A Visual Compendium of Principal Modifications within the Nucleic Acid Sugar Phosphate Backbone
Abstract
1. Introduction
2. Natural Nucleic Acids and Modification Directions
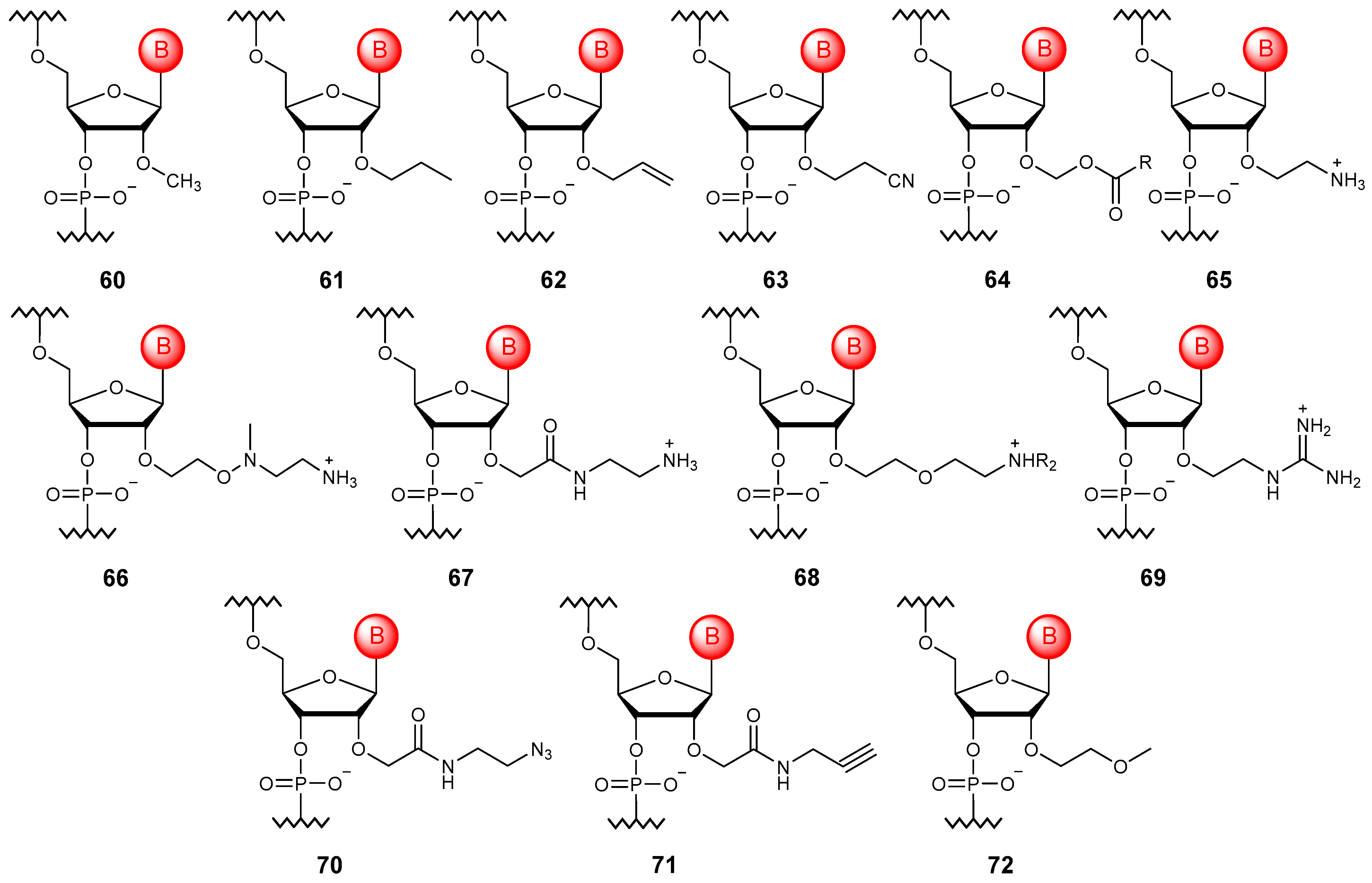
3. Phosphate Modifications
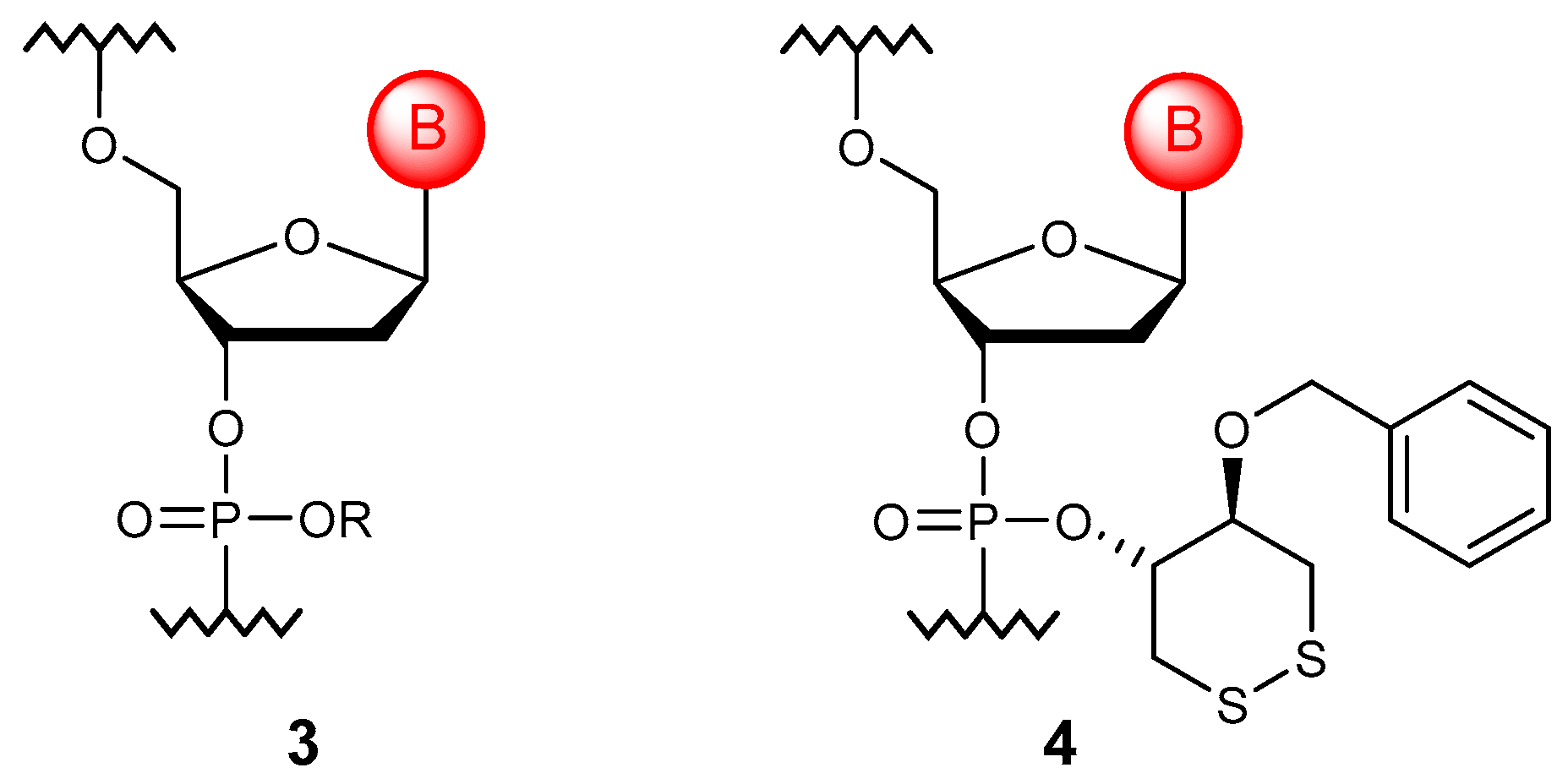
3.1. Phosphotriesters
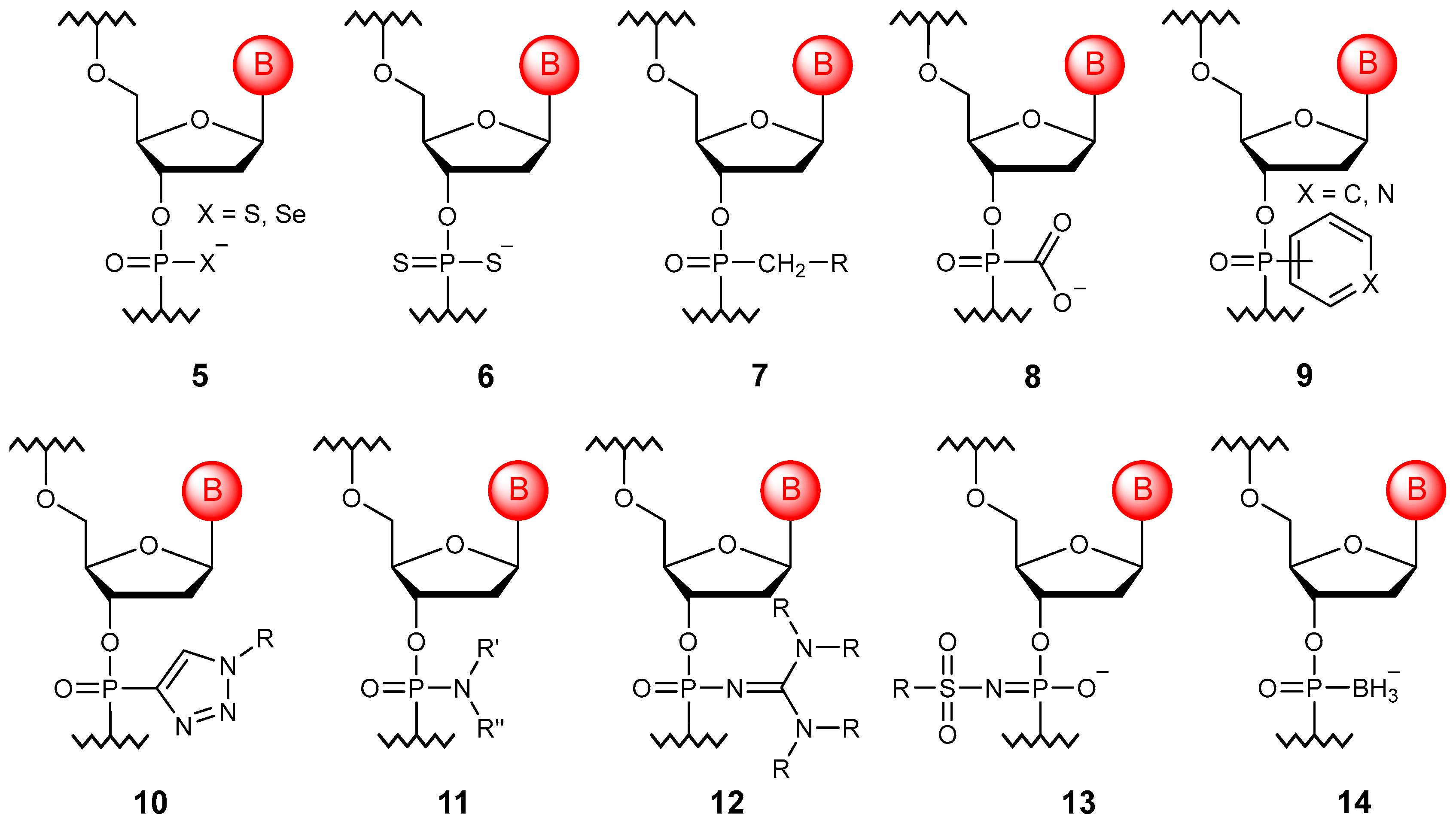
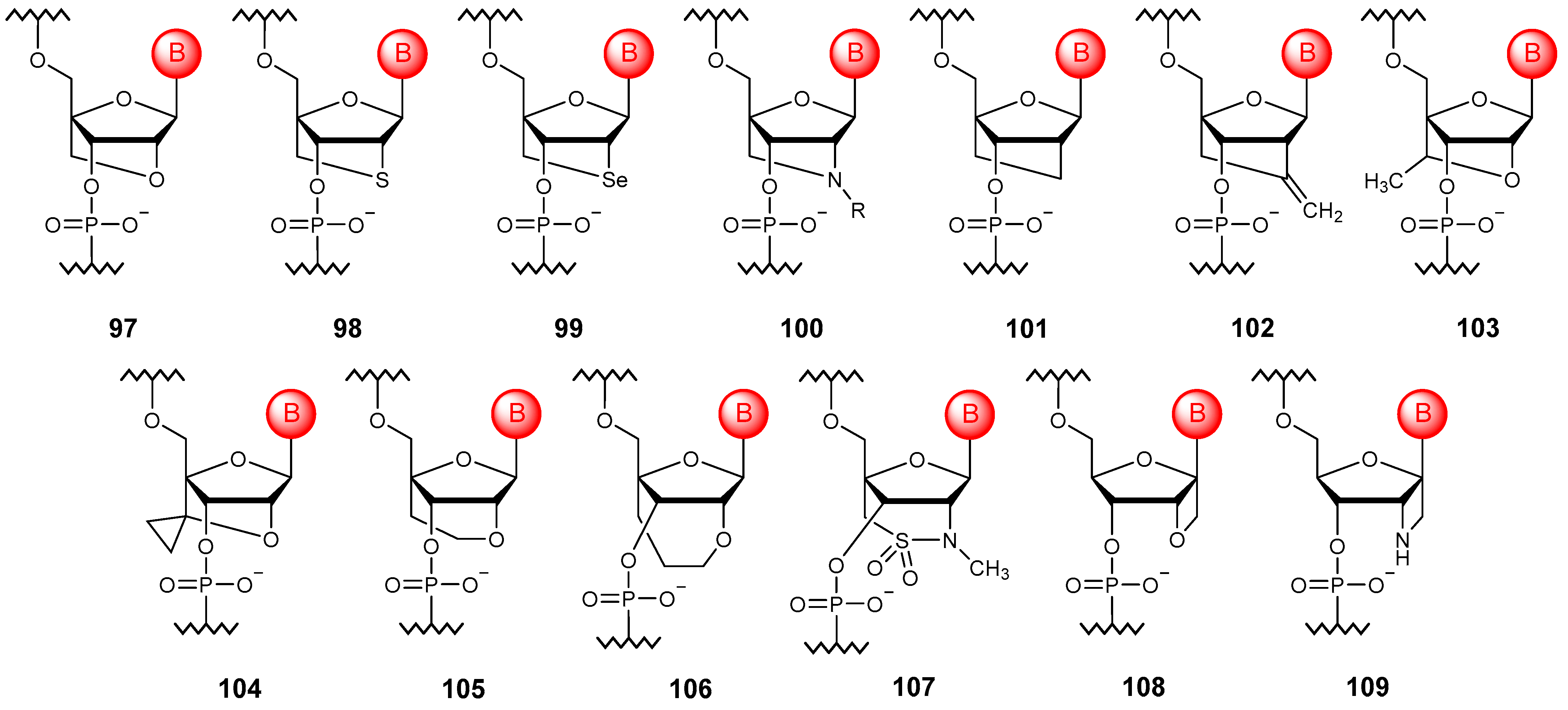
3.2. Substitution of Non-Bridging Oxygen Atoms
3.3. Substitution of Bridging Oxygen Atoms
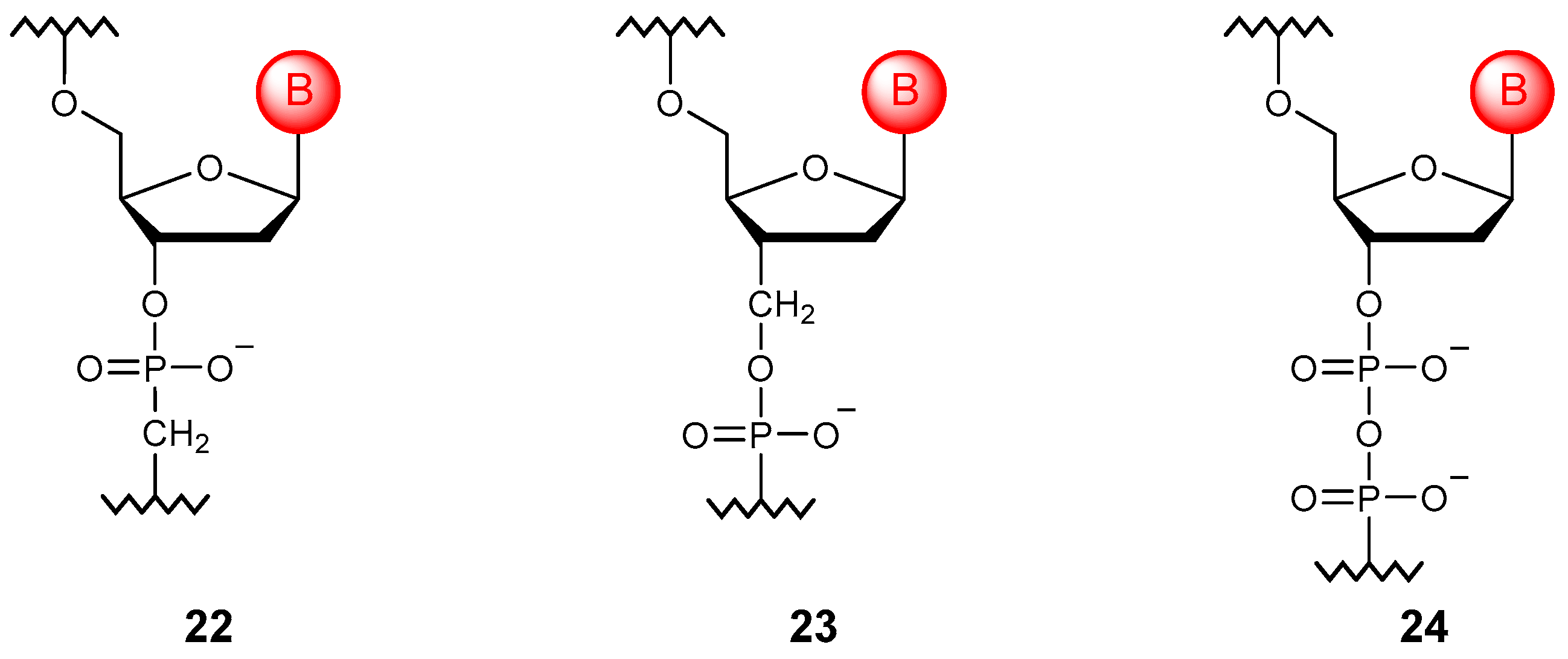
3.4. Phosphate Linkage Extension
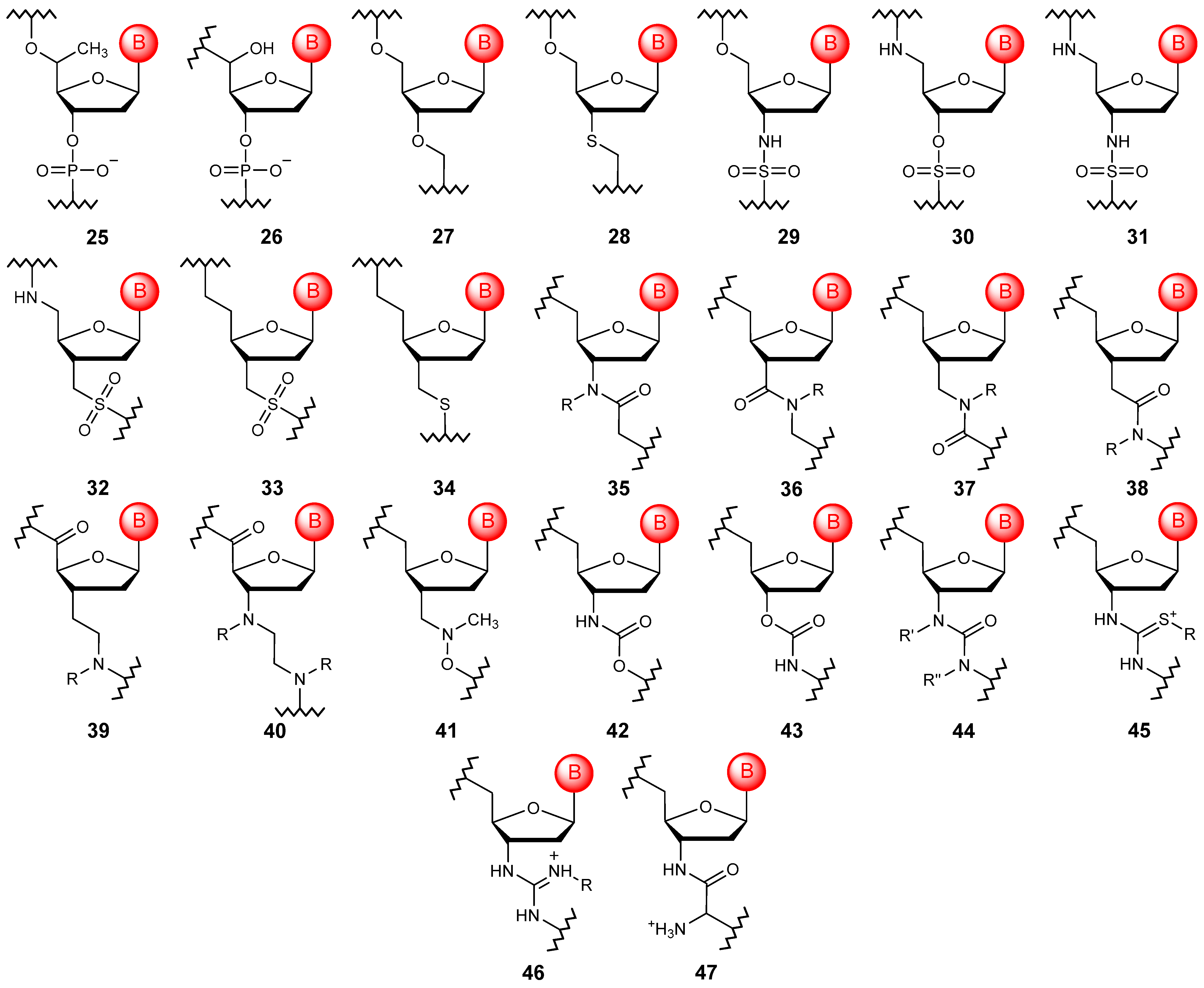
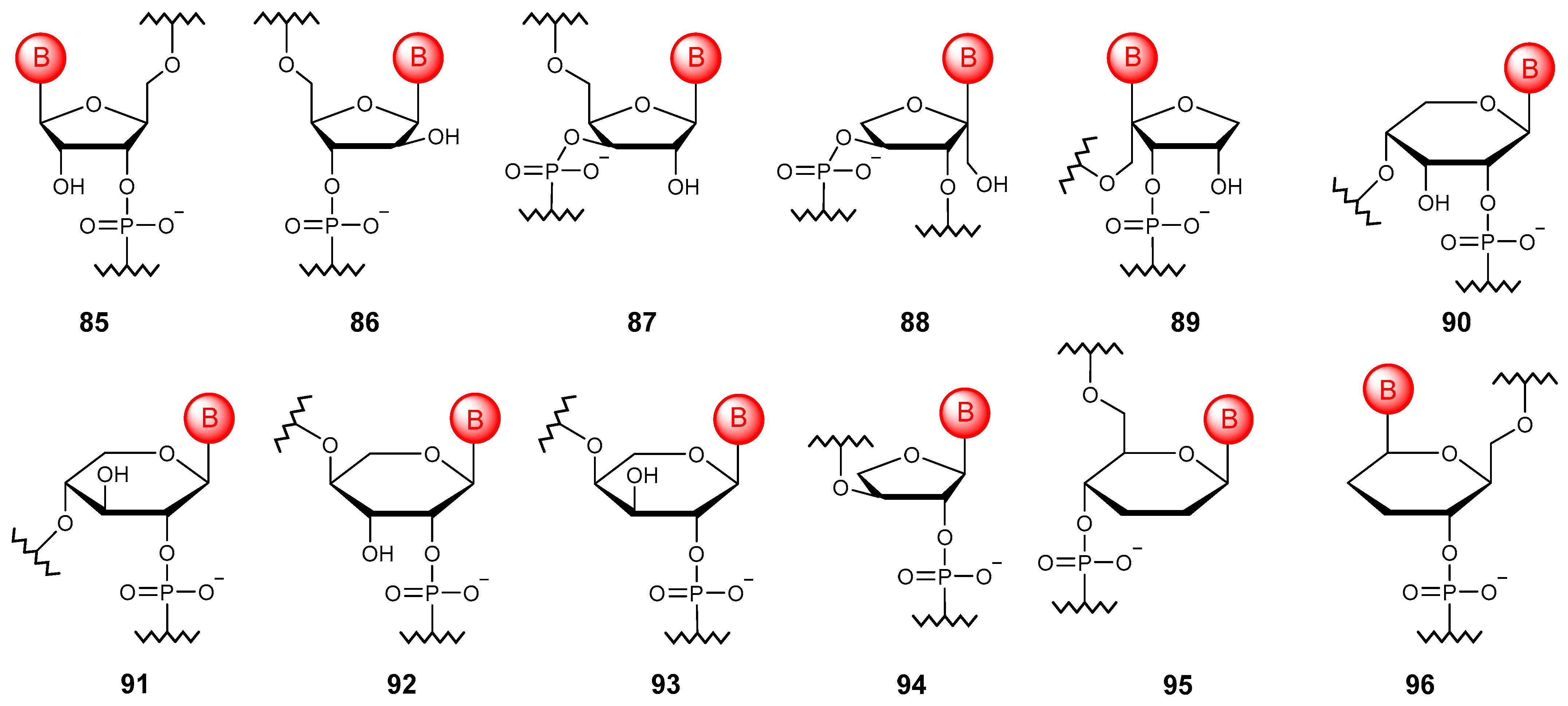
4. Sugar Linking Backbone Modifications
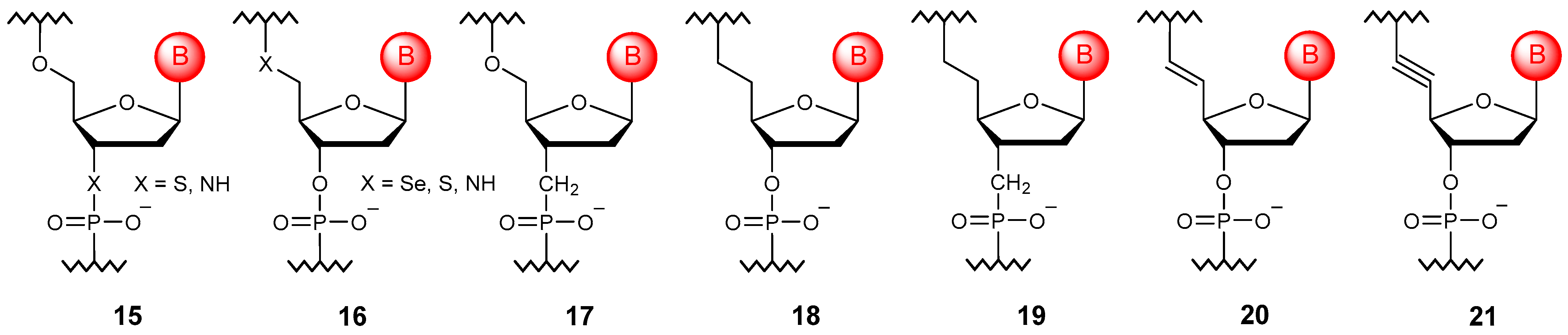
4.1. Acyclic Linkages
4.2. Cyclic Linkages
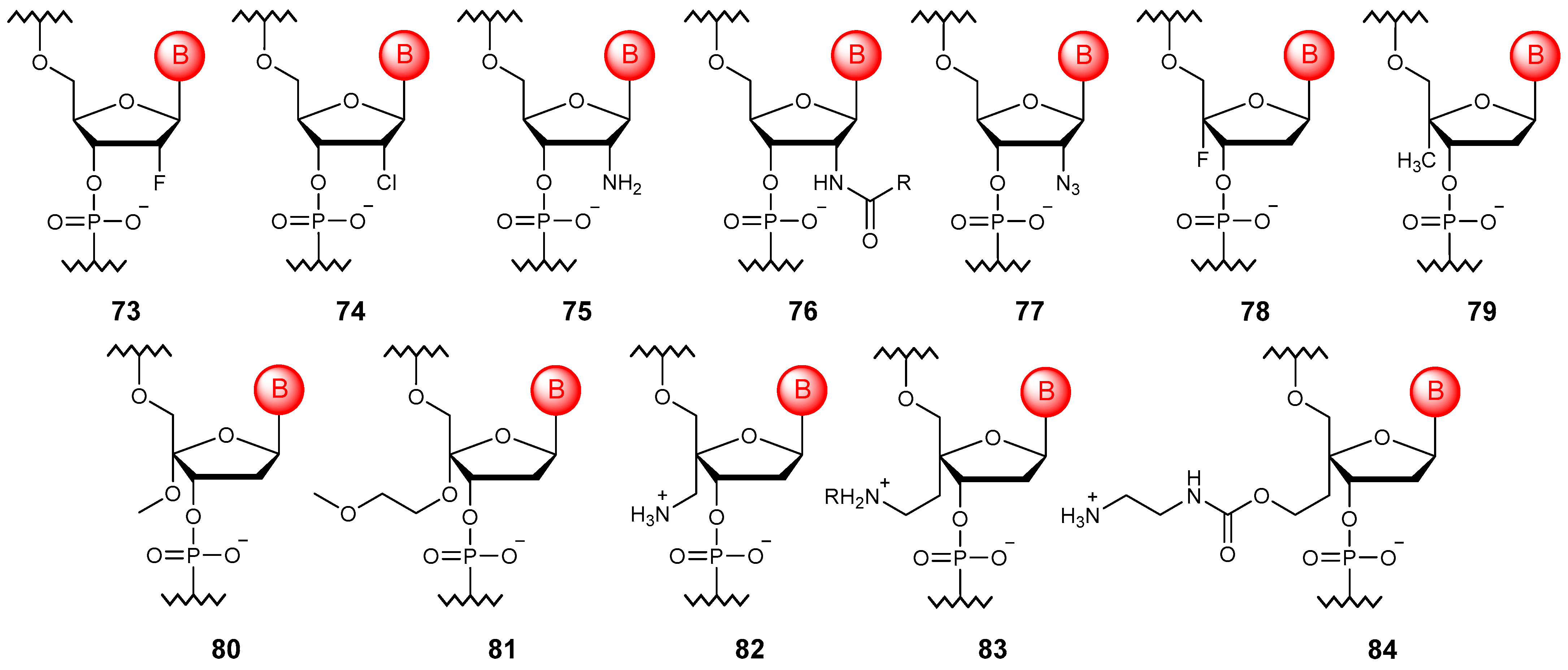
5. Sugar Modifications
5.1. Esterification of Free Hydroxy Groups in RNA
5.2. The Introduction of Substituents into the Ribose Ring
5.3. Alternative Sugar Moieties
5.3.1. Ribose Isomers
5.3.2. Other Sugars
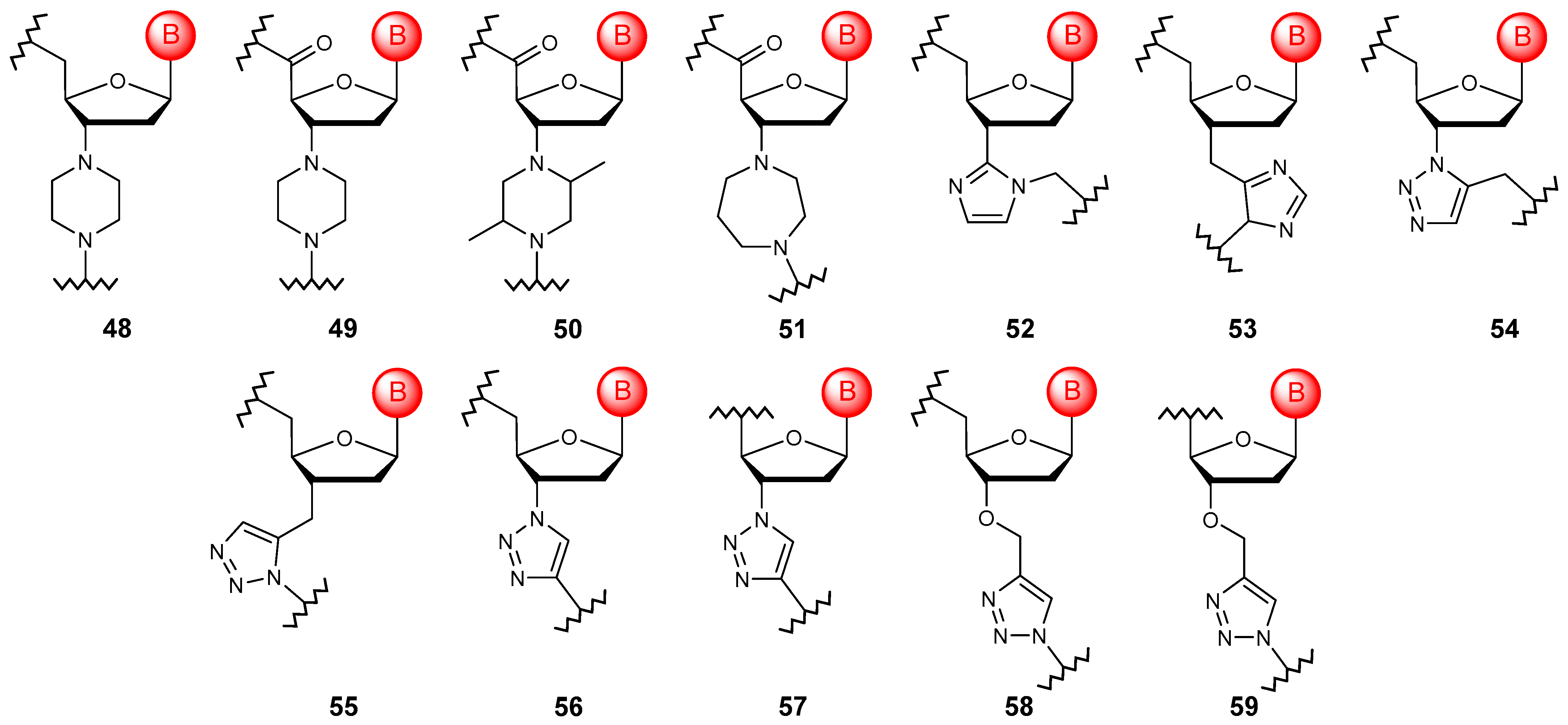
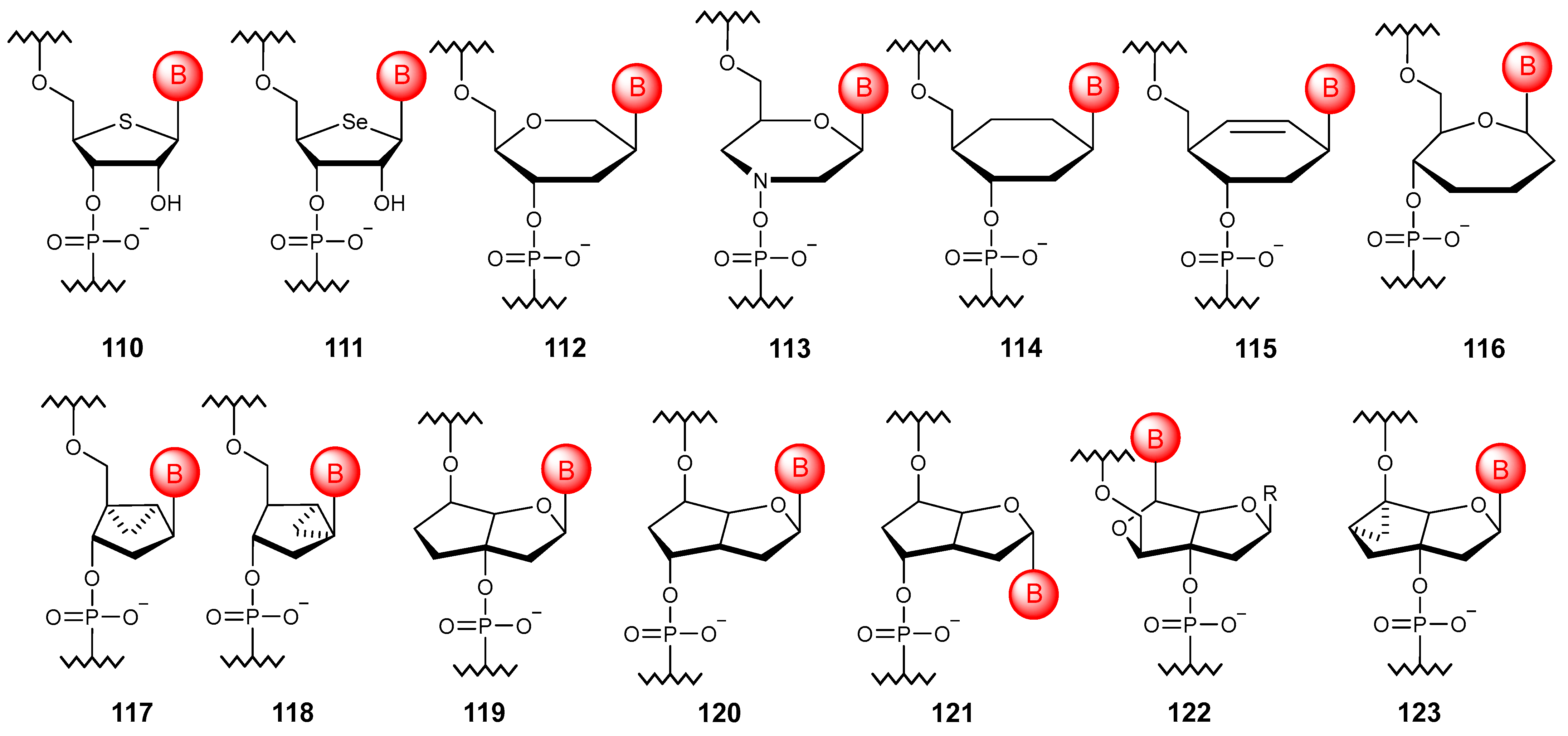
5.4. Bridged Ribose Ring Modifications
5.5. Substitution with Non-Sugar Cycles
5.5.1. Monocyclic Substitution
5.5.2. Bicyclic and Tricyclic Substitutions
5.6. Acyclic Substitutions
6. Alternative Scaffolds for Nitrogenous Bases
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Breaker, R.R. Natural and engineered nucleic acids as tools to explore biology. Nature 2004, 432, 838–845. [Google Scholar] [CrossRef]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef]
- Ivanov, G.S.; Tribulovich, V.G.; Pestov, N.B.; David, T.I.; Amoah, A.S.; Korneenko, T.V.; Barlev, N.A. Artificial genetic polymers against human pathologies. Biol. Direct 2022, 17, 39. [Google Scholar] [CrossRef]
- Friedrich, M.; Aigner, A. Therapeutic siRNA: State-of-the-art and future perspectives. BioDrugs 2022, 36, 549–571. [Google Scholar] [CrossRef]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug. Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef]
- Salem, A.K.; Weiner, G.J. CpG oligonucleotides as immunotherapeutic adjuvants: Innovative applications and delivery strategies. Adv. Drug. Deliv. Rev. 2009, 61, 193–194. [Google Scholar] [CrossRef]
- Hendel, A.; Bak, R.O.; Clark, J.T.; Kennedy, A.B.; Ryan, D.E.; Roy, S.; Steinfeld, I.; Lunstad, B.D.; Kaiser, R.J.; Wilkens, A.B.; et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015, 33, 985–989. [Google Scholar] [CrossRef]
- Cromwell, C.R.; Sung, K.; Park, J.; Krysler, A.R.; Jovel, J.; Kim, S.K.; Hubbard, B.P. Incorporation of bridged nucleic acids into CRISPR RNAs improves Cas9 endonuclease specificity. Nat. Commun. 2018, 9, 1448. [Google Scholar] [CrossRef]
- Gheibi-Hayat, S.M.; Jamialahmadi, K. Antisense oligonucleotide (AS-ODN) technology: Principle, mechanism and challenges. Biotechnol. Appl. Biochem. 2021, 68, 1086–1094. [Google Scholar] [CrossRef]
- Dhuri, K.; Bechtold, C.; Quijano, E.; Pham, H.; Gupta, A.; Vikram, A.; Bahal, R. Antisense oligonucleotides: An emerging area in drug discovery and development. J. Clin. Med. 2020, 9, 2004. [Google Scholar] [CrossRef]
- McCown, P.J.; Ruszkowska, A.; Kunkler, C.N.; Breger, K.; Hulewicz, J.P.; Wang, M.C.; Springer, N.A.; Brown, J.A. Naturally occurring modified ribonucleosides. Wiley Interdiscip. Rev. RNA 2020, 11, e1595. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Tena, M.; Chen, S.-K.; Winssinger, N. Supernatural: Artificial nucleobases and backbones to program hybridization-based assemblies and circuits. Bioconjugate Chem. 2023, 34, 111–123. [Google Scholar] [CrossRef]
- Lee, K.H.; Hamashima, K.; Kimoto, M.; Hirao, I. Genetic alphabet expansion biotechnology by creating unnatural base pairs. Curr. Opin. Biotechnol. 2018, 51, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.R.; Mohanan, K.; Luvino, D.; Floquet, N.; Baraguey, C.; Smietana, M.; Vasseur, J.-J. Expanding the borononucleotide family: Synthesis of borono-analogues of dCMP, dGMP and dAMP. Org. Biomol. Chem. 2009, 7, 4369–4377. [Google Scholar] [CrossRef]
- Miller, P.S.; Barrett, J.C.; Ts’o, P.O.P. Alkyl phosphotriesters of dinucleotides and oligonucleotides. 4. Synthesis of oligodeoxyribonucleotide ethyl phosphotriesters and their specific complex formation with transfer ribonucleic acid. Biochemistry 1974, 13, 4887–4896. [Google Scholar] [CrossRef]
- Miller, P.S.; Fang, K.N.; Kondo, N.S.; Ts’o, P.O.P. Conformation and interaction of dinucleoside mono- and diphosphates. V. Syntheses and properties of adenine and thymine nucleoside alkyl phosphotriesters, the neutral analogs of dinucleoside monophosphates. J. Am. Chem. Soc. 1971, 93, 6657–6665. [Google Scholar] [CrossRef] [PubMed]
- Stec, W.J.; Zon, G.; Gallo, K.A.; Byrd, R.A.; Uznanski, B.; Guga, P. Synthesis and absolute configuration of P-chiral O-isopropyl oligonucleotide triesters. Tetrahedron Lett. 1985, 26, 2191–2194. [Google Scholar] [CrossRef]
- Letsinger, R.L.; Bach, S.A.; Eadie, J.S. Effects of pendant groups at phosphorus on binding properties of d-ApA analogues. Nucleic Acids Res. 1986, 14, 3487–3499. [Google Scholar] [CrossRef]
- Wenninger, D.; Hinz, M.; Hahner, S.; Hillenkamp, F.; Seliger, H. Enzymatic and hybridization properties of oligonucleotide analogues containing novel phosphotriester internucleotide linkage. Nucleosides Nucleotides 1998, 17, 2117–2125. [Google Scholar] [CrossRef]
- Lancelot, G.; Guesnet, J.L.; Asseline, U.; Thuong, N.T. NMR studies of complex formation between the modified oligonucleotide d(T*TCTGT) covalently linked to an acridine derivative and its complementary sequence d(GCACAGAA). Biochemistry 1988, 27, 1265–1273. [Google Scholar] [CrossRef]
