Improved Classification Performance of Bacteria in Interference Using Raman and Fourier-Transform Infrared Spectroscopy Combined with Machine Learning
Abstract
1. Introduction
2. Results
2.1. Peak Assignments of Spectrum
2.2. Classification Models
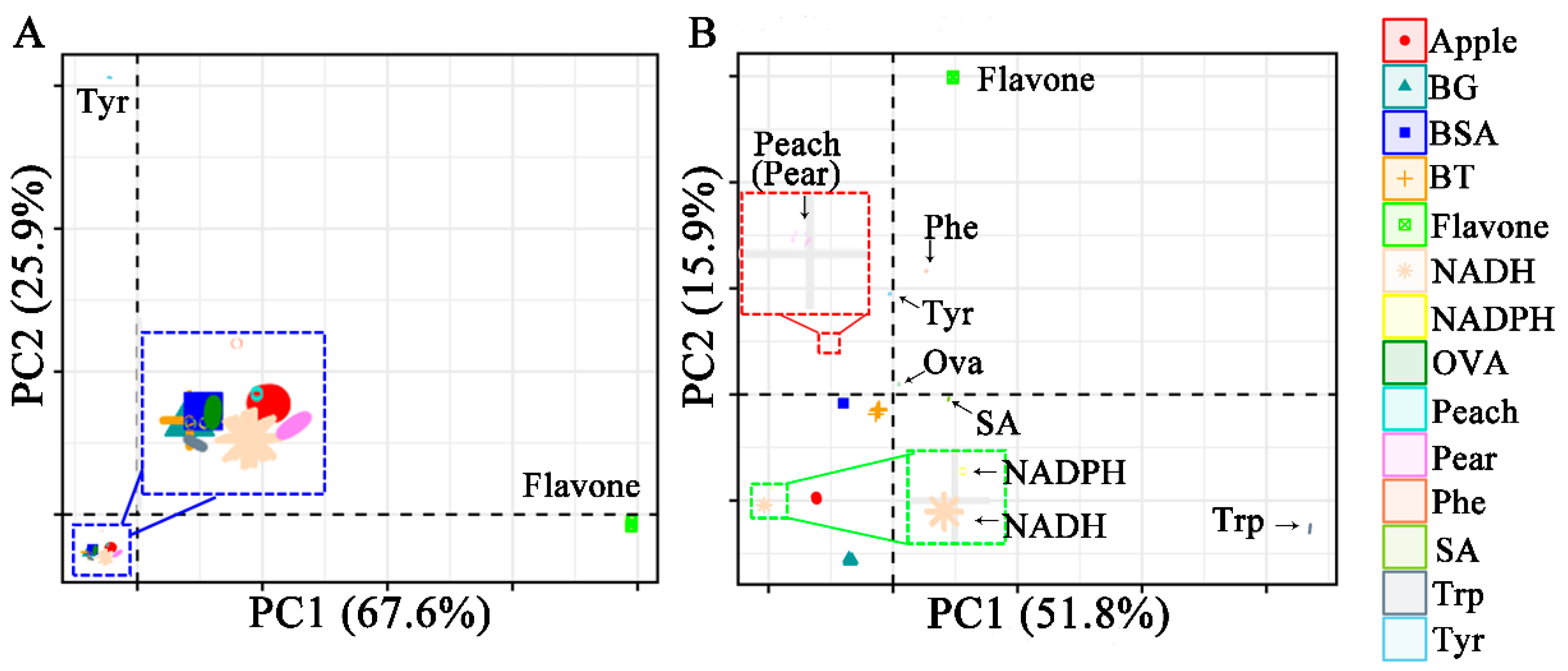
2.2.1. Principal Component Analysis
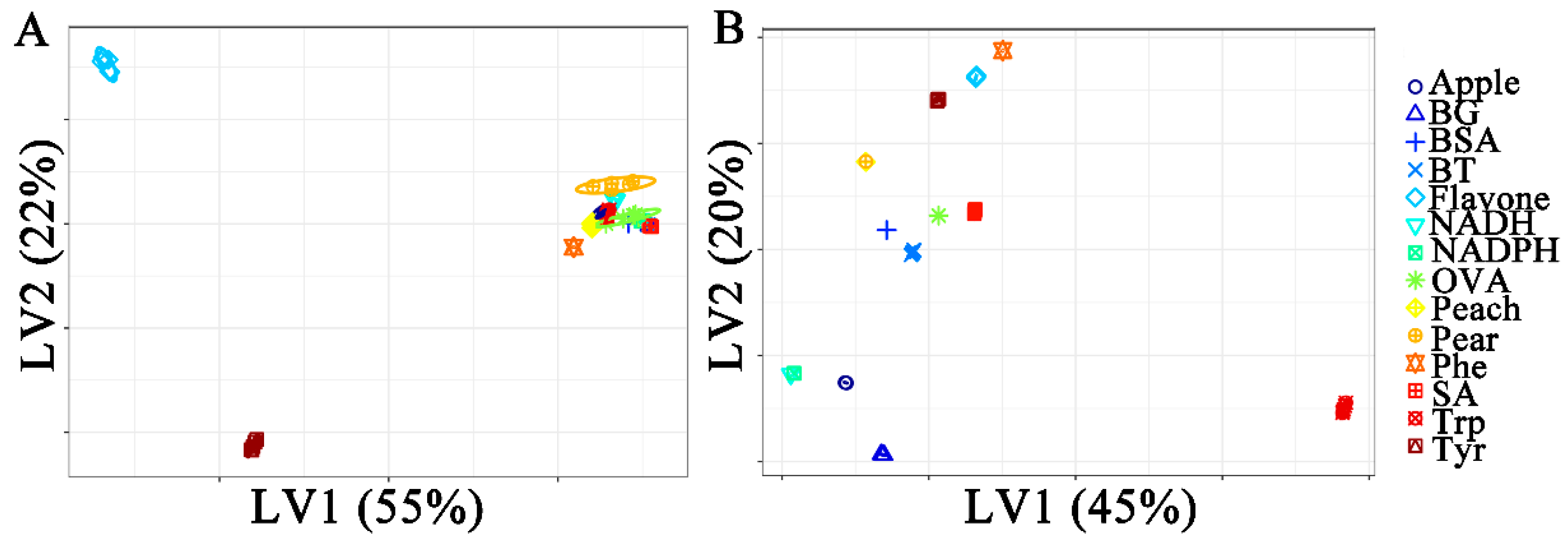
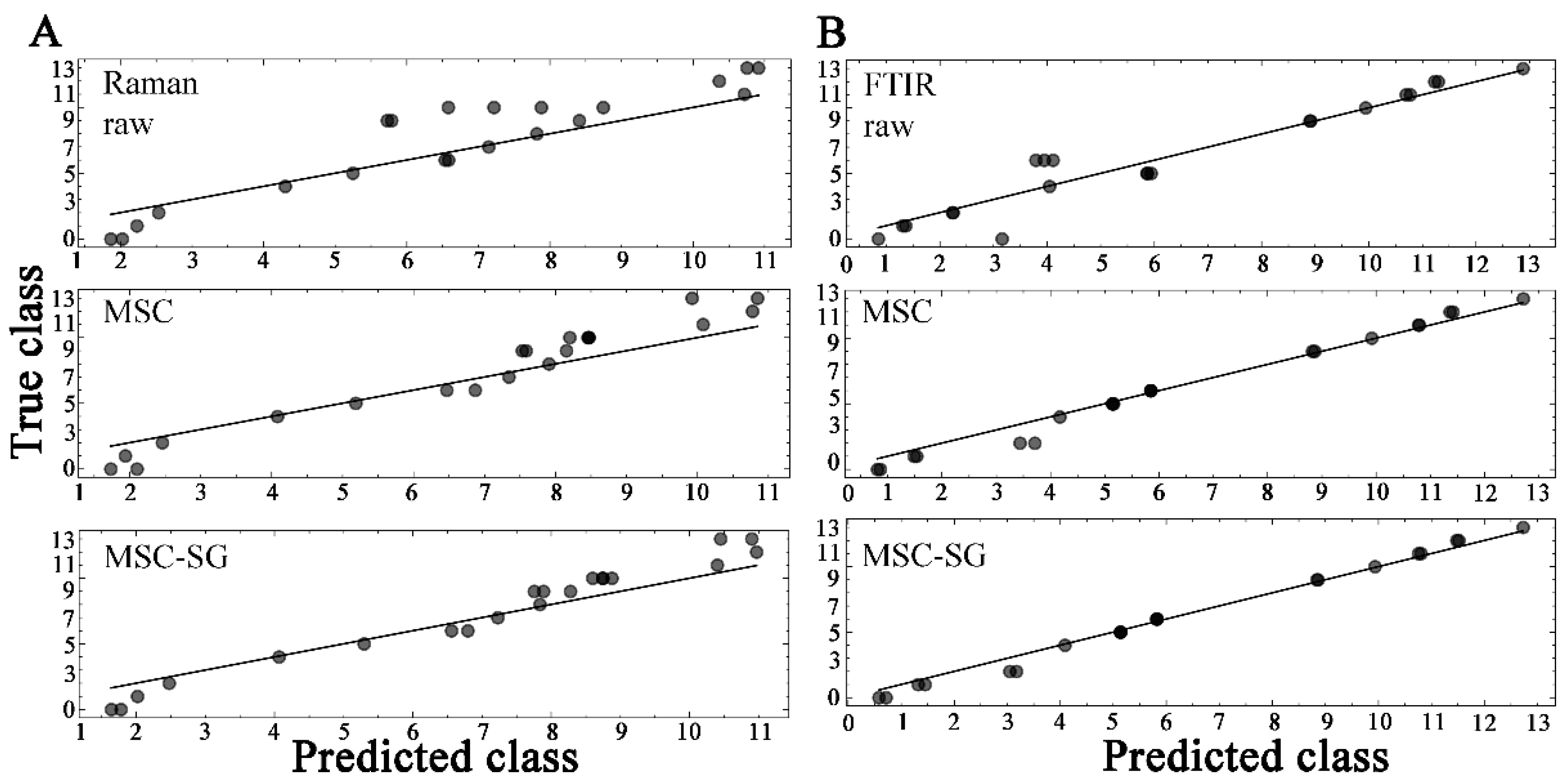
2.2.2. Partial Least Squares Discriminant Analysis
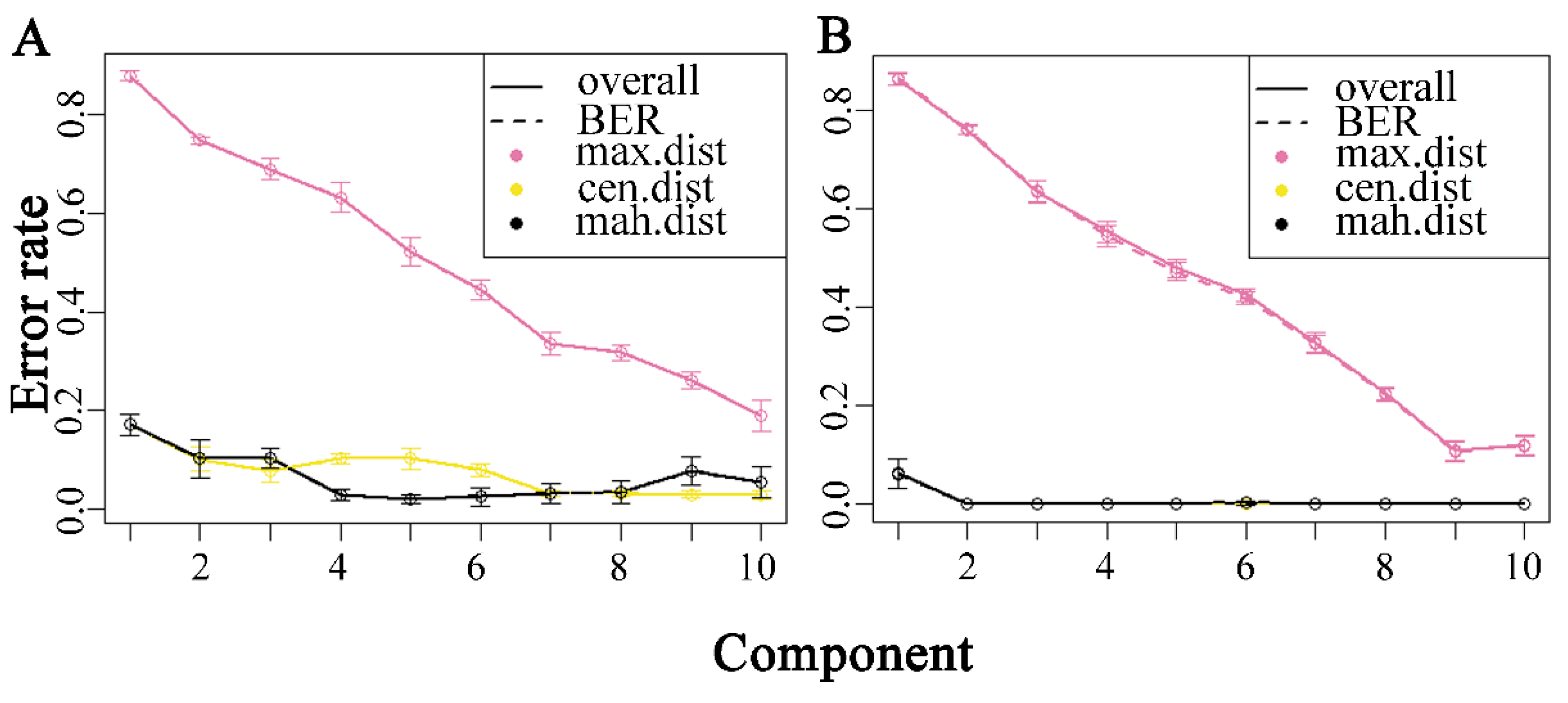
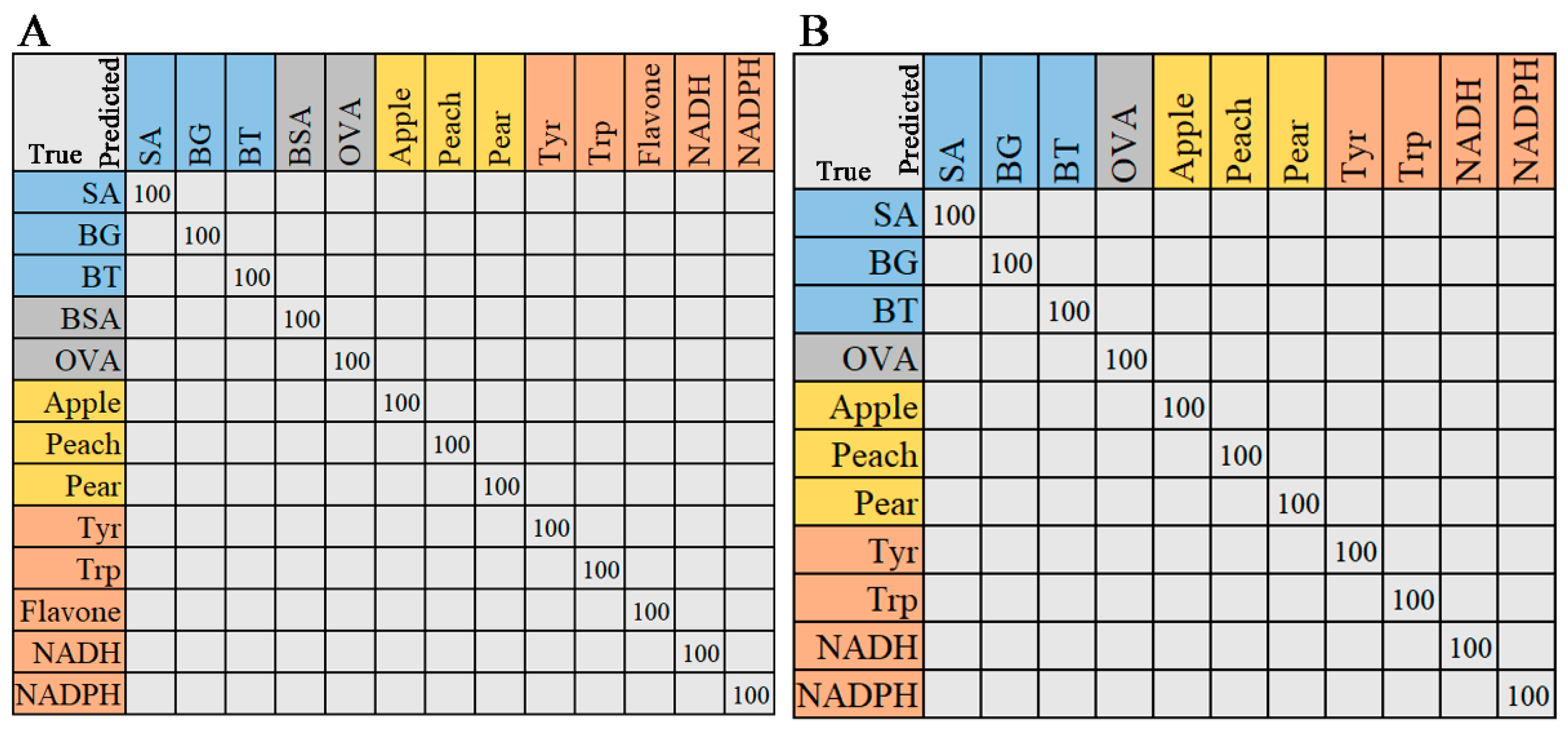
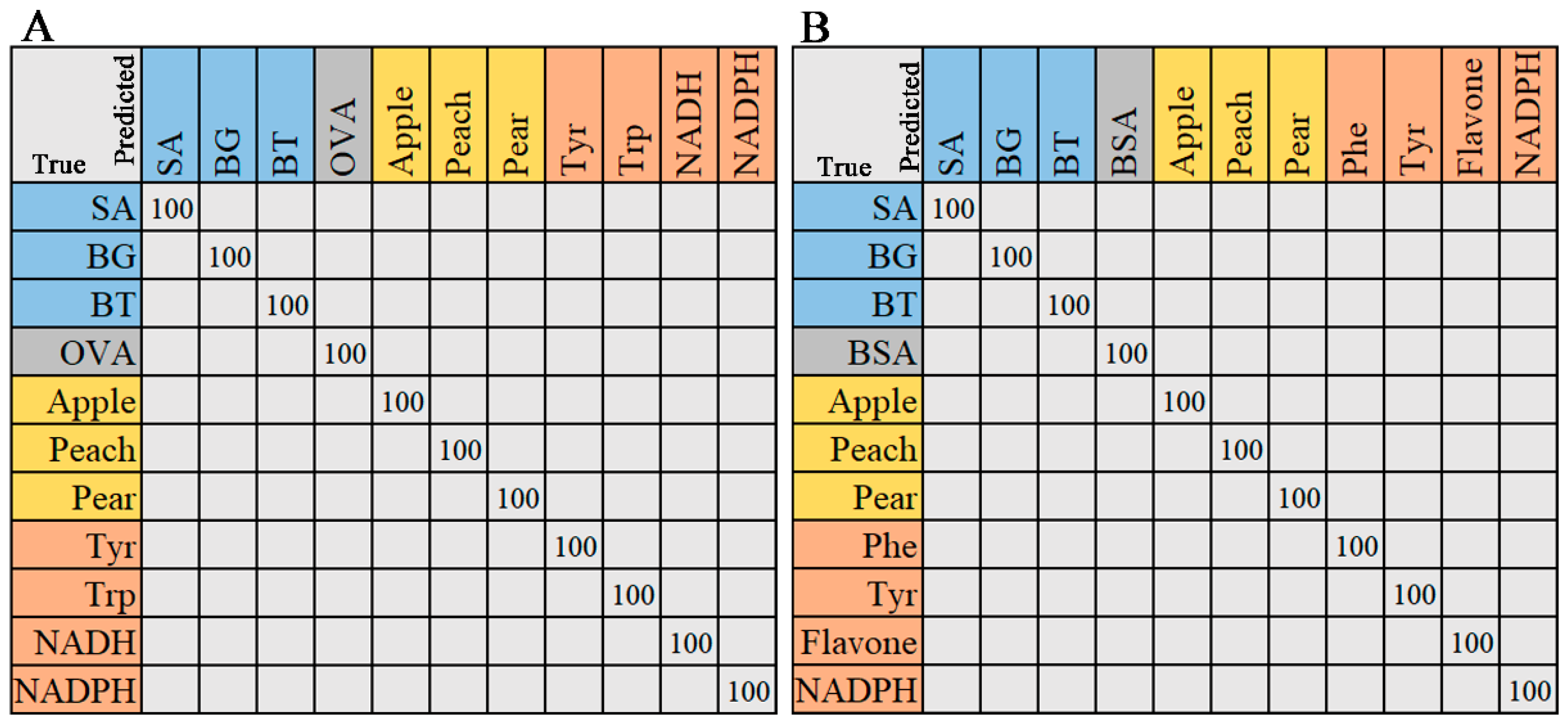
2.2.3. Random Forest
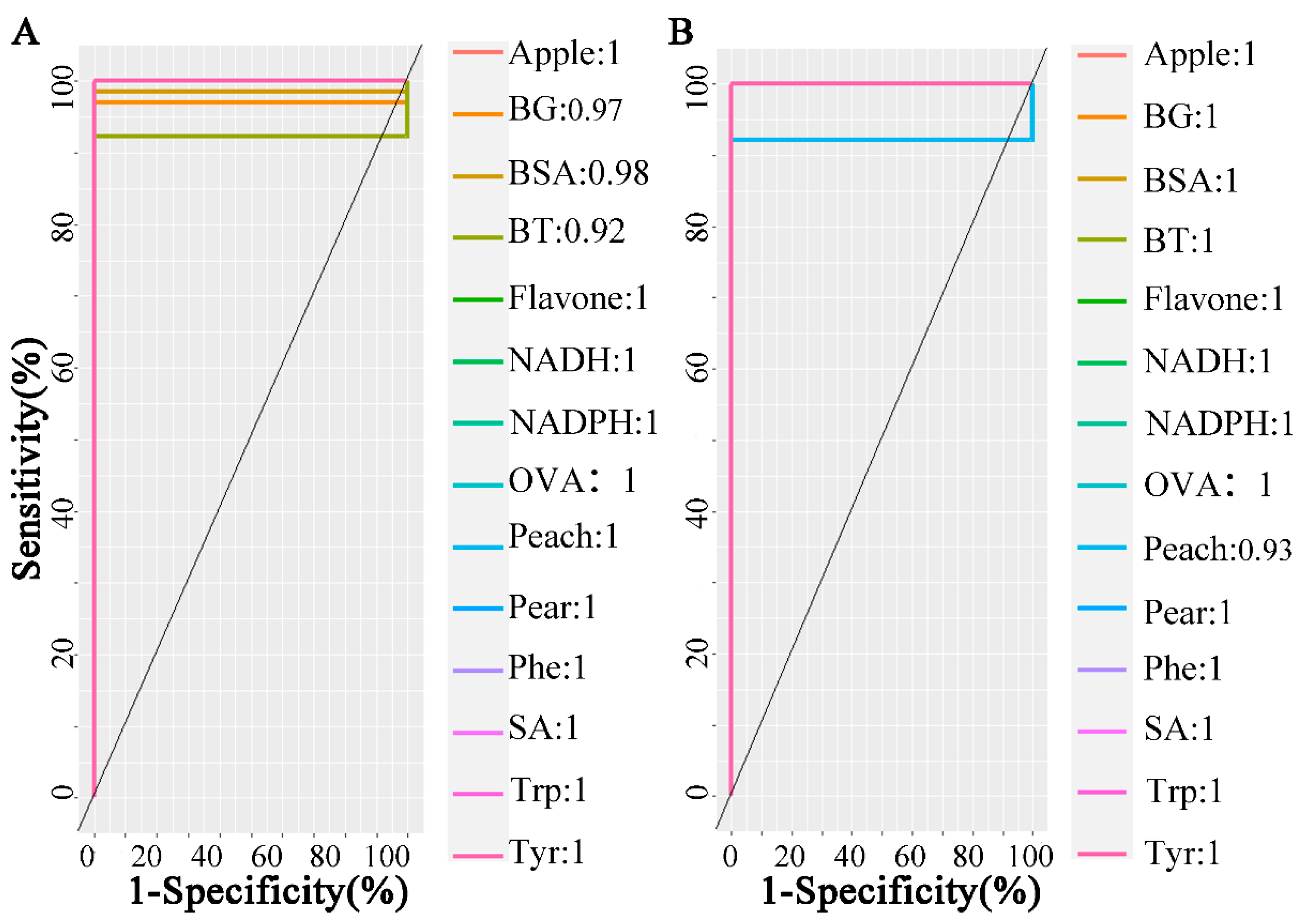
2.2.4. Support Vector Machine
2.2.5. Classification Performance of Fused Spectral Features
3. Discussion
4. Materials and Methods
4.1. Biological Samples
4.2. Attenuated Total Reflectance Fourier-Transform Infrared Spectral Measurements
4.3. Raman Spectral Measurements
4.4. Data Treatment
4.5. Performance Evaluation Metrics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lei, Y.; Tian, Z.; Sun, H.; Zhu, Z.; Liang, W.; Li, A. Self-cleaning and flexible filters based on aminopyridine conjugated microporous polymers nanotubes for bacteria sterilization and efficient PM2.5 capture. Sci. Total Environ. 2021, 766, 142594. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Yang, Y.; Xu, C.; Ye, Y.; Huang, L.; Sun, L.; Cai, Y.; Zhou, W.; Ge, Y.; Li, Y. Vertical gold nanowires-based surface-enhanced Raman scattering for direct detection of ocular bacteria. Sensor Actuators B Chem. 2023, 380, 133381. [Google Scholar] [CrossRef]
