The Influence of Clustered DNA Damage Containing Iz/Oz and OXOdG on the Charge Transfer through the Double Helix: A Theoretical Study
Abstract
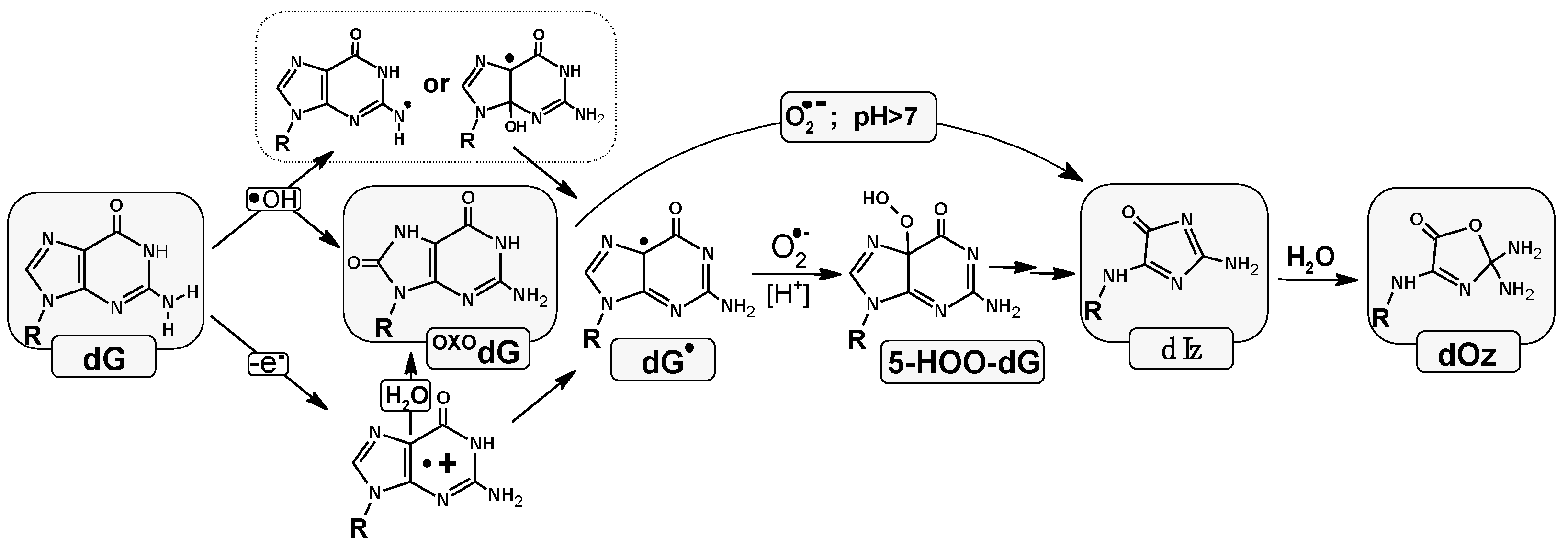
1. Introduction
2. Results
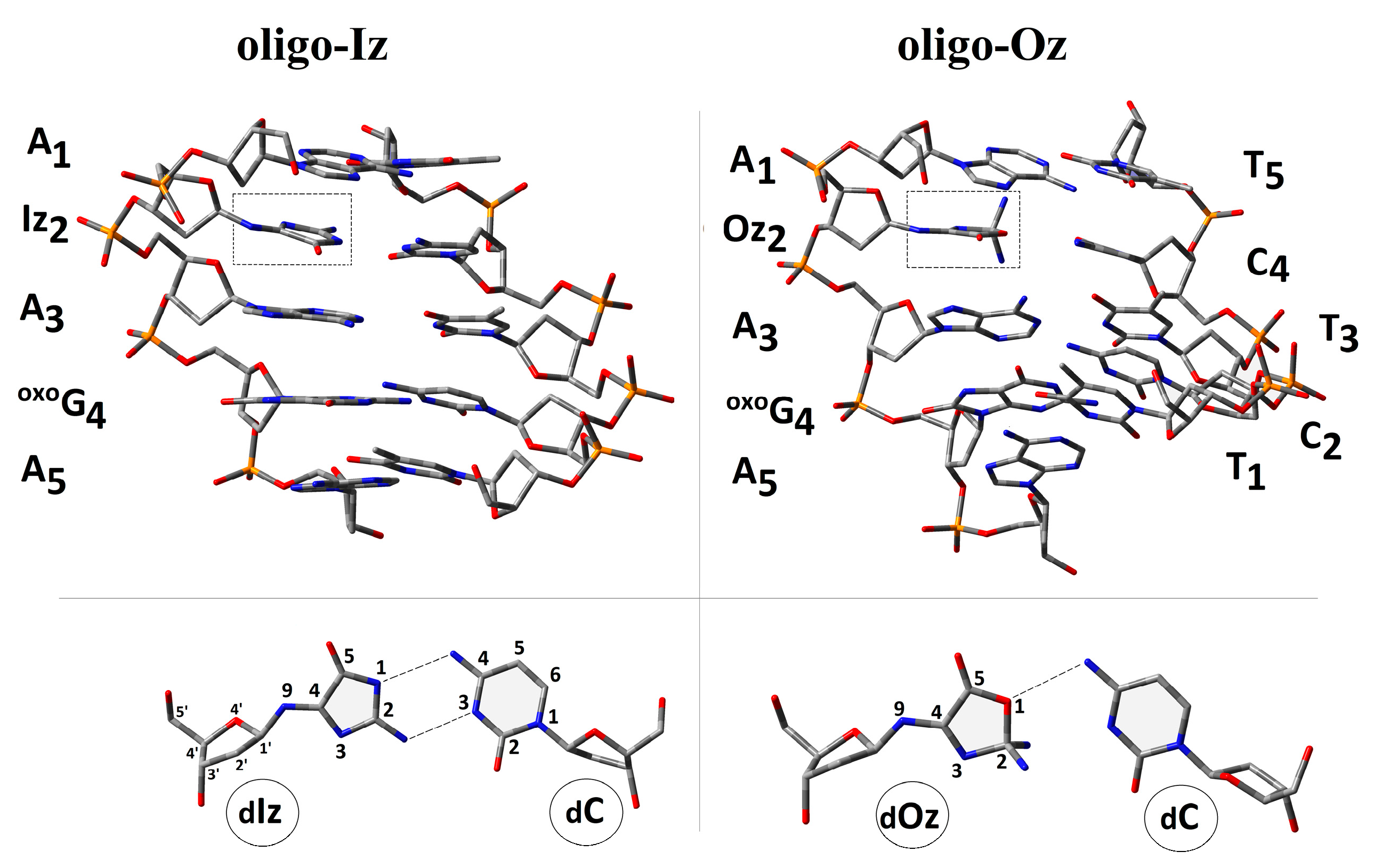
2.1. Influence of Iz and Oz on Double Helix Spatial Geometry
2.2. Influence of Iz and Oz on Double Hydrogen Bonds and Stacking Energies
2.3. Influence of Iz and Oz on Double Helix Electronic Properties
2.4. Influence of Iz or Oz on Charge Transfer through oligo-Iz and oligo-Oz
3. Discussion
4. Materials and Methods
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, T.A. Genomes, 2nd ed.; Wiley-Liss: Oxford, UK, 2002; ISBN 10: 0-471-25046-5. [Google Scholar]
- Nissanka, N.; Minczuk, M.; Moraes, C.T. Mechanisms of Mitochondrial DNA Deletion Formation. Trends Genet. 2019, 35, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Sudhir Ambekar, S. DNA: Damage and Repair Mechanisms in Humans. Glob. J. Pharm. Pharm. Sci. 2017, 3, 555613. [Google Scholar] [CrossRef][Green Version]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.D.; Dizdaroglu, M.; Cooke, M.S. Oxidative DNA damage and disease: Induction, repair and significance. Mutat. Res. /Rev. Mutat. Res. 2004, 567, 1–61. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef] [PubMed]
- Poletto, M.; Legrand, A.J.; Dianov, G.L. DNA Base Excision Repair: The Achilles’ Heel of Tumour Cells and their Microenvironment? Curr. Pharm. Des. 2020, 23, 4758–4772. [Google Scholar] [CrossRef]
- Farnell, D.A. Nucleotide Excision Repair in the Three Domains of Life. West. Undergrad. Res. J. Health Nat. Sci. 2011, 2, 1–6. [Google Scholar] [CrossRef]
- Jacobs, A.L.; Schär, P. DNA glycosylases: In DNA repair and beyond. Chromosoma 2012, 121, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Steenken, S.; Jovanovic, S.V. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997, 119, 617–618. [Google Scholar] [CrossRef]
- Khanduri, D.; Adhikary, A.; Sevilla, M.D. Highly Oxidizing Excited States of One-Electron-Oxidized Guanine in DNA: Wavelength and pH Dependence. J. Am. Chem. Soc. 2011, 12, 4527–4537. [Google Scholar] [CrossRef]
- Barry Halliwell, A.; Adhikary, M.; Dingfelder, M.D. Hydroxyl radical is a significant player in oxidative DNA damage in vivo. Chem. Soc. Rev. 2021, 50, 8355–8360. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 157. [Google Scholar] [CrossRef] [PubMed]