- Monfregola, L.; Caruthers, M.H. Solid-Phase Synthesis, hybridizing ability, uptake, and nuclease resistant profiles of position-selective cationic and hydrophobic phosphotriester oligonucleotides. J. Org. Chem. 2015, 80, 9147–9158. [Google Scholar] [CrossRef]
- Tosquellas, G.; Alvarez, K.; Dell’Aquila, C.; Morvan, F.; Vasseur, J.-J.; Imbach, J.-L.; Rayner, B. The pro-oligonucleotide approach: Solid phase synthesis and preliminary evaluation of model pro-dodecathymidylates. Nucleic Acids Res. 1998, 26, 2069–2074. [Google Scholar] [CrossRef]
- Hayashi, J.; Samezawa, Y.; Ochi, Y.; Wada, S.; Urata, H. Syntheses of prodrug-type phosphotriester oligonucleotides responsive to intracellular reducing environment for improvement of cell membrane permeability and nuclease resistance. Bioorg. Med. Chem. Lett. 2017, 27, 3135–3138. [Google Scholar] [CrossRef]
- Sugimoto, N.; Hayashi, J.; Funaki, R.; Wada, S.; Wada, F.; Harada-Shiba, M.; Urata, H. Prodrug-type phosphotriester oligonucleotides with linear disulfide promoieties responsive to reducing environment. ChemBioChem 2023, 24, e202300526. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Olesiak, M.; Padar, P.; McCuen, H.; Caruthers, M.H. Reduction of metal ions by boranephosphonate DNA. Org. Biomol. Chem. 2012, 10, 9130–9133. [Google Scholar] [CrossRef]
- Heinonen, P.; Lönnberg, H. Synthesis of phosphate-branched oligonucleotides. Bioconjugate Chem. 2004, 15, 1158–1160. [Google Scholar] [CrossRef] [PubMed]
- Jahns, H.; Roos, M.; Imig, J.; Baumann, F.; Wang, Y.; Gilmour, R.; Hall, J. Stereochemical bias introduced during RNA synthesis modulates the activity of phosphorothioate siRNAs. Nat. Commun. 2015, 6, 6317. [Google Scholar] [CrossRef] [PubMed]
- Tram, K.; Wang, X.; Yan, H. Facile synthesis of oligonucleotide phosphoroselenoates. Org. Lett. 2007, 9, 5103–5106. [Google Scholar] [CrossRef]
- Liang, C.; Allen, L.C. Sulfur does not form double bonds in phosphorothioate anions. J. Am. Chem. Soc. 1987, 109, 6449–6453. [Google Scholar] [CrossRef]
- Rahman, S.M.A.; Baba, T.; Kodama, T.; Islam, M.A.; Obika, S. Hybridizing ability and nuclease resistance profile of backbone modified cationic phosphorothioate oligonucleotides. Bioorg. Med. Chem. 2012, 20, 4098–4102. [Google Scholar] [CrossRef]
- Knouse, K.W.; de Gruyter, J.N.; Schmidt, M.A.; Zheng, B.; Vantourout, J.C.; Kingston, C.; Mercer, S.E.; Mcdonald, I.M.; Olson, R.E.; Zhu, Y.; et al. Unlocking P(V): Reagents for chiral phosphorothioate synthesis. Science 2018, 361, 1234–1238. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, N.; Butler, D.C.D.; Svrzikapa, N.; Mohapatra, S.; Zlatev, I.; Sah, D.W.Y.; Meena; Standley, S.M.; Lu, G.; Apponi, L.H.; et al. Control of phosphorothioate stereochemistry substantially increases the efficacy of antisense oligonucleotides. Nat. Biotechnol. 2017, 35, 845–851. [Google Scholar] [CrossRef] [PubMed]
- Kamaike, K.; Hirose, K.; Kayama, Y.; Kawashima, E. Synthesis of oligonucleoside phosphorodithioates on a solid support by the H-phosphonothioate method. Tetrahedron 2006, 62, 11814–11820. [Google Scholar] [CrossRef]
- Subach, M.F.; Khrenova, M.G.; Zvereva, M.I. Modern methods of aptamer chemical modification and principles of aptamer library selection. Mosc. Univ. Chem. Bull. 2024, 79, 79–85. [Google Scholar] [CrossRef]
- Xu, D.; Rivas-Bascón, N.; Padial, N.M.; Knouse, K.W.; Zheng, B.; Vantourout, J.C.; Schmidt, M.A.; Eastgate, M.D.; Baran, P.S. Enantiodivergent formation of C–P bonds: Synthesis of P-chiral phosphines and methyl-phosphonate oligonucleotides. J. Am. Chem. Soc. 2020, 142, 5785–5792. [Google Scholar] [CrossRef] [PubMed]
- Arangundy-Franklin, S.; Taylor, A.I.; Porebski, B.T.; Genna, V.; Peak-Chew, S.; Vaisman, A.; Woodgate, R.; Orozco, M.; Holliger, P. A synthetic genetic polymer with an uncharged backbone chemistry based on alkyl phosphonate nucleic acids. Nat. Chem. 2019, 11, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Fathi, R.; Huang, Q.; Syi, J.L.; Delaney, W.; Cook, A.F. (Aminomethyl)phosphonate derivatives of oligonucleotides. Bioconjugate Chem. 1994, 5, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Fathi, R.; Huang, Q.; Coppola, G.; Delaney, W.; Teasdale, R.; Krieg, A.M.; Cook, A.F. Oligonucleotides with novel, cationic backbone substituents: Aminoethylphosphonates. Nucleic Acids Res. 1994, 22, 5416–5424. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sheehan, D.; Lunstad, B.; Yamada, C.M.; Stell, B.G.; Caruthers, M.H.; Dellinger, D.J. Biochemical properties of phosphonoacetate and thiophosphonoacetate oligodeoxyribonucleotides. Nucleic Acids Res. 2003, 31, 4109–4118. [Google Scholar] [CrossRef]
- Yamada, C.M.; Dellinger, D.J.; Caruthers, M.H. Synthesis and biochemical evaluation of phosphonoformate oligodeoxyribonucleotides. J. Am. Chem. Soc. 2006, 128, 5251–5261. [Google Scholar] [CrossRef]
- Mag, M.; Muth, J.; Jahn, K.; Peyman, A.; Kretzschmar, G.; Engels, J.W.; Uhlmann, E. Synthesis and binding properties of oligodeoxynucleotides containing phenylphosphon(othio)ate linkages. Bioorg. Med. Chem. 1997, 5, 2213–2220. [Google Scholar] [CrossRef]
- Zmudzka, K.; Johansson, T.; Wojcik, M.; Janicka, M.; Nowak, M.; Stawinski, J.; Nawrot, B. Novel DNA analogues with 2-, 3- and 4-pyridylphosphonate internucleotide bonds: Synthesis and hybridization properties. New J. Chem. 2003, 27, 1698–1705. [Google Scholar] [CrossRef]