- Romano, S.; Di Salvo, M.; Rispoli, G.; Alifano, P.; Perrone, M.R.; Tala, A. Airborne bacteria in the Central Mediterranean: Structure and role of meteorology and air mass transport. Sci. Total Environ. 2019, 697, 134020. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Di, H.; Li, Y.; Yuan, Y.; Hua, D.; Wang, L.; Chen, D. Detection of aerosol mass concentration profiles using single-wavelength Raman Lidar within the planetary boundary layer. J. Quant. Spectrosc. Radiat. Transf. 2021, 272, 107833. [Google Scholar] [CrossRef]
- Schütze, C.; Lau, S.; Reiche, N.; Sauer, U.; Borsdorf, H.; Dietrich, P. Ground-based Remote Sensing with Open-path Fourier- transform Infrared (OP-FTIR) Spectroscopy for Large-scale Monitoring of Greenhouse Gases. Energy Procedia 2013, 37, 4276–4282. [Google Scholar] [CrossRef]
- Chen, Q.; Li, J.; Hua, X.; Jiang, X.; Mu, Z.; Wang, M.; Wang, J.; Shan, M.; Yang, X.; Fan, X. Identification of species and sources of atmospheric chromophores by fluorescence excitation-emission matrix with parallel factor analysis. Sci. Total Environ. 2020, 718, 137322. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, S.; Arigela, R.; Thyagarajan, S.; Raghunathan, R. Comparison and evaluation of enumeration methods for measurement of fungal spore emission. J. Aerosol Sci. 2022, 165, 106033. [Google Scholar] [CrossRef]
- Sengupta, A.; Laucks, M.L.; Dildine, N.; Drapala, E.; Davis, E.J. Bioaerosol characterization by surface-enhanced Raman spectroscopy (SERS). J. Aerosol Sci. 2005, 36, 651–664. [Google Scholar] [CrossRef]
- Ben-David, A.; Ren, H. Detection, identification, and estimation of biological aerosols and vapors with a Fourier-transform infrared spectrometer. Appl. Opt. 2003, 42, 4887–4900. [Google Scholar] [CrossRef]
- Doughty, D.C.; Hill, S.C. Automated aerosol Raman spectrometer for semi-continuous sampling of atmospheric aerosol. J. Quant. Spectrosc. Radiat. Transf. 2017, 188, 103–117. [Google Scholar] [CrossRef]
