dijkhuizen, L. Structure-function relationships of family GH70 glucansucrase and 4,6-alpha-glucanotransferase enzymes, and their evolutionary relationships with family GH13 enzymes. Cell Mol. Life Sci. 2016, 73, 2681–2706. [Google Scholar] [CrossRef]
- Claverie, M.; Cioci, G.; Vuillemin, M.; Monties, N.; Roblin, P.; Lippens, G.; Remaud-Simeon, M.; Moulis, C. Investigations on the Determinants Responsible for Low Molar Mass Dextran Formation by DSR-M Dextransucrase. ACS Catal. 2017, 7, 7106–7119. [Google Scholar] [CrossRef]
- Gao, X.; Liu, Z.; Cui, W.; Zhou, L.; Tian, Y.; Zhou, Z. Enhanced thermal stability and hydrolytic ability of Bacillus subtilis aminopeptidase by removing the thermal sensitive domain in the non-catalytic region. PLoS ONE 2014, 9, e92357. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Tang, L.; Liang, Y.; Jiao, S.; Yu, H.; Luo, H. Novel Chaperones RrGroEL and RrGroES for Activity and Stability Enhancement of Nitrilase in Escherichia coli and Rhodococcus ruber. Molecules 2020, 25, 1002. [Google Scholar] [CrossRef]
- Tang, L.; Yang, J.; Chen, J.; Zhang, J.; Yu, H.; Shen, Z. Design of salt-bridge cyclization peptide tags for stability and activity enhancement of enzymes. Process Biochem. 2019, 81, 39–47. [Google Scholar] [CrossRef]
- Zavrel, T.; Ocenasova, P.; Sinetova, M.A.; Cerveny, J. Determination of Storage (Starch/Glycogen) and Total Saccharides Content in Algae and Cyanobacteria by a Phenol-Sulfuric Acid Method. Bio Protoc. 2018, 8, e2966. [Google Scholar] [CrossRef] [PubMed]
- Sarwat, F.; Qader, S.A.U.; Aman, A.; Ahmed, N. Production & characterization of a unique dextran from an indigenous Leuconostoc mesenteroides CMG713. Int. J. Biol. Sci. 2008, 4, 379–386. [Google Scholar]
- Santos, M.; Teixeira, J.; Rodrigues, A. Production of dextransucrase, dextran and fructose from sucrose using Leuconostoc mesenteroides NRRL B512(f)2000. Biochem. Eng. J. 2000, 4, 177–188. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hu, Y.; Zhu, C.; Zhu, B.; Wang, Y. Cloning, sequencing and expression of a dextransucrase gene (dexYG) from Leuconostoc mesenteroides. Biotechnol. Lett. 2008, 30, 1441–1446. [Google Scholar] [CrossRef]
- Sumner, J.B.; Howell, S.F. A Method for Determination of Saccharase Activity. J. Biol. Chem. 1935, 108, 51–54. [Google Scholar] [CrossRef]
- Neubauer, H.; Bauche, A.; Mollet, B. Molecular characterization and expression analysis of the dextransucrase DsrD of Leuconostoc mesenteroides Lcc4 in homologous and heterologous Lactococcus lactis cultures. Microbiology 2003, 149, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Monchois, V.; Willemot, R.-M.; Remaud-Simeon, M.; Croux, C.; Monsan, P.J.G. Cloning and sequencing of a gene coding for a novel dextransucrase from Leuconostoc mesenteroides NRRL B-1299 synthesizing only α (1–6) and α (1–3) linkages. Gene 1996, 182, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Yamaner, C.I.; Sezen, I.Y.; Tanriseven, A. Selection of psychrotrophic Leuconostoc spp. from native fruits, and studies on their dextransucrases. Food Sci. Biotechnol. 2010, 19, 175–184. [Google Scholar] [CrossRef]
