Insights into Actin Isoform-Specific Interactions with Myosin via Computational Analysis
Abstract
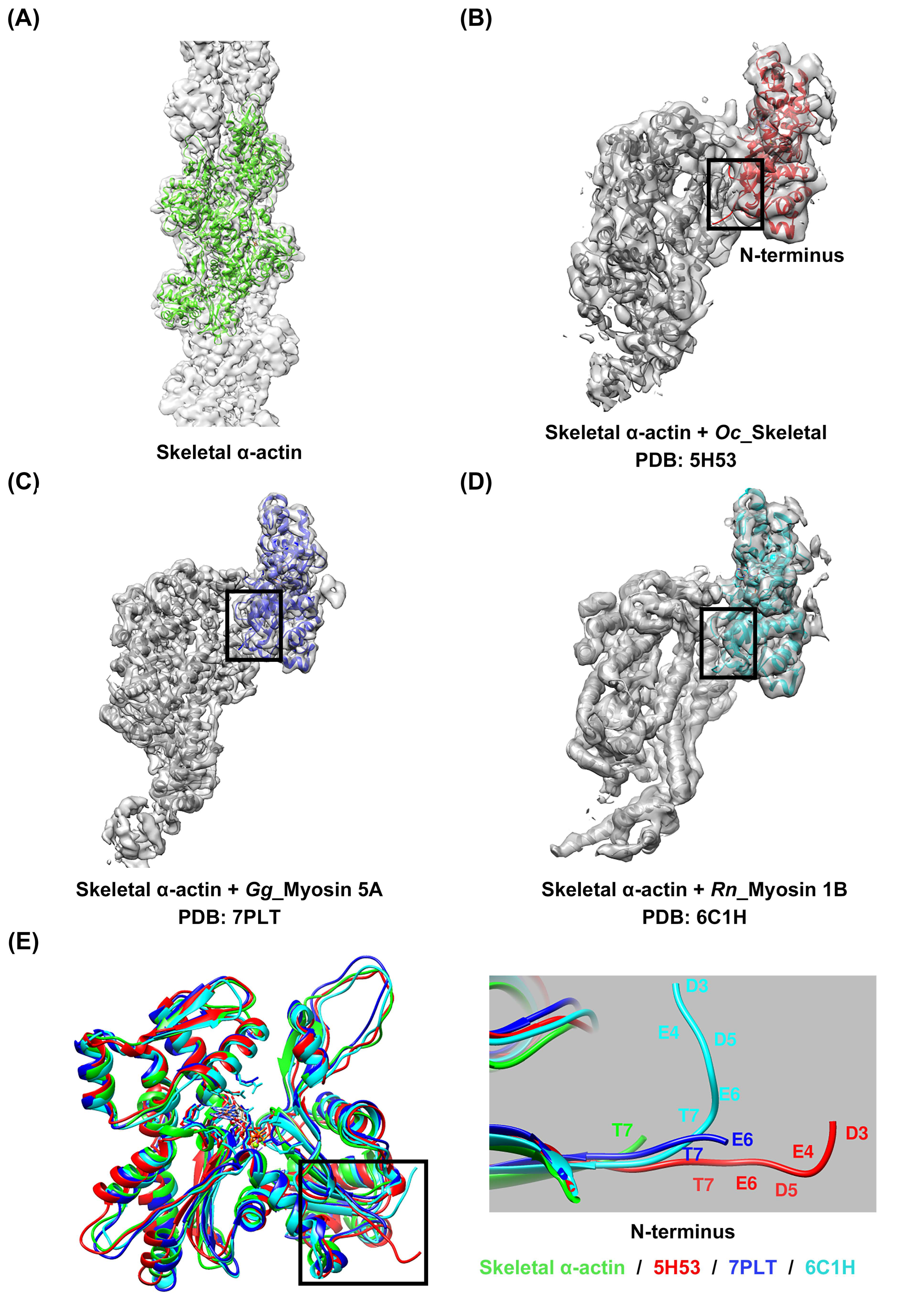
1. Introduction
2. Results
2.1. Variations in Sequence and Structure Depending on Actin Isoforms
2.2. Diversity in Sequence and Length of Myosin Loop 2 Interacting with Actin N-Terminus
2.3. Prediction of Specific Interactions between Actin and Loop 2 of the Actomyosin-7A Complex
3. Discussion
4. Materials and Methods
4.1. Multiple Sequence Alignment and Analysis
4.2. Cryo-EM Sample Preparation and Image Processing
4.3. Model Building and Refinement
4.4. Model Prediction and Visualization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Pollard, T.D.; Cooper, J.A. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 1986, 55, 987–1035. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W.; Mannherz, H.G.; Suck, D.; Pai, E.F.; Holmes, K.C. Atomic structure of the actin:DNase I complex. Nature 1990, 347, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Otterbein, L.R.; Graceffa, P.; Dominguez, R. The crystal structure of uncomplexed actin in the ADP state. Science 2001, 293, 708–711. [Google Scholar] [CrossRef] [PubMed]
- Carlier, M.F. Actin polymerization and ATP hydrolysis. Adv. Biophys. 1990, 26, 51–73. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.Z.; Pollard, T.D. Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. Proc. Natl. Acad. Sci. USA 2019, 116, 4265–4274. [Google Scholar] [CrossRef]
- Dominguez, R. Nucleotide-dependent conformational changes in the actin filament: Subtler than expected. Proc. Natl. Acad. Sci. USA 2019, 116, 3959–3961. [Google Scholar] [CrossRef] [PubMed]
- Zigmond, S.H. Signal transduction and actin filament organization. Curr. Opin. Cell Biol. 1996, 8, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Cantiello, H.F. Role of actin filament organization in cell volume and ion channel regulation. J. Exp. Zool. 1997, 279, 425–435. [Google Scholar] [CrossRef]
- Kim, C.H.; Lisman, J.E. A role of actin filament in synaptic transmission and long-term potentiation. J. Neurosci. 1999, 19, 4314–4324. [Google Scholar] [CrossRef]
- Schoenenberger, C.A.; Mannherz, H.G.; Jockusch, B.M. Actin: From structural plasticity to functional diversity. Eur. J. Cell Biol. 2011, 90, 797–804. [Google Scholar] [CrossRef]
- Schevzov, G.; Curthoys, N.M.; Gunning, P.W.; Fath, T. Functional diversity of actin cytoskeleton in neurons and its regulation by tropomyosin. Int. Rev. Cell Mol. Biol. 2012, 298, 33–94. [Google Scholar] [CrossRef] [PubMed]
- Storti, R.V.; Coen, D.M.; Rich, A. Tissue-specific forms of actin in the developing chick. Cell 1976, 8, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Tondeleir, D.; Vandamme, D.; Vandekerckhove, J.; Ampe, C.; Lambrechts, A. Actin isoform expression patterns during mammalian development and in pathology: Insights from mouse models. Cell Motil. Cytoskeleton. 2009, 66, 798–815. [Google Scholar] [CrossRef] [PubMed]