- Krishna, H.; Caruthers, M.H. Alkynyl phosphonate DNA: A versatile “click”able backbone for DNA-based biological applications. J. Am. Chem. Soc. 2012, 134, 11618–11631. [Google Scholar] [CrossRef]
- Jager, A.; Levy, M.J.; Hecht, S.M. Oligonucleotide N-alkylphosphoramidates: Synthesis and binding to polynucleotides. Biochemistry 1988, 27, 7237–7246. [Google Scholar] [CrossRef]
- Kupryushkin, M.S.; Zharkov, T.D.; Ilina, E.S.; Markov, O.V.; Kochetkova, A.S.; Akhmetova, M.M.; Lomzov, A.A.; Pyshnyi, D.V.; Lavrik, O.I.; Khodyreva, S.N. Triazinylamidophosphate oligonucleotides: Synthesis and study of their interaction with cells and DNA-binding proteins. Russ. J. Bioorganic Chem. 2021, 47, 719–733. [Google Scholar] [CrossRef]
- Michel, T.; Martinand-Mari, C.; Debart, F.; Lebleu, B.; Robbins, I.; Vasseur, J.-J. Cationic phosphoramidate-oligonucleotides efficiently target single-stranded DNA and RNA and inhibit hepatitis C virus IRES-mediated translation. Nucleic Acids Res. 2003, 31, 5282–5290. [Google Scholar] [CrossRef]
- Deglane, G.; Abes, S.; Michel, T.; Prévot, P.; Vives, E.; Debart, F.; Barvik, I.; Lebleu, B.; Vasseur, J.-J. Impact of the guanidinium group on hybridization and cellular uptake of cationic oligonucleotides. ChemBioChem 2006, 7, 684–692. [Google Scholar] [CrossRef]
- Kupryushkin, M.S.; Pyshnyi, D.V.; Stetsenko, D.A. Phosphoryl guanidines: A new type of nucleic acid analogues. Acta Naturae 2014, 6, 116–118. [Google Scholar] [CrossRef]
- Monian, P.; Shivalila, C.; Lu, G.; Shimizu, M.; Boulay, D.; Bussow, K.; Byrne, M.; Bezigian, A.; Chatterjee, A.; Chew, D.; et al. Endogenous ADAR-mediated RNA editing in non-human primates using stereopure chemically modified oligonucleotides. Nat. Biotechnol. 2022, 40, 1093–1102. [Google Scholar] [CrossRef]
- Chelobanov, B.P.; Burakova, E.A.; Prokhorova, D.V.; Fokina, A.A.; Stetsenko, D.A. New oligodeoxynucleotide derivatives containing N-(methanesulfonyl)-phosphoramidate (mesyl phosphoramidate) internucleotide group. Russ. J. Bioorg. Chem. 2017, 43, 664–668. [Google Scholar] [CrossRef]
- Patutina, O.A.; Gaponova Miroshnichenko, S.K.; Sen’kova, A.V.; Savin, I.A.; Gladkikh, D.V.; Burakova, E.A.; Fokina, A.A.; Maslov, M.A.; Shmendel’, E.V.; Wood, M.J.A.; et al. Mesyl phosphoramidate backbone modified antisense oligonucleotides targeting miR-21 with enhanced in vivo therapeutic potency. Proc. Natl. Acad. Sci. USA 2020, 117, 32370–32379. [Google Scholar] [CrossRef]
- Hara, R.I.; Saito, T.; Kogure, T.; Hamamura, Y.; Uchiyama, N.; Nukaga, Y.; Iwamoto, N.; Wada, T. Stereocontrolled synthesis of boranophosphate DNA by an oxazaphospholidine approach and evaluation of its properties. J. Org. Chem. 2019, 84, 7971–7983. [Google Scholar] [CrossRef]
- Takahashi, Y.; Kakuta, K.; Namioka, Y.; Igarashi, A.; Sakamoto, T.; Hara, R.I.; Sato, K.; Wada, T. Synthesis of P-Modified DNA from boranophosphate DNA as a precursor via acyl phosphite intermediates. J. Org. Chem. 2023, 88, 10617–10631. [Google Scholar] [CrossRef]
- Conlon, P.F.; Eguaogie, O.; Wilson, J.J.; Sweet, J.S.T.; Steinhoegl, J.; Englert, K.; Hancox, O.G.A.; Law, C.J.; Allman, S.A.; Tucker, J.H.R.; et al. Solid-phase synthesis and structural characterisation of phosphoroselenolate-modified DNA: A backbone analogue which does not impose conformational bias and facilitates SAD X-ray crystallography. Chem Sci. 2019, 10, 10948–10957. [Google Scholar] [CrossRef]
- Duschmalé, J.; Hansen, H.F.; Duschmalé, M.; Koller, E.; Albaek, N.; Møller, M.R.; Jensen, K.; Koch, T.; Wengel, J.; Bleicher, K. In vitro and in vivo properties of therapeutic oligonucleotides containing non-chiral 3′ and 5′ thiophosphate linkages. Nucleic Acids Res. 2020, 48, 63–74. [Google Scholar] [CrossRef]
- Barsky, D.; Colvin, M.E.; Zon, G.; Gryaznov, S.M. Hydration effects on the duplex stability of phosphoramidate DNA-RNA oligomers. Nucleic Acids Res. 1997, 25, 830–835. [Google Scholar] [CrossRef][Green Version]
- Ding, D.; Gryaznov, S.M.; Wilson, W.D. NMR solution structure of the N3′ --> P5′ phosphoramidate duplex d(CGCGAATTCGCG)2 by the iterative relaxation matrix approach. Biochemistry 1998, 37, 12082–12093. [Google Scholar] [CrossRef]
- Lelyveld, V.S.; Zhang, W.; Szostak, J.W. Synthesis of phosphoramidate-linked DNA by a modified DNA polymerase. Proc. Natl. Acad. Sci. USA 2020, 117, 7276–7283. [Google Scholar] [CrossRef]
- An, H.; Wang, T.; Maier, M.A.; Manoharan, M.; Ross, B.S.; Cook, P.D. Synthesis of novel 3′-C-methylene thymidine and 5-methyluridine/cytidine H-phosphonates and phosphonamidites for new backbone modification of oligonucleotides. J. Org. Chem. 2001, 66, 2789–2801. [Google Scholar] [CrossRef]
- Szabó, T.; Kers, A.; Stawinski, J. A new approach to the synthesis of the 5′-deoxy-5′-methylphosphonate linked thymidine oligonucleotide analogues. Nucleic Acids Res. 1995, 23, 893–900. [Google Scholar] [CrossRef]
- Hutter, D.; Blaettler, M.O.; Benner, S.A. From phosphate to bis(methylene) sulfone: Non-ionic backbone linkers in DNA. Helv. Chim. Acta 2002, 85, 2777–2806. [Google Scholar] [CrossRef]
- Parmar, R.; Willoughby, J.L.; Liu, J.; Foster, D.J.; Brigham, B.; Theile, C.S.; Charisse, K.; Akinc, A.; Guidry, E.; Pei, Y.; et al. 5′-(E)-Vinylphosphonate: A stable phosphate mimic can improve the RNAi activity of siRNA-GalNAc conjugates. ChemBioChem 2016, 17, 985–989. [Google Scholar] [CrossRef]
- Horiba, M.; Yamaguchi, T.; Obika, S. Synthesis and properties of oligonucleotides having ethynylphosphonate linkages. J. Org. Chem. 2020, 85, 1794–1801. [Google Scholar] [CrossRef]
- Dikmen, Z.G.; Wright, W.E.; Shay, J.W.; Gryaznov, S.M. Telomerase targeted oligonucleotide thio-phosphoramidates in T24-luc bladder cancer cells. J. Cell. Biochem. 2008, 104, 444–452. [Google Scholar] [CrossRef]