- Keith, B.; Simon, M.C. Hypoxia-Inducible Factors, Stem Cells, and Cancer. Cell 2007, 129, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Scanlan, L.D.; Coskun, S.H.; Jaruga, P.; Hanna, S.K.; Sims, C.M.; Almeida, J.L.; Catoe, D.; Coskun, E.; Golan, R.; Dizdaroglu, M.; et al. Measurement of Oxidatively Induced DNA Damage in Caenorhabditis elegans with High-Salt DNA Extraction and Isotope-Dilution Mass Spectrometry. Anal. Chem. 2019, 91, 12149–12155. [Google Scholar] [CrossRef] [PubMed]
- Matter, B.; Malejka-giganti, D.; Csallany, A.S.; Tretyakova, N. Quantitative analysis of the oxidative DNA lesion, 2,2-diamino-4-(2-deoxy-b-D-erythro-in vitro and in vivo by isotope dilution-capillary HPLC-ESI-MS/MS. Nucleic Acids Res. 2006, 34, 5449–5460. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat. Res.—Fundam. Mol. Mech. Mutagen. 2005, 591, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Kamiya, H. Mutations induced by 8-hydroxyguanine (8-oxo-7,8-dihydroguanine), a representative oxidized base, in mammalian cells. Genes Environ. 2017, 39, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Kairupan, C.; Scott, R.J. Base excision repair and the role of MUTYH. Hered. Cancer Clin. Pract. 2007, 5, 199–209. [Google Scholar] [CrossRef]
- Choi, S.; Cooley, R.B.; Hakemian, A.S.; Larrabee, Y.C.; Bunt, R.C.; Maupas, S.D.; Muller, J.G.; Burrows, C.J. Mechanism of Two-Electron Oxidation of Deoxyguanosine 5′-Monophosphate by a Platinum(IV) Complex. J. Am. Chem. Soc. 2004, 126, 591–598. [Google Scholar] [CrossRef]
- Kino, K.; Kawada, T.; Hirao-Suzuki, M.; Morikawa, M.; Miyazawa, H. Products of oxidative guanine damage form base pairs with guanine. Int. J. Mol. Sci. 2020, 21, 7645. [Google Scholar] [CrossRef]
- Morikawa, M.; Kino, K.; Oyoshi, T.; Suzuki, M.; Kobayashi, T.; Miyazawa, H. Product analysis of photooxidation in isolated quadruplex DNA; 8-oxo-7,8-dihydroguanine and its oxidation product at 3′-G are formed instead of 2,5-diamino-4H-imidazol-4-one. RSC Adv. 2013, 3, 25694–25697. [Google Scholar] [CrossRef]
- Suzuki, M.; Kino, K.; Kawada, T.; Morikawa, M.; Kobayashi, T.; Miyazawa, H. Analysis of Nucleotide Insertion Opposite 2,2,4-Triamino-5(2H)-oxazolone by Eukaryotic B- and Y-Family DNA Polymerases. Chem. Res. Toxicol. 2015, 28, 1307–1316. [Google Scholar] [CrossRef]
- Cadet, J.; Angelov, D.; Wagner, J.R. Hydroxyl radical is predominantly involved in oxidatively generated base damage to cellular DNA exposed to ionizing radiation. Int. J. Radiat. Biol. 2022, 98, 1684–1690. [Google Scholar] [CrossRef]
- Duarte, V. In vitro DNA synthesis opposite oxazolone and repair of this DNA damage using modified oligonucleotides. Nucleic Acids Res. 2000, 28, 1555–1563. [Google Scholar] [CrossRef] [PubMed]