- Dawoud, T.M.; Alshehrei, F.; Siddiqui, K.; Ameen, F.; Akhtar, J.; Arif, A. Purification, Characterization and N-terminal Protein Sequencing of the Enzyme Dextransucrase Produced by Leuconostoc mesenteroides. Biosci. Biotechnol. Res. Asia 2021, 18, 287–295. [Google Scholar] [CrossRef]
- Zhang, Y.-X.; Yang, J.-W.; Wu, Y.-Y.; Hu, X.-Q.; Zhang, H.-B. The stability improvement of dextransucrase by artificial extension modification of the V domain of the enzyme. Enzym. Microb. Technol. 2021, 151, 109919. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Q.; Zhang, H.B.; Li, Y.; Hu, X.Q.; Yang, J.W. The thermoduric effects of site-directed mutagenesis of proline and lysine on dextransucrase from Leuconostoc mesenteroides 0326. Int. J. Biol. Macromol. 2018, 107, 1641–1649. [Google Scholar] [CrossRef]
- Monchois, V.; Willemot, R.-M.; Monsan, P. Glucansucrases: Mechanism of action and structure–function relationships. FEMS Microbiol. Rev. 1999, 23, 131–151. [Google Scholar] [CrossRef]
- Bozonnet, S.; Dols-Laffargue, M.; Fabre, E.; Pizzut, S.; Remaud-Simeon, M.; Monsan, P.; Willemot, R.M. Molecular characterization of DSR-E, an alpha-1,2 linkage-synthesizing dextransucrase with two catalytic domains. J. Bacteriol. 2002, 184, 5753–5761. [Google Scholar] [CrossRef]
- Monchois, V.; Remaud-Simeon, M.; Russell, R.R.; Monsan, P.; Willemot, R.M. Characterization of Leuconostoc mesenteroides NRRL B-512F dextransucrase (DSRS) and identi®cation of amino-acid residues playing a key role in enzyme activity. Appl. Microbiol. Biotechnol. 1997, 48, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Van Hijum, S.A.; Kralj, S.; Ozimek, L.K.; Dijkhuizen, L.; van Geel-Schutten, I.G. Structure-function relationships of glucansucrase and fructansucrase enzymes from lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2006, 70, 157–176. [Google Scholar] [CrossRef] [PubMed]
- Funane, K.; Mizuno, K.; Takahara, H.; Kobayashi, M. Gene Encoding a Dextransucrase-like Protein in Leuconostoc mesenteroides NRRL B-512F. Biosci. Biotechnol. Biochem. 2000, 64, 29–38. [Google Scholar] [CrossRef]
- Monchois, V.; Remaud-Simeon, M.; Monsan, P.; Willemot, R.M. Cloning and sequencing of a gene coding for an extracellular dextransucrase(DSRB) from Leuconostoc mesenteroides NRRL B-1299 synthesizing only a alpha (1–6) glucan. FEMS Microbiol. Lett. 1998, 159, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Yalin, Y.; Jin, L.; Jianhua, W.; Da, T.; Zigang, T. Expression and characterization of dextransucrase gene dsrX from Leuconostoc mesenteroides in Escherichia coli. J. Biotechnol. 2008, 133, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Malten, M.; Hollmann, R.; Deckwer, W.D.; Jahn, D. Production and secretion of recombinant Leuconostoc mesenteroides dextransucrase DsrS in Bacillus megaterium. Biotechnol. Bioeng. 2005, 89, 206–218. [Google Scholar] [CrossRef]
- Ul-Qader, S.A.; Aman, A.; Bano, S.; Syed, N.; Azhar, A. The effect of calcium ions and temperature on the production, activity, & stability of dextransucrase from the newly isolated strain Leuconostoc mesenteroides PCSIR-4. Rom. J. Biochem. 2008, 45, 159–168. [Google Scholar]




| N-Terminal Truncation Primers | Sequence (5′to 3′) |
|---|---|
| pET-30a (+)-ΔN 20 LmDexA | TAGTCATGAGCTTTTGCATTAACCGCCTC |