- Páv, O.; Košiová, I.; Barvík, I.; Pohl, R.; Buděšínský, M.; Rosenberg, I. Synthesis of oligoribonucleotides with phosphonate-modified linkages. Org. Biomol. Chem. 2011, 9, 6120–6126. [Google Scholar] [CrossRef]
- Sipova, H.; Springer, T.; Rejman, D.; Simak, O.; Petrova, M.; Novak, P.; Rosenbergova, S.; Pav, O.; Liboska, R.; Barvik, I.; et al. 5′-O-Methylphosphonate nucleic acids—New modified DNAs that increase the Escherichia coli RNase H cleavage rate of hybrid duplexes. Nucleic Acids Res. 2014, 42, 5378–5389. [Google Scholar] [CrossRef]
- Ahmadibeni, Y.; Parang, K. Synthesis and evaluation of modified oligodeoxynucleotides containing diphosphodiester internucleotide linkages. Angew. Chem. 2007, 46, 4739–4743. [Google Scholar] [CrossRef]
- Saha, A.K.; Waychunas, C.; Caulfield, T.J.; Upson, D.A.; Hobbs, C.; Yawman, A.M. 5′-Methyl-DNA—A new oligonucleotide analog: Synthesis and biochemical properties. J. Org. Chem. 1995, 60, 788–789. [Google Scholar] [CrossRef]
- Seth, P.P.; Allerson, C.R.; Siwkowski, A.; Vasquez, G.; Berdeja, A.; Migawa, M.T.; Gaus, H.; Prakash, T.P.; Bhat, B.; Swayze, E.E. Configuration of the 5′-methyl group modulates the biophysical and biological properties of locked nucleic acid (LNA) oligonucleotides. J. Med. Chem. 2010, 53, 8309–8318. [Google Scholar] [CrossRef]
- Prakash, T.P.; Lima, W.F.; Murray, H.M.; Li, W.; Kinberger, G.A.; Chappell, A.E.; Gaus, H.; Seth, P.P.; Bhat, B.; Crooke, S.T.; et al. Identification of metabolically stable 5′-phosphate analogs that support single-stranded siRNA activity. Nucleic Acids Res. 2015, 43, 2993–3011. [Google Scholar] [CrossRef]
- Králíková, Š.; Buděšínský, M.; Rosenberg, I. α-Hydroxyphosphonate oligonucleotides: A promising DNA type? Nucleos. Nucleot. Nucl. 2003, 22, 1061–1064. [Google Scholar] [CrossRef]
- Rozners, E.; Katkevica, D.; Strömberg, R. Oligoribonucleotide analogues containing a mixed backbone of phosphodiester and formacetal internucleoside linkages, together with vicinal 2′-O-methyl groups. ChemBioChem 2007, 8, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shaw, J.T.; Matteucci, M.D. Synthesis and hybridization property of an oligonucleotide containing a 3′-thioformcetal linked pentathymidylate. Bioorg. Med. Chem. Lett. 1999, 9, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Fettes, K.J.; Howard, N.; Hickman, D.T.; Adah, S.; Player, M.R.; Torrence, P.F.; Micklefield, J. Synthesis and nucleic-acid-binding properties of sulfamide- and 3′-N-sulfamate-modified DNA. J. Chem. Soc. Perkin Trans. 1 2002, 485–495. [Google Scholar] [CrossRef]
- Edward, M.H.; Mindy, R.K.; George, L.T. Oligonucleotides with a nuclease-resistant sulfur-based linkage. J. Org. Chem. 1992, 57, 4569–4570. [Google Scholar] [CrossRef]
- Micklefield, J.; Fettes, K.J. Sulfamide replacement of the phosphodiester linkage in dinucleotides: Synthesis and conformational analysis. Tetrahedron 1998, 54, 2129–2142. [Google Scholar] [CrossRef]
- Kurt, B.Z.; Dhara, D.; El-Sagheer, A.H.; Brown, T. Synthesis and properties of oligonucleotides containing LNA-sulfamate and sulfamide backbone linkages. Org. Lett. 2024, 26, 4137–4141. [Google Scholar] [CrossRef] [PubMed]
- Korotkovs, V.; Reichenbach, L.F.; Pescheteau, C.; Burley, G.A.; Liskamp, R.M.J. Molecular construction of sulfonamide antisense oligonucleotides. J. Org. Chem. 2019, 84, 10635–10648. [Google Scholar] [CrossRef]
- Huang, Z.; Benner, S.A. Oligodeoxyribonucleotide analogues with bridging dimethylene sulfide, sulfoxide, and sulfone groups. Toward a second-generation model of nucleic acid structure. J. Org. Chem. 2002, 67, 3996–4013. [Google Scholar] [CrossRef]
- Viswanadham, G.; Petersen, G.V.; Wengel, J. Incorporation of amide linked thymidine dimers into oligodeoxynucleotides. Bioorg. Med. Chem. Lett. 1996, 6, 987–990. [Google Scholar] [CrossRef]
- Rozners, E.; Katkevica, D.; Bizdena, E.; Strömberg, R. Synthesis and properties of RNA analogues having amides as interuridine linkages at selected positions. J. Am. Chem. Soc. 2003, 125, 12125–12136. [Google Scholar] [CrossRef]
- Tanui, P.; Kennedy, S.D.; Lunstad, B.D.; Haas, A.; Leake, D.; Rozners, E. Synthesis, biophysical studies and RNA interference activity of RNA having three consecutive amide linkages. Org. Biomol. Chem. 2014, 12, 1207–1210. [Google Scholar] [CrossRef] [PubMed]
- Baker, Y.R.; Thorpe, C.; Chen, J.; Poller, L.M.; Cox, L.; Kumar, P.; Lim, W.F.; Lie, L.; McClorey, G.; Epple, S.; et al. An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides. Nat. Commun. 2022, 13, 4036. [Google Scholar] [CrossRef]
- Morvan, F.; Sanghvi, Y.S.; Perbost, M.; Vasseur, J.J.; Bellon, L. Oligonucleotide mimics for antisense therapeutics: Solution phase and automated solid-support synthesis of MMI linked oligomers. J. Am. Chem. Soc. 1996, 118, 255–256. [Google Scholar] [CrossRef]
- Thorpe, C.; Epple, S.; Woods, B.; El-Sagheer, A.H.; Brown, T. Synthesis and biophysical properties of carbamate-locked nucleic acid (LNA) oligonucleotides with potential antisense applications. Org. Biomol. Chem. 2019, 17, 5341–5348. [Google Scholar] [CrossRef]
- Waldner, A.; Demesmaeker, A.; Lebreton, J.; Fritsch, V.; Wolf, R.M. Ureas as backbone replacements for the phosphodiester linkage in oligonucleotides. Synlett 1994, 1, 57–61. [Google Scholar] [CrossRef]
- Arya, D.P.; Bruice, T.C. Solid-phase synthesis of oligomeric deoxynucleic-thiourea (DNT) and deoxynucleic S-methylthiourea (DNmt): A neutral/polycationic analogue of DNA. Bioorg. Med. Chem. Lett. 2000, 10, 691–693. [Google Scholar] [CrossRef]