- Vandekerckhove, J.; Weber, K. At least six different actins are expressed in a higher mammal: An analysis based on the amino acid sequence of the amino-terminal tryptic peptide. J. Mol. Biol. 1978, 126, 783–802. [Google Scholar] [CrossRef] [PubMed]
- Gunning, P.; Ponte, P.; Kedes, L.; Eddy, R.; Shows, T. Chromosomal location of the co-expressed human skeletal and cardiac actin genes. Proc. Natl. Acad. Sci. USA 1984, 81, 1813–1817. [Google Scholar] [CrossRef] [PubMed]
- Otey, C.A.; Kalnoski, M.H.; Bulinski, J.C. Identification and quantification of actin isoforms in vertebrate cells and tissues. J. Cell Biochem. 1987, 34, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Rubenstein, P.A. The functional importance of multiple actin isoforms. Bioessays 1990, 12, 309–315. [Google Scholar] [CrossRef]
- Arora, A.S.; Huang, H.L.; Singh, R.; Narui, Y.; Suchenko, A.; Hatano, T.; Heissler, S.M.; Balasubramanian, M.K.; Chinthalapudi, K. Structural insights into actin isoforms. eLife 2023, 12, e82015. [Google Scholar] [CrossRef] [PubMed]
- Herman, I.M. Actin isoforms. Curr. Opin. Cell Biol. 1993, 5, 48–55. [Google Scholar] [CrossRef]
- Khaitlina, S.Y. Functional specificity of actin isoforms. Int. Rev. Cytol. 2001, 202, 35–98. [Google Scholar] [CrossRef]
- Perrin, B.J.; Ervasti, J.M. The actin gene family: Function follows isoform. Cytoskeleton 2010, 67, 630–634. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, P.J.; Gooch, J.T.; Mannherz, H.G.; Weeds, A.G. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature 1993, 364, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Morton, W.M.; Ayscough, K.R.; McLaughlin, P.J. Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat. Cell Biol. 2000, 2, 376–378. [Google Scholar] [CrossRef]
- Grintsevich, E.E.; Ge, P.; Sawaya, M.R.; Yesilyurt, H.G.; Terman, J.R.; Zhou, Z.H.; Reisler, E. Catastrophic disassembly of actin filaments via Mical-mediated oxidation. Nat. Commun. 2017, 8, 2183. [Google Scholar] [CrossRef] [PubMed]
- Rebowski, G.; Boczkowska, M.; Drazic, A.; Ree, R.; Goris, M.; Arnesen, T.; Dominguez, R. Mechanism of actin N-terminal acetylation. Sci. Adv. 2020, 6, eaay8793. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Namba, K. Structure of actomyosin rigour complex at 5.2 Å resolution and insights into the ATPase cycle mechanism. Nat. Commun. 2017, 8, 13969. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, C.; Hu, Z.; Huang, Z.; Warrington, J.A.; Taylor, D.W.; Trybus, K.M.; Lowey, S.; Taylor, K.A. The structure of the actin-smooth muscle myosin motor domain complex in the rigor state. J. Struct. Biol. 2017, 200, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Gurel, P.S.; Kim, L.Y.; Ruijgrok, P.V.; Omabegho, T.; Bryant, Z.; Alushin, G.M. Cryo-EM structures reveal specialization at the myosin VI-actin interface and a mechanism of force sensitivity. eLife 2017, 6, e31125. [Google Scholar] [CrossRef] [PubMed]
- Mentes, A.; Huehn, A.; Liu, X.; Zwolak, A.; Dominguez, R.; Shuman, H.; Ostap, E.M.; Sindelar, C.V. High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing. Proc. Natl. Acad. Sci. USA 2018, 115, 1292–1297. [Google Scholar] [CrossRef]