- Gong, Z.; Pan, Y.L.; Videen, G.; Wang, C. Optical trapping-Raman spectroscopy (OT-RS) with embedded microscopy imaging for concurrent characterization and monitoring of physical and chemical properties of single particles. Anal. Chim. Acta 2018, 1020, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Jabbour, R.E.; Guicheteau, J.A.; Christesen, S.D.; Emge, D.K.; Fountain, A.W.; Bottiger, J.R.; Emmons, E.D.; Snyder, A.P. Bioaerosol analysis with Raman chemical imaging microspectroscopy. Anal. Chem. 2009, 81, 6981–6990. [Google Scholar] [CrossRef] [PubMed]
- McKenna, O.E.; Posselt, G.; Briza, P.; Lackner, P.; Schmitt, A.O.; Gadermaier, G.; Wessler, S.; Ferreira, F. Multi-Approach Analysis for the Identification of Proteases within Birch Pollen. Int. J. Mol. Sci. 2017, 18, 1433. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Rani, A.; Goyal, A. Insights into the immune manipulation mechanisms of pollen allergens by protein domain profiling. Comput. Biol. Chem. 2017, 70, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Gottardini, E.; Rossi, S.; Cristofolini, F.; Benedetti, L. Use of Fourier transform infrared (FT-IR) spectroscopy as a tool for pollen identification. Aerobiologia 2007, 23, 211–219. [Google Scholar] [CrossRef]
- Jin, H.; Wang, J.; Jin, S.; Jiang, L.; Zou, Y. Raman spectroscopy of potential bio-hazards commonly found in bio-aerosols. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 243, 118753. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Zhao, N.; Yin, G.; Gan, T.; Yang, R.; Chen, X.; Chen, M.; Duan, J. Artificial neural networks combined multi-wavelength transmission spectrum feature extraction for sensitive identification of waterborne bacteria. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 251, 119423. [Google Scholar] [CrossRef] [PubMed]