- Henderson, P.T.; Delaney, J.C.; Gu, F.; Tannenbaum, S.R.; Essigmann, J.M. Oxidation of 7,8-dihydro-8-oxoguanine affords lesions that are potent sources of replication errors in vivo. Biochemistry 2002, 41, 914–921. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, M.; Fleming, A.M.; Burrows, C.J.; Wallace, S.S. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J. Biol. Chem. 2013, 288, 27263–27272. [Google Scholar] [CrossRef]
- Karahalil, B.; Hogue, B.A.; Souza-Pinto, N.C.; Bohr, V.A. Base excision repair capacity in mitochondria and nuclei: Tissue-specific variations. FASEB J. 2002, 16, 1895–1902. [Google Scholar] [CrossRef] [PubMed]
- Romano, C.A. DNA-mediated charge transfer between [4Fe-4S] cluster glycosylases. Ph.D. Dissertation, California Institute of Technology, Pasadena, CA, USA, 2011. [Google Scholar]
- White, M.F.; Dillingham, M.S. Iron-sulphur clusters in nucleic acid processing enzymes. Curr. Opin. Struct. Biol. 2012, 22, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Lomax, M.E.; Folkes, L.K.; Neill, P.O. Biological Consequences of Radiation-induced DNA Damage: Relevance to Radiotherapy Statement of Search Strategies Used and Sources of Information Why Radiation Damage is More Effective than Endogenous Damage at Killing Cells Ionising Radiation-induced Do. Clin. Oncol. 2013, 25, 578–585. [Google Scholar] [CrossRef]
- Bukowska, B.; Karwowski, B.T. The Clustered DNA Lesions—Types, Pathways of Repair and Relevance to Human Health. Curr. Med. Chem. 2018, 25, 2722–2735. [Google Scholar] [CrossRef]
- Karwowski, B.T. The Influence of Single, Tandem, and Clustered DNA Damage on the Electronic Properties of the Double Helix: A Theoretical Study. Molecules 2020, 25, 3126. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Zhang, Y.; Liu, C.; Zhang, M.; Han, S. Application of radiosensitizers in cancer radiotherapy. Int. J. Nanomed. 2021, 16, 1083–1102. [Google Scholar] [CrossRef]
- Huang, R.; Zhou, P. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target. Ther. 2020, 5, 60. [Google Scholar] [CrossRef]
- Mignon, P.; Loverix, S.; Steyaert, J.; Geerlings, P. Influence of the π–π interaction on the hydrogen bonding capacity of stacked DNA/RNA bases. Nucleic Acids Res. 2005, 33, 1779–1789. [Google Scholar] [CrossRef] [PubMed]
- Boal, A.K.; Yavin, E.; Lukianova, O.A.; O’Shea, V.L.; David, S.S.; Barton, J.K. DNA-bound redox activity of DNA repair glycosylases containing [4Fe-4S] clusters. Biochemistry 2005, 44, 8397–8407. [Google Scholar] [CrossRef]