- Blaskó, A.; Dempcy, R.O.; Minyat, E.E.; Bruice, T.C. Association of Short-Strand DNA Oligomers with Guanidinium-Linked Nucleosides. A Kinetic and Thermodynamic Study. J. Am. Chem. Soc. 1996, 118, 7892–7899. [Google Scholar] [CrossRef]
- Meng, M.; Schmidtgall, B.; Ducho, C. Enhanced stability of DNA oligonucleotides with partially zwitterionic backbone structures in biological media. Molecules 2018, 23, 2941. [Google Scholar] [CrossRef]
- Petersen, G.V.; Wengel, J. Synthesis and characterization of short oligonucleotide segments containing nonnatural internucleoside amine- and amide linkages. Nucleosides Nucleotides Nucleic Acids 1995, 14, 925–928. [Google Scholar] [CrossRef]
- Beban, M.; Miller, P.S. Preparation of an imidazole-conjugated oligonucleotide. Bioconjugate Chem. 2000, 11, 599–603. [Google Scholar] [CrossRef]
- Matt, P.V.; Altmann, K.-H. Replacement of the phosphodiester linkage in oligonucleotides by heterocycles: The effect of triazole- and imidazole-modified backbones on DNA/RNA duplex stability. Bioorg. Med. Chem. Lett. 1997, 7, 1553–1556. [Google Scholar] [CrossRef]
- Nuzzi, A.; Massi, A.; Dondoni, A. Model studies toward the synthesis of thymidine oligonucleotides with triazole internucleosidic linkages via iterative Cu(I)-promoted azide–alkyne ligation chemistry. QSAR Comb. Sci. 2007, 26, 1191–1199. [Google Scholar] [CrossRef]
- Sanzone, A.P.; El-Sagheer, A.H.; Brown, T.; Tavassoli, A. Assessing the biocompatibility of click-linked DNA in Escherichia coli. Nucleic Acids Res. 2012, 40, 10567–10575. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; El-Sagheer, A.H.; Truong, L.; Brown, T. Locked nucleic acid (LNA) enhances binding affinity of triazole-linked DNA towards RNA. Chem. Commun. 2017, 53, 8910–8913. [Google Scholar] [CrossRef]
- Epple, S.; Modi, A.; Baker, Y.R.; Wȩgrzyn, E.; Traore, D.; Wanat, P.; Tyburn, A.E.S.; Shivalingam, A.; Taemaitree, L.; El-Sagheer, A.H.; et al. A new 1,5-disubstituted triazole DNA backbone mimic with enhanced polymerase compatibility. J. Am. Chem. Soc. 2021, 143, 16293–16301. [Google Scholar] [CrossRef]
- Motorin, Y.; Helm, M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2011, 2, 611–631. [Google Scholar] [CrossRef]
- Sabahi, A.; Guidry, J.; Inamati, G.B.; Manoharan, M.; Wittung-Stafshede, P. Hybridization of 2′-ribose modified mixed-sequence oligonucleotides: Thermodynamic and kinetic studies. Nucleic Acids Res. 2001, 29, 2163–2170. [Google Scholar] [CrossRef]
- Odadzic, D.; Bramsen, J.B.; Smicius, R.; Bus, C.; Kjems, J.; Engels, J.W. Synthesis of 2′-O-modified adenosine building blocks and application for RNA interference. Bioorg. Med. Chem. 2008, 16, 518–529. [Google Scholar] [CrossRef]
- Saneyoshi, H.; Seio, K.; Sekine, M. A general method for the synthesis of 2′-O-cyanoethylated oligoribonucleotides having promising hybridization affinity for DNA and RNA and enhanced nuclease resistance. J. Org. Chem. 2005, 70, 10453–10460. [Google Scholar] [CrossRef]
- Martin, A.R.; Lavergne, T.; Vasseur, J.-J.; Debart, F. Assessment of new 2′-O-acetalester protecting groups for regular RNA synthesis and original 2′-modified proRNA. Bioorg. Med. Chem. Lett. 2009, 19, 4046–4049. [Google Scholar] [CrossRef] [PubMed]
- Odadzic, D.; Engels, J.W. Different strategies for the synthesis of 2′-O-aminoethyl adenosine building blocks. Nucleos. Nucleot. Nucl. 2007, 26, 873–877. [Google Scholar] [CrossRef]
- Prakash, T.P.; Kawasaki, A.M.; Lesnik, E.A.; Sioufi, N.; Manoharan, M. Synthesis of 2′-O-[2-[(N,N-dialkylamino)oxy]ethyl]-modified oligonucleotides: Hybridization affinity, resistance to nuclease, and protein binding characteristics. Tetrahedron 2003, 59, 7413–7422. [Google Scholar] [CrossRef]
- Milton, S.; Honcharenko, D.; Rocha, C.S.J.; Moreno, P.M.D.; Edvard Smith, C.I.; Strömberg, R. Nuclease resistant oligonucleotides with cell penetrating properties. Chem. Commun. 2015, 51, 4044–4047. [Google Scholar] [CrossRef] [PubMed]
- Prhavc, M.; Prakash, T.P.; Minasov, G.; Cook, P.D.; Egli, M.; Manoharan, M. 2′-O-[2-[2-(N,N-Dimethylamino)ethoxy]ethyl] modified oligonucleotides: symbiosis of charge interaction factors and stereoelectronic effects. Org. Lett. 2003, 5, 2017–2020. [Google Scholar] [CrossRef]
- Prakash, T.P.; Püschl, A.; Lesnik, E.; Mohan, V.; Tereshko, V.; Egli, M.; Manoharan, M. 2′-O-[2-(Guanidinium)ethyl]-modified oligonucleotides: stabilizing effect on duplex and triplex structures. Org. Lett. 2004, 6, 1971–1974. [Google Scholar] [CrossRef]
- Wenska, M.; Milton, S.; Stromberg, R. Clickable 2′-O-alkyladenosine building blocks. Nucleic Acids Symp. Ser. 2007, 51, 149–150. [Google Scholar] [CrossRef]
- Egli, M.; Minasov, G.; Tereshko, V.; Pallan, P.S.; Teplova, M.; Inamati, G.B.; Lesnik, E.A.; Owens, S.R.; Ross, B.S.; Prakash, T.P.; et al. Probing the influence of stereoelectronic effects on the biophysical properties of oligonucleotides: Comprehensive analysis of the RNA affinity, nuclease resistance, and crystal structure of ten 2′-O-ribonucleic acid modifications. Biochemistry 2005, 44, 9045–9057. [Google Scholar] [CrossRef]
- Egli, M.; Manoharan, M. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res. 2023, 51, 2529–2573. [Google Scholar] [CrossRef] [PubMed]
- Guschlbauer, W.; Jankowski, K. Nucleoside conformation is determined by the electronegativity of the sugar substituent. Nucleic Acids Res. 1980, 8, 1421–1433. [Google Scholar] [CrossRef]