- Maya-Manzano, J.M.; Tummon, F.; Abt, R.; Allan, N.; Bunderson, L.; Clot, B.; Crouzy, B.; Daunys, G.; Erb, S.; Gonzalez-Alonso, M. Towards European automatic bioaerosol monitoring: Comparison of 9 automatic pollen observational instruments with classic Hirst-type traps. Sci. Total Environ. 2023, 866, 161220. [Google Scholar] [CrossRef] [PubMed]
- Nabatchian, A.; Abdel-Raheem, E.; Ahmadi, M. Illumination invariant feature extraction and mutual-information-based local matching for face recognition under illumination variation and occlusion. Pattern Recognit. 2011, 44, 2576–2587. [Google Scholar] [CrossRef]
- Wang, Y.; Ni, H.; Li, H.; Chen, J.; Zhang, D.; Fu, L. Plasmonic microneedle arrays for rapid extraction, SERS detection, and inactivation of bacteria. Chem. Eng. J. 2022, 442, 136140. [Google Scholar] [CrossRef]
- Frain, M.; Schmidt, D.P.; Pan, Y.-L.; Chang, R.K. Selective Deflection and Localization of Flowing Aerosols onto a Substrate. Aerosol Sci. Technol. 2006, 40, 218–225. [Google Scholar] [CrossRef]
- Lu, W.; Li, H.; Qiu, H.; Wang, L.; Feng, J.; Fu, Y.V. Identification of pathogens and detection of antibiotic susceptibility at single-cell resolution by Raman spectroscopy combined with machine learning. Front. Microbiol. 2023, 13, 1076965. [Google Scholar] [CrossRef]
- Tang, J.-W.; Li, J.-Q.; Yin, X.-C.; Xu, W.-W.; Pan, Y.-C.; Liu, Q.-H.; Gu, B.; Zhang, X.; Wang, L. Rapid Discrimination of Clinically Important Pathogens through Machine Learning Analysis of Surface Enhanced Raman Spectra. Front. Microbiol. 2022, 13, 843417. [Google Scholar] [CrossRef]