- Boal, A.K.; Yavin, E.; Barton, J.K. DNA repair glycosylases with a [4Fe-4S] cluster: A redox cofactor for DNA-mediated charge transport? J. Inorg. Biochem. 2007, 101, 1913–1921. [Google Scholar] [CrossRef][Green Version]
- Karwowski, B. How Clustered DNA Damage Can Change the Electronic Properties of ds-DNA, Differences between GAG, GAOXOG, OXOGAOXOG. Biomolecules 2023, 13, 517. [Google Scholar] [CrossRef]
- Dapprich, S.; Komáromi, I.; Byun, K.S.; Morokuma, K.; Frisch, M.J. A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J. Mol. Struct. THEOCHEM 1999, 461–462, 1–21. [Google Scholar] [CrossRef]
- Miertus, S.; Tomasi, J. Approximate evaluations of the electrostatic free energy and internal energy changes in solution processes. Chem. Phys. 1982, 65, 239–245. [Google Scholar] [CrossRef]
- Cancès, E.; Mennucci, B.; Tomasi, J. A new integral equation formalism for the polarizable continuum model: Theoretical background and applications to Isotropic and anisotropic dielectrics. J. Chem. Phys. 1997, 107, 3032–3041. [Google Scholar] [CrossRef]
- Olson, W.K.; Bansal, M.; Burley, S.K.; Dickerson, R.E.; Gerstein, M.; Harvey, S.C.; Heinemann, U.; Lu, X.J.; Neidle, S.; Shakked, Z.; et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001, 313, 229–237. [Google Scholar] [CrossRef]
- Kim, H.J.; Ro, Y.; Hong, B.; Ji, H.G. Formation of au nanowires by DNA molecules as a template. J. Korean Phys. Soc. 2009, 55, 1892–1895. [Google Scholar] [CrossRef]
- Sontz, P.A.; Mui, T.P.; Fuss, J.O.; Tainer, J.A.; Barton, J.K. DNA charge transport as a first step in coordinating the detection of lesions by repair proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 1856–1861. [Google Scholar] [CrossRef] [PubMed]
- Arnittali, M.; Rissanou, A.N.; Harmandaris, V. Structure of Biomolecules Through Molecular Dynamics Simulations. Procedia Comput. Sci. 2019, 156, 69–78. [Google Scholar] [CrossRef]
- Marenich, A.V.; Jerome, S.V.; Cramer, C.J.; Truhlar, D.G. Charge Model 5: An Extension of Hirshfeld Population Analysis for the Accurate Description of Molecular Interactions in Gaseous and Condensed Phases. J. Chem. Theory Comput. 2012, 8, 527–541. [Google Scholar] [CrossRef]
- Voityuk, A.A. Estimates of electronic coupling for excess electron transfer in DNA. J. Chem. Phys. 2005, 123, 34903. [Google Scholar] [CrossRef]
- Ram Kumar Pandian, S.; Yuan, C.J.; Lin, C.C.; Wang, W.H.; Chang, C.C. DNA-based nanowires and nanodevices. Adv. Phys. X 2017, 2, 22–34. [Google Scholar] [CrossRef]