- Pospich, S.; Sweeney, H.L.; Houdusse, A.; Raunser, S. High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism. eLife 2021, 10, e73724. [Google Scholar] [CrossRef]
- Gong, R.; Jiang, F.; Moreland, Z.G.; Reynolds, M.J.; de Los Reyes, S.E.; Gurel, P.; Shams, A.; Heidings, J.B.; Bowl, M.R.; Bird, J.E.; et al. Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia. Sci. Adv. 2022, 8, eabl4733. [Google Scholar] [CrossRef] [PubMed]
- Stokasimov, E.; Rubenstein, P.A. Actin isoform-specific conformational differences observed with hydrogen/deuterium exchange and mass spectrometry. J. Biol. Chem. 2009, 284, 25421–25430. [Google Scholar] [CrossRef] [PubMed]
- Cook, R.K.; Root, D.; Miller, C.; Reisler, E.; Rubenstein, P.A. Enhanced stimulation of myosin subfragment 1 ATPase activity by addition of negatively charged residues to the yeast actin NH2 terminus. J. Biol. Chem. 1993, 268, 2410–2415. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Diensthuber, R.P.; Chizhov, I.; Claus, P.; Heissler, S.M.; Preller, M.; Taft, M.H.; Manstein, D.J. Distinct functional interactions between actin isoforms and nonsarcomeric myosins. PLoS ONE 2013, 8, e70636. [Google Scholar] [CrossRef] [PubMed]
- Warrick, H.M.; Spudich, J.A. Myosin structure and function in cell motility. Annu. Rev. Cell Biol. 1987, 3, 379–421. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.E.; Bridgman, P.C. Myosin function in nervous and sensory systems. J. Neurobiol. 2004, 58, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Coluccio, L.M. Myosins and Disease. Adv. Exp. Med. Biol. 2020, 1239, 245–316. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.; Kitamoto, J.; Williams, D.S. Abnormal phagocytosis by retinal pigmented epithelium that lacks myosin VIIa, the Usher syndrome 1B protein. Proc. Natl. Acad. Sci. USA 2003, 100, 6481–6486. [Google Scholar] [CrossRef] [PubMed]
- Weil, D.; Blanchard, S.; Kaplan, J.; Guilford, P.; Gibson, F.; Walsh, J.; Mburu, P.; Varela, A.; Levilliers, J.; Weston, M.D.; et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 1995, 374, 60–61. [Google Scholar] [CrossRef]
- Hasson, T.; Heintzelman, M.B.; Santos-Sacchi, J.; Corey, D.P.; Mooseker, M.S. Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc. Natl. Acad. Sci. USA 1995, 92, 9815–9819. [Google Scholar] [CrossRef]
- Hasson, T.; Gillespie, P.G.; Garcia, J.A.; MacDonald, R.B.; Zhao, Y.; Yee, A.G.; Mooseker, M.S.; Corey, D.P. Unconventional myosins in inner-ear sensory epithelia. J. Cell Biol. 1997, 137, 1287–1307. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ondek, B.; Williams, D.S. Mutant myosin VIIa causes defective melanosome distribution in the RPE of shaker-1 mice. Nat. Genet. 1998, 19, 117–118. [Google Scholar] [CrossRef]
- Korn, E.D. Coevolution of head, neck, and tail domains of myosin heavy chains. Proc. Natl. Acad. Sci. USA 2000, 97, 12559–12564. [Google Scholar] [CrossRef] [PubMed]