| pET-30a (+)-ΔN 39 LmDexA | CAGTCATGACAGAACACTACGGTTACCGA |
| pET-30a (+)-ΔN 99 LmDexA | CAGTCATGACAATCTGCTGATAATAATGTG |
| pET-30a (+)-ΔN148LmDexA | CAGTCATGATTAGCGGCAAGTACGTTGAA |
| pET-30a (+)-ΔN190LmDexA | TAGTCATGATCAAAGGACAGTATGTCACAAT |
| pET-30a (+)-ΔN280LmDexA | TAGTCATGATGATTGATGGTCAAATAATGAC |
| Strain | Dextransucrase | Protein Size (aa) | Expression Plasmid | Expression Strain | Crude Enzyme Activity | References |
|---|---|---|---|---|---|---|
| NRRL B-512F | dsrS | 1527 | pTrc99A | DH1 | 0.2 U/mL | [41] |
| dsrS | 1527 | pBad/Thio-TOPO | One Shot Top 10 | 5.85 U/mL | [42] | |
| dsrS | 1527 | pET-23d | BL21(DE3). | 0.85 U/mL | [43] | |
| dsrT | 1015 | pET-23d | BL21(DE3) | 0.17 U/mL | [43] | |
| dsrT5 | 1499 | pET-23d | BL21(DE3) | 1.9 U/mL | [43] | |
| NRRL B-1299 | dsrB | 1508 | pTrc 99A | DH1 | 2.0 mU/mL | [44] |
| dsrE | 2835 | pBad/Thio-TOPO | One Shot Top 10 | 0.58 U/mL | [20] | |
| DsrM | 2043 | pET-55-DEST | BL21(DE3) | 2.28 U/mL | [2] | |
| DSDP | 1279 | pENTR/D-TOPO | BL21(DE3) | 0.75 U/mL | [2] | |
| Dsr-MΔ1 | 1411 | pENTR/D-TOPO | BL21(DE3) | 60 U/mL | [23] | |
| Dsr-MΔ2 | 1263 | pENTR/D-TOPO | BL21(DE3) | 67 U/mL | [23] | |
| 0326 | dex-YG | 1501 | pET-28a(+) | BL21(DE3) | 36 U/mL | [31] |
| NN710 | LmDexA | 1562 | pET-30a(+) | Rosetta (DE3) | 2.49 U/mL | This work |
| ΔN190LmDexA | 1372 | pET-30a(+) | Rosetta (DE3) | 107.7 U/mL | This work | |
| CGMCC 1.544 | dsrX | 1522 | pET-28a(+) | BL21(DE3) | 8.8 U/mL | [45] |
| NRRL B-512F | dsrS | 1527 | pMM1520 | B. megaterium | 65 mU/mL | [46] |
| MS941 (ΔnprM) | 28.6 U/mL | [46] | ||||
| Lcc4 | dsrD | 1527 | pNZ124 | L. lactis MG1363 | 0.8 U/mL | [33] |
| Crude Enzyme Solution | Volume (mL) | Protein Concentration (mg/mL) | Specific Activity (U/mg) | Total Activity (U) | Purification (Fold) | Yield % (Total Activity) |
|---|---|---|---|---|---|---|
| ΔN190LmDexA | 4.0 | 16.55 | 6.51 | 430.8 | 1 | 100 |
| NiΔN190LmDexA | 2.0 | 0.6 | 126.13 | 151.78 | 19.37 | 35.23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuo, X.; Pan, L.; Zhang, W.; Zhu, J.; Qin, Y.; Xu, X.; Wang, Q. The Discovery, Molecular Cloning, and Characterization of Dextransucrase LmDexA and Its Active Truncated Mutant from Leuconostoc mesenteroides NN710. Molecules 2024, 29, 3242. https://doi.org/10.3390/molecules29133242
Zuo X, Pan L, Zhang W, Zhu J, Qin Y, Xu X, Wang Q. The Discovery, Molecular Cloning, and Characterization of Dextransucrase LmDexA and Its Active Truncated Mutant from Leuconostoc mesenteroides NN710. Molecules. 2024; 29(13):3242. https://doi.org/10.3390/molecules29133242
Chicago/Turabian StyleZuo, Xiaoqiong, Lixia Pan, Wenchao Zhang, Jing Zhu, Yan Qin, Xiuying Xu, and Qingyan Wang. 2024. "The Discovery, Molecular Cloning, and Characterization of Dextransucrase LmDexA and Its Active Truncated Mutant from Leuconostoc mesenteroides NN710" Molecules 29, no. 13: 3242. https://doi.org/10.3390/molecules29133242
APA StyleZuo, X., Pan, L., Zhang, W., Zhu, J., Qin, Y., Xu, X., & Wang, Q. (2024). The Discovery, Molecular Cloning, and Characterization of Dextransucrase LmDexA and Its Active Truncated Mutant from Leuconostoc mesenteroides NN710. Molecules, 29(13), 3242. https://doi.org/10.3390/molecules29133242