- Lewis, F.D.; Liu, J.; Weigel, W.; Rettig, W.; Kurnikov, I.V.; Beratan, D.N. Donor-bridge-acceptor energetics determine the distance dependence of electron tunneling in DNA. Proc. Natl. Acad. Sci. USA 2002, 99, 12536–12541. [Google Scholar] [CrossRef]
- Genereux, J.C.; Barton, J.K. Mechanisms for DNA charge transport. Chem. Rev. 2010, 110, 1642–1662. [Google Scholar] [CrossRef]
- Marcus, R.A. Electron transfer reactions in chemistry. Theory and experiment. Rev. Mod. Phys. 1993, 65, 599–610. [Google Scholar] [CrossRef]
- Rawtani, D.; Kuntmal, B.; Agrawal, Y. Charge transfer in DNA and its diverse modelling approaches. Front. Life Sci. 2016, 9, 214–225. [Google Scholar] [CrossRef][Green Version]
- Bolton, J.R.; Archer, M.D. Basic Electron-Transfer Theory. In Electron Transfer in Inorganic, Organic, and Biological Systems; Bolton, J.R., Mataga, N., McLendon, G., Eds.; American Chemical Society: Washington, DC, USA, 1991; Volume 228, pp. 7–23. [Google Scholar]
- Rösch, N.; Voityuk, A.A. Quantum Chemical Calculation of Donor–Acceptor Coupling for Charge Transfer in DNA. In Long-Range Charge Transfer in DNA II; Springer: Berlin/Heidelberg, Germany, 2012; pp. 37–72. [Google Scholar] [CrossRef]
- Khan, A. Reorganization energy, activation energy, and mechanism of hole transfer process in DNA: A theoretical study. J. Chem. Phys. 2008, 128, 075101. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M. Oxidatively Induced DNA Damage and Its Repair. Ref. Modul. Life Sci. 2017, 1–5. [Google Scholar] [CrossRef]
- Olinski, R.; Siomek, A.; Rozalski, R.; Gackowski, D.; Foksinski, M.; Guz, J.; Dziaman, T.; Szpila, A.; Tudek, B. Oxidative damage to DNA and antioxidant status in aging and age-related diseases. Acta Biochim. Pol. 2007, 54, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Demple, B.; Harrison, L. Repair of oxidative damage to DNA: Enzymology and biology. Annu. Rev. Biochem. 1994, 63, 915–948. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.S. Biological consequences of free radical-damaged dna bases. Free Radic. Biol. Med. 2002, 33, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hang, B. Base excision repair. In DNA Repair, Genetic Instability, and Cancer, 1st ed.; Wei, Q., Li, L., Chen, D.J., Eds.; World Scientific: Singapore, 2007; pp. 23–64. [Google Scholar] [CrossRef]
- Shikazono, N.; Pearson, C.; Neill, P.O.; Thacker, J. The roles of specific glycosylases in determining the mutagenic consequences of clustered DNA base damage. Nucleic Acids Res. 2006, 34, 3722–3730. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.S. DNA glycosylases search for and remove oxidized DNA bases. Environ. Mol. Mutagen. 2013, 54, 691–704. [Google Scholar] [CrossRef]