- Ono, T.; Scalf, M.; Smith, L.M. 2′-Fluoro modified nucleic acids: Polymerase-directed synthesis, properties and stability to analysis by matrix-assisted laser desorption/ionization mass spectrometry. Nucleic Acids Res. 1997, 25, 4581–4588. [Google Scholar] [CrossRef]
- Chen, T.; Romesberg, F.E. Enzymatic synthesis, amplification, and application of DNA with a functionalized backbone. Angew. Chem. Int. Ed. 2017, 56, 14046–14051. [Google Scholar] [CrossRef]
- Danielsen, M.B.; Lou, C.; Lisowiec-Wachnicka, J.; Pasternak, A.; Jorgensen, T.; Wengel, J. Gapmer antisense oligonucleotides containing 2′,3′-dideoxy-2′-fluoro-3′-C-hydroxymethyl-β-d-lyxofuranosyl nucleotides display site-specific RNase H cleavage and induce gene silencing. Chem. Eur. J. 2020, 26, 1368–1379. [Google Scholar] [CrossRef]
- Pham, J.W.; Radhakrishnan, I.; Sontheimer, E.J. Thermodynamic and structural characterization of 2′-nitrogen-modified RNA duplexes. Nucleic Acids Res. 2004, 32, 3446–3455. [Google Scholar] [CrossRef][Green Version]
- Fauster, K.; Hartl, M.; Santner, T.; Aigner, M.; Kreutz, C.; Bister, K.; Ennifar, E.; Micura, R. 2′-Azido RNA, a versatile tool for chemical biology: Synthesis, X-ray structure, siRNA applications, click labeling. ACS Chem. Biol. 2012, 7, 581–589. [Google Scholar] [CrossRef]
- Li, Q.; Chen, J.; Trajkovski, M.; Zhou, Y.; Fan, C.; Lu, K.; Tang, P.; Su, X.; Plavec, J.; Xi, Z.; et al. 4′-Fluorinated RNA: Synthesis, structure, and applications as a sensitive 19F NMR probe of RNA structure and function. J. Am. Chem. Soc. 2020, 142, 4739–4748. [Google Scholar] [CrossRef]
- Liboska, R.; Snášel, J.; Barvík, I.; Buděšínský, M.; Pohl, R.; Točík, Z.; Pav, O.; Rejman, D.; Novak, P.; Rosenberg, I. 4′-Alkoxy oligodeoxynucleotides: A novel class of RNA mimics. Org. Biomol. Chem. 2011, 9, 8261–8267. [Google Scholar] [CrossRef]
- Kanazaki, M.; Ueno, Y.; Shuto, S.; Matsuda, A. Highly nuclease-resistant phosphodiester-type oligodeoxynucleotides containing 4′α-C-aminoalkylthymidines form thermally stable duplexes with DNA and RNA. A candidate for potent antisense molecules. J. Am. Chem. Soc. 2000, 122, 2422–2432. [Google Scholar] [CrossRef]
- Malek-Adamian, E.; Patrascu, M.B.; Jana, S.K.; Martínez-Montero, S.; Moitessier, N.; Damha, M.J. Adjusting the structure of 2′-modified nucleosides and oligonucleotides via C4′-α-F or C4′-α-OMe substitution: Synthesis and conformational analysis. J. Org. Chem. 2018, 83, 9839–9849. [Google Scholar] [CrossRef]
- Koizumi, K.; Maeda, Y.; Kano, T.; Yoshida, H.; Sakamoto, T.; Yamagishi, K.; Ueno, Y. Synthesis of 4′-C-aminoalkyl-2′-O-methyl modified RNA and their biological properties. Bioorg. Med. Chem. 2018, 26, 3521–3534. [Google Scholar] [CrossRef]
- Vater, A.; Klussmann, S. Turning mirror-image oligonucleotides into drugs: The evolution of Spiegelmer® therapeutics. Drug Discov. Today 2015, 20, 147–155. [Google Scholar] [CrossRef]
- Watts, J.K.; Martín-Pintado, N.; Gómez-Pinto, I.; Schwartzentruber, J.; Portella, G.; Orozco, M.; González, C.; Damha, M.J. Differential stability of 2′F-ANA*RNA and ANA*RNA hybrid duplexes: Roles of structure, pseudohydrogen bonding, hydration, ion uptake and flexibility. Nucleic Acids Res. 2010, 38, 2498–2511. [Google Scholar] [CrossRef]
- Maiti, M.; Maiti, M.; Knies, C.; Dumbre, S.; Lescrinier, E.; Rosemeyer, H.; Ceulemans, A.; Herdewijn, P. Xylonucleic acid: Synthesis, structure, and orthogonal pairing properties. Nucleic Acids Res. 2015, 43, 7189–7200. [Google Scholar] [CrossRef]
- Efthymiou, T.; Gavette, J.; Stoop, M.; De Riccardis, F.; Froeyen, M.; Herdewyn, P.; Krishnamurthy, R. Chimeric XNA—An unconventional design for orthogonal informational systems. Chem. Eur. J. 2018, 24, 12811–12819. [Google Scholar] [CrossRef]
- Beier, M.; Reck, F.; Wagner, T.; Krishnamurthy, R.; Eschenmoser, A. Chemical etiology of nucleic acid structure: Comparing pentopyranosyl-(2′→4′) oligonucleotides with RNA. Science 1999, 283, 699–703. [Google Scholar] [CrossRef]
- Schöning, K.; Scholz, P.; Guntha, S.; Wu, X.; Krishnamurthy, R.; Eschenmoser, A. Chemical etiology of nucleic acid structure: The alpha-threofuranosyl-(3′-->2′) oligonucleotide system. Science 2000, 290, 1347–1351. [Google Scholar] [CrossRef]
- D’Alonzo, D.; Amato, J.; Schepers, G.; Froeyen, M.; Van Aerschot, A.; Herdewijn, P.; Guaragna, A. Enantiomeric selection properties of β-homoDNA: Enhanced pairing for heterochiral complexes. Angew. Chem. Int. Ed. Engl. 2013, 52, 6662–6665. [Google Scholar] [CrossRef]
- Nauwelaerts, K.; Lescrinier, E.; Herdewijn, P. Structure of the α-homo-DNA:RNA duplex and the function of twist and slide to catalogue nucleic acid duplexes. Chem. Eur. J. 2007, 13, 90–98. [Google Scholar] [CrossRef]
- Torigoe, H.; Hari, Y.; Sekiguchi, M.; Obika, S.; Imanishi, T. 2′-O,4′-C-Methylene bridged nucleic acid modification promotes pyrimidine motif triplex DNA formation at physiological pH: Thermodynamic and kinetic studies. J. Biol. Chem. 2001, 276, 2354–2360. [Google Scholar] [CrossRef]
- Koshkin, A.A.; Singh, S.K.; Nielsen, P.; Rajwanshi, V.K.; Kumar, R.; Meldgaard, M.; Olsen, C.E.; Wengel, J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 1998, 54, 3607–3630. [Google Scholar] [CrossRef]
- Obika, S.; Nanbu, D.; Hari, Y.; Morio, K.; In, Y.; Ishida, T.; Imanishi, T. Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3′-endo sugar puckering. Tetrahedron Lett. 1997, 38, 8735–8738. [Google Scholar] [CrossRef]
- Singh, S.K.; Koshkin, A.A.; Wengel, J.; Nielsen, P. LNA (locked nucleic acids): Synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998, 1998, 455–456. [Google Scholar] [CrossRef]