- Cope, M.J.; Whisstock, J.; Rayment, I.; Kendrick-Jones, J. Conservation within the myosin motor domain: Implications for structure and function. Structure 1996, 4, 969–987. [Google Scholar] [CrossRef] [PubMed]
- Lymn, R.W.; Taylor, E.W. Transient state phosphate production in the hydrolysis of nucleoside triphosphates by myosin. Biochemistry 1970, 9, 2975–2983. [Google Scholar] [CrossRef]
- Lymn, R.W.; Taylor, E.W. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry 1971, 10, 4617–4624. [Google Scholar] [CrossRef] [PubMed]
- Okimoto, N.; Yamanaka, K.; Ueno, J.; Hata, M.; Hoshino, T.; Tsuda, M. Theoretical studies of the ATP hydrolysis mechanism of myosin. Biophys. J. 2001, 81, 2786–2794. [Google Scholar] [CrossRef]
- von der Ecken, J.; Heissler, S.M.; Pathan-Chhatbar, S.; Manstein, D.J.; Raunser, S. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature 2016, 534, 724–728. [Google Scholar] [CrossRef]
- Risi, C.; Schäfer, L.U.; Belknap, B.; Pepper, I.; White, H.D.; Schröder, G.F.; Galkin, V.E. High-Resolution Cryo-EM Structure of the Cardiac Actomyosin Complex. Structure 2021, 29, 50–60.e4. [Google Scholar] [CrossRef]
- Behrmann, E.; Müller, M.; Penczek, P.A.; Mannherz, H.G.; Manstein, D.J.; Raunser, S. Structure of the rigor actin-tropomyosin-myosin complex. Cell 2012, 150, 327–338. [Google Scholar] [CrossRef]
- Lorenz, M.; Holmes, K.C. The actin-myosin interface. Proc. Natl. Acad. Sci. USA 2010, 107, 12529–12534. [Google Scholar] [CrossRef] [PubMed]
- Onishi, H.; Mikhailenko, S.V.; Morales, M.F. Toward understanding actin activation of myosin ATPase: The role of myosin surface loops. Proc. Natl. Acad. Sci. USA 2006, 103, 6136–6141. [Google Scholar] [CrossRef]
- Furch, M.; Remmel, B.; Geeves, M.A.; Manstein, D.J. Stabilization of the actomyosin complex by negative charges on myosin. Biochemistry 2000, 39, 11602–11608. [Google Scholar] [CrossRef]
- Joel, P.B.; Trybus, K.M.; Sweeney, H.L. Two conserved lysines at the 50/20-kDa junction of myosin are necessary for triggering actin activation. J. Biol. Chem. 2001, 276, 2998–3003. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.T.; Spudich, J.A. The sequence of the myosin 50–20K loop affects Myosin’s affinity for actin throughout the actin-myosin ATPase cycle and its maximum ATPase activity. Biochemistry 1999, 38, 3785–3792. [Google Scholar] [CrossRef] [PubMed]
- Krementsova, E.B.; Hodges, A.R.; Lu, H.; Trybus, K.M. Processivity of chimeric class V myosins. J. Biol. Chem. 2006, 281, 6079–6086. [Google Scholar] [CrossRef] [PubMed]
- Clobes, A.M.; Guilford, W.H. Loop 2 of myosin is a force-dependent inhibitor of the rigor bond. J. Muscle Res. Cell Motil. 2014, 35, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.J.; Park, Y.H.; Ryu, B.; Jung, H.S. Sequence Alignment-Based Prediction of Myosin 7A: Structural Implications and Protein Interactions. Int. J. Mol. Sci. 2024, 25, 3365. [Google Scholar] [CrossRef]
- van den Boom, F.; Düssmann, H.; Uhlenbrock, K.; Abouhamed, M.; Bähler, M. The Myosin IXb motor activity targets the myosin IXb RhoGAP domain as cargo to sites of actin polymerization. Mol. Biol. Cell 2007, 18, 1507–1518. [Google Scholar] [CrossRef]