- Dikec, J.; Pacheco, M.; Dujourdy, L.; Sandt, C.; Winckler, P.; Perrier-Cornet, J.M. Influence of hydration on calcium dipicolinate (CaDPA) during UVb and UVc exposure studied via Raman, FTIR and O-PTIR spectroscopy. J. Photochem. Photobiol. A Chem. 2023, 443, 114823. [Google Scholar] [CrossRef]
- Sun, J.; Xu, X.; Feng, S.; Zhang, H.; Xu, L.; Jiang, H.; Sun, B.; Meng, Y.; Chen, W. Rapid identification of salmonella serovars by using Raman spectroscopy and machine learning algorithm. Talanta 2023, 253, 123807. [Google Scholar] [CrossRef] [PubMed]
- Moros, J.; Garrigues, S.; de la Guardia, M. Evaluation of nutritional parameters in infant formulas and powdered milk by Raman spectroscopy. Anal. Chim. Acta 2007, 593, 30–38. [Google Scholar] [CrossRef]
- Notingher, I.; Green, C.; Dyer, C.; Perkins, E.; Hopkins, N.; Lindsay, C.; Hench, L.L. Discrimination between ricin and sulphur mustard toxicity in vitro using Raman spectroscopy. J. R. Soc. Interface 2004, 1, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Stone, N.; Kendall, C.; Smith, J.; Crow, P.; Barr, H. Raman spectroscopy for identification of epithelial cancers. Faraday Discuss. 2004, 126, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.C.C.; Aguiar, E.M.G.; Silva, A.T.F.; Santos, L.L.D.; Cardoso-Sousa, L.; Araujo, T.G.; Santos, D.W.; Goulart, L.R.; Sabino-Silva, R.; Maia, Y.C.P. Attenuated Total Reflection-Fourier Transform Infrared (ATR-FTIR) Spectroscopy Analysis of Saliva for Breast Cancer Diagnosis. J. Oncol. 2020, 2020, 4343590. [Google Scholar] [CrossRef]