- Kumar, R.; Singh, S.K.; Koshkin, A.A.; Rajwanshi, V.K.; Meldgaard, M.; Wengel, J. The first analogues of LNA (Locked Nucleic Acids): Phosphorothioate-LNA and 2′-thio-LNA. Bioorg. Med. Chem. Lett. 1998, 8, 2219–2222. [Google Scholar] [CrossRef]
- Morihiro, K.; Kodama, T.; Kentefu, Y.M.; Veedu, R.N.; Obika, S. Selenomethylene locked nucleic acid enables reversible hybridization in response to redox changes. Angew. Chem. Int. Ed. Engl. 2013, 52, 5074–5078. [Google Scholar] [CrossRef]
- Sawamoto, H.; Arai, Y.; Yamakoshi, S.; Obika, S.; Kawanishi, E. Synthetic method for 2′-amino-LNA bearing any of the four nucleobases via a transglycosylation reaction. Org. Lett. 2018, 20, 1928–1931. [Google Scholar] [CrossRef]
- Danielsen, M.B.; Christensen, N.J.; Jorgensen, T.; Jensen, K.J.; Wengel, J.; Lou, C. Polyamine-functionalized 2′-amino-LNA in oligonucleotides: Facile synthesis of new monomers and high-affinity binding towards ssDNA and dsDNA. Chem. Eur. J. 2021, 27, 1416–1422. [Google Scholar] [CrossRef]
- Ejlersen, M.; Christensen, N.J.; Sorensen, K.K.; Jensen, K.J.; Wengel, J.; Lou, C. Synergy of two highly specific biomolecular recognition events: Aligning an AT-hook peptide in DNA minor grooves via covalent conjugation to 2′-amino-LNA. Bioconjugate Chem. 2018, 29, 1025–1029. [Google Scholar] [CrossRef]
- Shrestha, A.R.; Kotobuki, Y.; Hari, Y.; Obika, S. Guanidine bridged nucleic acid (GuNA): An effect of a cationic bridged nucleic acid on DNA binding affinity. Chem. Commun. 2014, 50, 575–577. [Google Scholar] [CrossRef]
- Lou, C.; Samuelsen, S.V.; Christensen, N.J.; Vester, B.; Wengel, J. Oligonucleotides containing aminated 2′-amino-LNA nucleotides: Synthesis and strong binding to complementary DNA and RNA. Bioconjugate Chem. 2017, 28, 1214–1220. [Google Scholar] [CrossRef]
- Ries, A.; Kumar, R.; Lou, C.; Kosbar, T.; Vengut-Climent, E.; Jorgensen, T.; Morales, J.C.; Wengel, J. Synthesis and biophysical investigations of oligonucleotides containing galactose-modified DNA, LNA, and 2′-amino-LNA monomers. J. Org. Chem. 2016, 81, 10845–10856. [Google Scholar] [CrossRef]
- Xu, J.; Liu, Y.; Dupouy, C.; Chattopadhyaya, J. Synthesis of conformationally locked carba-LNAs through intramolecular free-radical addition to C=N. Electrostatic and steric implication of the carba-LNA substituents in the modified oligos for nuclease and thermodynamic stabilities. J. Org. Chem. 2009, 74, 6534–6554. [Google Scholar] [CrossRef] [PubMed]
- Seth, P.P.; Allerson, C.R.; Berdeja, A.; Siwkowski, A.; Pallan, P.S.; Gaus, H.; Prakash, T.P.; Watt, A.T.; Egli, M.; Swayze, E.E. An exocyclic methylene group acts as a bio-isostere of the 2′-oxygen atom in LNA. J. Am. Chem. Soc. 2010, 132, 14942–14950. [Google Scholar] [CrossRef] [PubMed]
- Seth, P.P.; Siwkowski, A.; Allerson, C.R.; Vasquez, G.; Lee, S.; Prakash, T.P.; Wancewicz, E.V.; Witchell, D.; Swayze, E.E. Short antisense oligonucleotides with novel 2′-4′ conformationally restricted nucleoside analogues show improved potency without increased toxicity in animals. J. Med. Chem. 2009, 52, 10–13. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Horiba, M.; Obika, S. Synthesis and properties of 2′-O,4′-C-spirocyclopropylene bridged nucleic acid (scpBNA), an analogue of 2’,4′-BNA/LNA bearing a cyclopropane ring. Chem. Commun. 2015, 51, 9737–9740. [Google Scholar] [CrossRef] [PubMed]
- Morita, K.; Hasegawa, C.; Kaneko, M.; Tsutsumi, S.; Sone, J.; Ishikawa, T.; Imanishi, T.; Koizumi, M. 2′-O,4′-C-Ethylene-bridged nucleic acids (ENA): Highly nuclease-resistant and thermodynamically stable oligonucleotides for antisense drug. Bioorg. Med. Chem. Lett. 2002, 12, 73–76. [Google Scholar] [CrossRef] [PubMed]
- Morita, K.; Takagi, M.; Hasegawa, C.; Kaneko, M.; Tsutsumi, S.; Sone, J.; Ishikawa, T.; Imanishi, T.; Koizumi, M. Synthesis and properties of 2′-O,4′-C-ethylene-bridged nucleic acids (ENA) as effective antisense oligonucleotides. Bioorg. Med. Chem. 2003, 11, 2211–2226. [Google Scholar] [CrossRef] [PubMed]
- Mitsuoka, Y.; Fujimura, Y.; Waki, R.; Kugimiya, A.; Yamamoto, T.; Hari, Y.; Obika, S. Sulfonamide-bridged nucleic acid: Synthesis, high RNA selective hybridization, and high nuclease resistance. Org. Lett. 2014, 16, 5640–5643. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, Y.; Wu, Y.; Takahashi, Y.; Takakura, Y. Development of nucleic acid medicines based on chemical technology. Adv. Drug Deliv. Rev. 2023, 199, 114872. [Google Scholar] [CrossRef] [PubMed]
- Pradeepkumar, P.I.; Cheruku, P.; Plashkevych, O.; Acharya, P.; Gohil, S.; Chattopadhyaya, J. Synthesis, physicochemical and biochemical studies of 1′,2′-oxetane constrained adenosine and guanosine modified oligonucleotides, and their comparison with those of the corresponding cytidine and thymidine analogues. J. Am. Chem. Soc. 2004, 126, 11484–11499. [Google Scholar] [CrossRef] [PubMed]
- Honcharenko, D.; Varghese, O.P.; Plashkevych, O.; Barman, J.; Chattopadhyaya, J. Synthesis and structure of novel conformationally constrained 1′,2′-azetidine-fused bicyclic pyrimidine nucleosides: their incorporation into oligo-DNAs and thermal stability of the heteroduplexes. J. Org. Chem. 2006, 71, 299–314. [Google Scholar] [CrossRef]
- Hoshika, S.; Inoue, N.; Minakawa, N.; Matsuda, A. Investigation of physical and physiological properties of 4′-thioribonucleotide (4′-thioRNA). Nucleic Acids Res. 2003, 3, 209–210. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ota, M.; Takahashi, H.; Nogi, Y.; Kagotani, Y.; Saito-Tarashima, N.; Kondo, J.; Minakawa, N. Synthesis and properties of fully-modified 4′-selenoRNA, an endonuclease-resistant RNA analog. Bioorg. Med. Chem. 2022, 76, 117093. [Google Scholar] [CrossRef] [PubMed]