- Struchholz, S.; Elfrink, K.; Pieper, U.; Kalhammer, G.; Honnert, U.; Grützner, A.; Linke, W.A.; Liao, W.; Bähler, M. Functional role of the extended loop 2 in the myosin 9b head for binding F-actin. J. Biol. Chem. 2009, 284, 3663–3671. [Google Scholar] [CrossRef]
- Elfrink, K.; Liao, W.; Pieper, U.; Oeding, S.J.; Bähler, M. The loop2 insertion of type IX myosin acts as an electrostatic actin tether that permits processive movement. PLoS ONE 2014, 9, e84874. [Google Scholar] [CrossRef] [PubMed]
- Uyeda, T.Q.; Ruppel, K.M.; Spudich, J.A. Enzymatic activities correlate with chimaeric substitutions at the actin-binding face of myosin. Nature 1994, 368, 567–569. [Google Scholar] [CrossRef] [PubMed]
- Furch, M.; Geeves, M.A.; Manstein, D.J. Modulation of actin affinity and actomyosin adenosine triphosphatase by charge changes in the myosin motor domain. Biochemistry 1998, 37, 6317–6326. [Google Scholar] [CrossRef] [PubMed]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Zivanov, J.; Nakane, T.; Forsberg, B.O.; Kimanius, D.; Hagen, W.J.; Lindahl, E.; Scheres, S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 2018, 7, e42166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016, 193, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H.; Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 2012, 9, 853–854. [Google Scholar] [CrossRef] [PubMed]
- Zivanov, J.; Nakane, T.; Scheres, S.H.W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 2020, 7, 253–267. [Google Scholar] [CrossRef]
- Zivanov, J.; Nakane, T.; Scheres, S.H.W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 2019, 6, 5–17. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Casañal, A.; Lohkamp, B.; Emsley, P. Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci. 2020, 29, 1069–1078. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Poon, B.K.; Read, R.J.; Sobolev, O.V.; Terwilliger, T.C.; Urzhumtsev, A.; Adams, P.D. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 2018, 74, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv 2021, 2021, 463034. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, C.J.; Park, Y.H.; An, M.Y.; Ryu, B.; Jung, H.S. Insights into Actin Isoform-Specific Interactions with Myosin via Computational Analysis. Molecules 2024, 29, 2992. https://doi.org/10.3390/molecules29132992
Yu CJ, Park YH, An MY, Ryu B, Jung HS. Insights into Actin Isoform-Specific Interactions with Myosin via Computational Analysis. Molecules. 2024; 29(13):2992. https://doi.org/10.3390/molecules29132992
Chicago/Turabian StyleYu, Chan Jong, Yoon Ho Park, Mi Young An, Bumhan Ryu, and Hyun Suk Jung. 2024. "Insights into Actin Isoform-Specific Interactions with Myosin via Computational Analysis" Molecules 29, no. 13: 2992. https://doi.org/10.3390/molecules29132992
APA StyleYu, C. J., Park, Y. H., An, M. Y., Ryu, B., & Jung, H. S. (2024). Insights into Actin Isoform-Specific Interactions with Myosin via Computational Analysis. Molecules, 29(13), 2992. https://doi.org/10.3390/molecules29132992