- Guleken, Z.; Unubol, B.; Bilici, R.; Saribal, D.; Toraman, S.; Gunduz, O.; Erdem Kuruca, S. Investigation of the discrimination and characterization of blood serum structure in patients with opioid use disorder using IR spectroscopy and PCA-LDA analysis. J. Pharm. Biomed. Anal. 2020, 190, 113553. [Google Scholar] [CrossRef]
- Sheng, D.; Wu, Y.; Wang, X.; Huang, D.; Chen, X.; Liu, X. Comparison of serum from gastric cancer patients and from healthy persons using FTIR spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2013, 116, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Dou, J.; Dawuti, W.; Li, J.; Zhao, H.; Zhou, R.; Zhou, J.; Lin, R.; Lu, G. Rapid detection of serological biomarkers in gallbladder carcinoma using fourier transform infrared spectroscopy combined with machine learning. Talanta 2023, 259, 124457. [Google Scholar] [CrossRef] [PubMed]
- Sahu, S.K.; Pandey, M. An optimal hybrid multiclass SVM for plant leaf disease detection using spatial Fuzzy C-Means model. Expert Syst. Appl. 2023, 214, 118989. [Google Scholar] [CrossRef]
- Thirumala, K.; Pal, S.; Jain, T.; Umarikar, A.C. A classification method for multiple power quality disturbances using EWT based adaptive filtering and multiclass SVM. Neurocomputing 2019, 334, 265–274. [Google Scholar] [CrossRef]
- Wu, S.; Jia, D.K.; Liu, X.L.; Yan, F.G.; Li, Y.F. Application of continuous wavelet features and multi-class sphere SVM to chatter prediction. Adv. Mater. Res. 2011, 188, 675–680. [Google Scholar] [CrossRef]
- Galvin-King, P.; Haughey, S.A.; Elliott, C.T. Garlic adulteration detection using NIR and FTIR spectroscopy and chemometrics. J. Food Compos. Anal. 2021, 96, 103757. [Google Scholar] [CrossRef]
- Leng, T.; Li, F.; Xiong, L.; Xiong, Q.; Zhu, M.; Chen, Y. Quantitative detection of binary and ternary adulteration of minced beef meat with pork and duck meat by NIR combined with chemometrics. Food Control 2020, 113, 107203. [Google Scholar] [CrossRef]