- De Bouvere, B.; Kerreinans, L.; Hendrix, C.; De Winter, H.; Schepers, G.; Van Aerschot, A.; Herdewijn, P. Hexitol nucleic acids (HNA): Synthesis and properties. Nucleosides Nucleotides 1997, 16, 973–976. [Google Scholar] [CrossRef]
- Chen, S.; Le, B.; Rahimizadeh, K.; Shaikh, K.; Mohal, N.; Veedu, R. Synthesis of a morpholino nucleic acid (MNA)-uridine phosphoramidite, and exon skipping using MNA/2′-O-methyl mixmer antisense oligonucleotide. Molecules 2016, 21, 1582. [Google Scholar] [CrossRef] [PubMed]
- Maurinsh, Y.; Rosemeyer, H.; Esnouf, R.; Medvedovici, A.; Wang, J.; Ceulemans, G.; Lescrinier, E.; Hendrix, C.; Busson, R.; Sandra, P.; et al. Synthesis and pairing properties of oligonucleotides containing 3-hydroxy-4-hydroxymethyl-1-cyclohexanyl nucleosides. Chem. Eur. J. 1999, 5, 2139–2150. [Google Scholar] [CrossRef]
- Wang, J.; Verbeure, B.; Luyten, I.; Lescrinier, E.; Froeyen, M.; Hendrix, C.; Rosemeyer, H.; Seela, F.; Van Aerschot, A.; Herdewijn, P. Cyclohexene nucleic acids (CeNA): serum stable oligonucleotides that activate RNase H and increase duplex stability with complementary RNA. J. Am. Chem. Soc. 2000, 122, 8595–8602. [Google Scholar] [CrossRef]
- Sabatino, D.; Damha, M.J. Oxepane nucleic acids: Synthesis, characterization, and properties of oligonucleotides bearing a seven-membered carbohydrate ring. J. Am. Chem. Soc. 2007, 129, 8259–8270. [Google Scholar] [CrossRef]
- Maier, M.A.; Choi, Y.; Gaus, H.; Barchi, J., Jr.; Marquez, V.E.; Manoharan, M. Synthesis and characterization of oligonucleotides containing conformationally constrained bicyclo[3.1.0]hexane pseudosugar analogs. Nucleic Acids Res. 2004, 32, 3642–3650. [Google Scholar] [CrossRef]
- Bolli, M.; Trafelet, H.U.; Leumann, C. Watson–Crick base-pairing properties of bicyclo-DNA. Nucleic Acids Res. 1996, 24, 4660–4667. [Google Scholar] [CrossRef]
- Evéquoz, D.; Leumann, C.J. Probing the backbone topology of DNA: Synthesis and properties of 7′,5′-bicyclo-DNA. Chem. Eur. J. 2017, 23, 7953–7968. [Google Scholar] [CrossRef]
- Evéquoz, D.; Verhaart, I.E.C.; Vijver, D.; Renner, W.; Aartsma-Rus, A.; Leumann, C.J. 7′,5′-alpha-bicyclo-DNA: New chemistry for oligonucleotide exon splicing modulation therapy. Nucleic Acids Res. 2021, 49, 12089–12105. [Google Scholar] [CrossRef]
- Raunkjaer, M.; Sørensen, M.D.; Wengel, J. Synthesis and thermal denaturation studies of novel 2′-O,3′-C-linked bicyclic oligonucleotides with a methoxy or a piperazino group facing the major groove of nucleic acid duplexes. Org. Biomol. Chem. 2005, 3, 130–135. [Google Scholar] [CrossRef]
- Aupy, P.; Echevarría, L.; Relizani, K.; Goyenvalle, A. The use of tricyclo-DNA oligomers for the treatment of genetic disorders. Biomedicines 2017, 6, 2. [Google Scholar] [CrossRef]
- Langkjær, N.; Pasternak, A.; Wengel, J. UNA (unlocked nucleic acid): A flexible RNA mimic that allows engineering of nucleic acid duplex stability. Bioorg. Med. Chem. 2009, 17, 5420–5425. [Google Scholar] [CrossRef]
- Merle, Y.; Bonneil, E.; Merle, L.; Sági, J.; Szemzö, A. Acyclic oligonucleotide analogues. Int. J. Biol. Macromol. 1995, 17, 239–246. [Google Scholar] [CrossRef]
- Le, B.T.; Murayama, K.; Shabanpoor, F.; Asanuma, H.; Veedu, R.N. Antisense oligonucleotide modified with serinol nucleic acid (SNA) induces exon skipping in mdx myotubes. RSC Adv. 2017, 7, 34049–34052. [Google Scholar] [CrossRef]
- Murayama, K.; Kashida, H.; Asanuma, H. Acyclic L-threoninol nucleic acid (L-aTNA) with suitable structural rigidity cross-pairs with DNA and RNA. Chem. Commun. 2015, 51, 6500–6503. [Google Scholar] [CrossRef]
- Schlegel, M.K.; Peritz, A.E.; Kittigowittana, K.; Zhang, L.; Meggers, E. Duplex formation of the simplified nucleic acid GNA. ChemBioChem 2007, 8, 927–932. [Google Scholar] [CrossRef]
- Menchise, V.; De Simone, G.; Tedeschi, T.; Corradini, R.; Sforza, S.; Marchelli, R.; Capasso, D.; Saviano, M.; Pedone, C. Insights into peptide nucleic acid (PNA) structural features: The crystal structure of a D-lysine-based chiral PNA-DNA duplex. Proc. Natl. Acad. Sci. USA 2003, 100, 12021–12026. [Google Scholar] [CrossRef] [PubMed]
- Bartolami, E.; Gilles, A.; Dumy, P.; Ulrich, S. Synthesis of α-PNA containing a functionalized triazine as nucleobase analogue. Tetrahedron Lett. 2015, 56, 2319–2323. [Google Scholar] [CrossRef]
- Suparpprom, C.; Vilaivan, T. Perspectives on conformationally constrained peptide nucleic acid (PNA): Insights into the structural design, properties and applications. RSC Chem. Biol. 2022, 3, 648–697. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novikova, D.; Sagaidak, A.; Vorona, S.; Tribulovich, V. A Visual Compendium of Principal Modifications within the Nucleic Acid Sugar Phosphate Backbone. Molecules 2024, 29, 3025. https://doi.org/10.3390/molecules29133025
Novikova D, Sagaidak A, Vorona S, Tribulovich V. A Visual Compendium of Principal Modifications within the Nucleic Acid Sugar Phosphate Backbone. Molecules. 2024; 29(13):3025. https://doi.org/10.3390/molecules29133025
Chicago/Turabian StyleNovikova, Daria, Aleksandra Sagaidak, Svetlana Vorona, and Vyacheslav Tribulovich. 2024. "A Visual Compendium of Principal Modifications within the Nucleic Acid Sugar Phosphate Backbone" Molecules 29, no. 13: 3025. https://doi.org/10.3390/molecules29133025
APA StyleNovikova, D., Sagaidak, A., Vorona, S., & Tribulovich, V. (2024). A Visual Compendium of Principal Modifications within the Nucleic Acid Sugar Phosphate Backbone. Molecules, 29(13), 3025. https://doi.org/10.3390/molecules29133025