- Gao, F.; Zeng, G.; Wang, B.; Xiao, J.; Zhang, L.; Cheng, W.; Wang, H.; Li, H.; Shi, X. Discrimination of the geographic origins and varieties of wine grapes using high-throughput sequencing assisted by a random forest model. LWT-Food Sci. Technol. 2021, 145, 111333. [Google Scholar] [CrossRef]
- Lu, X.; Huang, Q.; Miller, W.G.; Aston, D.E.; Xu, J.; Xue, F.; Zhang, H.; Rasco, B.A.; Wang, S.; Konkel, M.E. Comprehensive detection and discrimination of Campylobacter species by use of confocal micro-Raman spectroscopy and multilocus sequence typing. J. Clin. Microbiol. 2012, 50, 2932–2946. [Google Scholar] [CrossRef]
- Ramesh, G.; Paul, W.; Valparambil Puthanveetil, V.; Raja, K.; Embekkat Kaviyil, J. Raman spectroscopy as a novel technique for the identification of pathogens in a clinical microbiology laboratory. Spectrosc. Lett. 2022, 55, 546–551. [Google Scholar] [CrossRef]
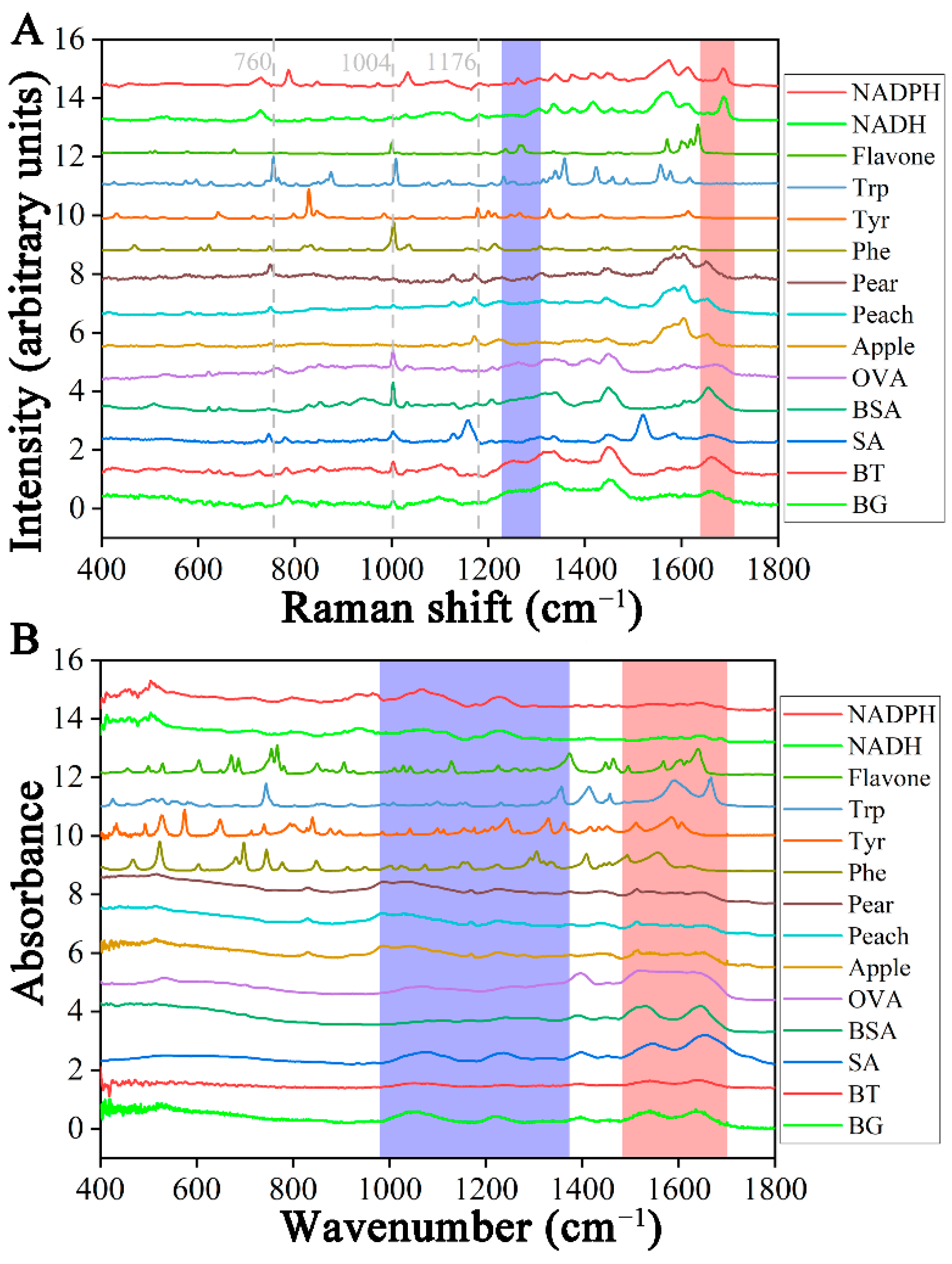

| Wavenumber (cm−1) | Assignment | Reference |
|---|---|---|
| Raman spectroscopy | ||
| 1655–1680 | Amide I (proteins), C=O stretching (lipids) | [27] |
| 1300–1230 | Amide III | [27] |
| 1209 | Tryptophan and phenylalanine ν(C–C6H5) mode | [27,28] |
| 1176 | C–H bending tyrosine (proteins) | [27] |
| 1004 | Phenylalanine | [27] |
| 760 | Ring breathing tryptophan (proteins) | [27] |
| ATR-FTIR spectroscopy | ||
| 1710–1475 | Amino acids and proteins | [29] |
| 1700–1600 | Amide I | [30] |
| 1600–1500 | Amide II | [31] |
| 1354–980 | Lipids, nucleic acids, and carbohydrate | [32] |
| Spectra | RMSEC | RMSEP | ||
|---|---|---|---|---|
| Raman | ||||
| raw | 0.743 | 1.816 | 0.965 | 0.792 |
| MSC | 0.751 | 1.402 | 0.965 | 0.876 |
| MSC-SG | 0.694 | 1.200 | 0.970 | 0.909 |
| ATR-FTIR | ||||
| raw | 0.388 | 1.141 | 0.990 | 0.925 |
| MSC | 0.322 | 0.619 | 0.993 | 0.978 |
| MSC-SG | 0.270 | 0.462 | 0.995 | 0.988 |
| Spectra | Average-Acc | RMSE | R2 | Overall-Acc |
|---|---|---|---|---|
| Raman | 1 | 0 | 1 | 1 |
| ATR-FTIR | 0.9090 | 0.3085 | 0.9995 | 0.9048 |
| Spectra | Average-acc | RMSE | R2 | Overall-Acc |
|---|---|---|---|---|
| RF | 1 | 0 | 1 | 1 |
| SVM | 1 | 0 | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.; Xu, J.; Du, B.; Yang, Q.; Liu, B.; Xu, J.; Tong, Z. Improved Classification Performance of Bacteria in Interference Using Raman and Fourier-Transform Infrared Spectroscopy Combined with Machine Learning. Molecules 2024, 29, 2966. https://doi.org/10.3390/molecules29132966
Zhang P, Xu J, Du B, Yang Q, Liu B, Xu J, Tong Z. Improved Classification Performance of Bacteria in Interference Using Raman and Fourier-Transform Infrared Spectroscopy Combined with Machine Learning. Molecules. 2024; 29(13):2966. https://doi.org/10.3390/molecules29132966
Chicago/Turabian StyleZhang, Pengjie, Jiwei Xu, Bin Du, Qianyu Yang, Bing Liu, Jianjie Xu, and Zhaoyang Tong. 2024. "Improved Classification Performance of Bacteria in Interference Using Raman and Fourier-Transform Infrared Spectroscopy Combined with Machine Learning" Molecules 29, no. 13: 2966. https://doi.org/10.3390/molecules29132966
APA StyleZhang, P., Xu, J., Du, B., Yang, Q., Liu, B., Xu, J., & Tong, Z. (2024). Improved Classification Performance of Bacteria in Interference Using Raman and Fourier-Transform Infrared Spectroscopy Combined with Machine Learning. Molecules, 29(13), 2966. https://doi.org/10.3390/molecules29132966