- Byrne, S.; Cunniffe, S.; O’Neill, P.; Lomax, M.E. 5,6-Dihydrothymine impairs the base excision repair pathway of a closely opposed AP site or single-strand break. Radiat. Res. 2009, 172, 537–549. [Google Scholar] [CrossRef]
- Friedman, J.I.; Stivers, J.T. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry 2010, 49, 4957–4967. [Google Scholar] [CrossRef]
- Tse, E.C.M.; Zwang, T.J.; Barton, J.K. The Oxidation State of [4Fe4S] Clusters Modulates the DNA-Binding Affinity of DNA Repair Proteins. J. Am. Chem. Soc. 2017, 136, 12784–12792. [Google Scholar] [CrossRef] [PubMed]
- Baranovskiy, A.G.; Babayeva, N.D.; Zhang, Y.; Blanco, L.; Pavlov, Y.I.; Tahirov, T.H. Comment on “The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science 2017, 357, 2954. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Burrows, C.J. Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Radic. Biol. Med. 2017, 107, 35–52. [Google Scholar] [CrossRef] [PubMed]
- Ming, X.; Matter, B.; Song, M.; Veliath, E.; Shanley, R.; Jones, R. Mapping Structurally Defined Guanine Oxidation Products along DNA Duplexes: Influence of Local Sequence Context and Endogenous Cytosine Methylation. J. Am. Chem. Soc. 2014, 136, 4223–4235. [Google Scholar] [CrossRef] [PubMed]
- Fujitsuka, M.; Majima, T. Charge transfer in DNA. Pure Appl. Chem. 2013, 85, 1367–1377. [Google Scholar] [CrossRef]
- Yin, J.; Zhang, N.; Wang, H. Liquid chromatography-mass spectrometry for analysis of DNA damages induced by environmental exposure. Trends in Analytical Chemistry 2019, 120, 1–11. [Google Scholar] [CrossRef]
- Neeley, W.L.; Delaney, S.; Alekseyev, Y.O.; Jarosz, D.F.; Delaney, J.C.; Walker, G.C.; Essigmann, J.M. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J. Biol. Chem. 2007, 282, 12741–12748. [Google Scholar] [CrossRef] [PubMed]
- Syed, A.; Tainer, J.A. Charge Transport Communication through DNA by Protein Fe − S Clusters: How Far Is Not Too Far? ACS Cent. Sci. 2019, 5, 7–9. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, A.H.S.; da Silva, A.E.; de Oliveira, I.M.; Henriques, J.A.P.; Agnez-Lima, L.F. MutY-glycosylase: An overview on mutagenesis and activities beyond the GO system. Mutat. Res.—Fundam. Mol. Mech. Mutagen. 2014, 769, 119–131. [Google Scholar] [CrossRef]
- Arnold, A.R.; Grodick, M.A.; Barton, J.K. DNA Charge Transport: From Chemical Principles to the Cell. Cell Chem. Biol. 2016, 23, 183–197. [Google Scholar] [CrossRef]
- Eccles, L.J.; Neill, P.O.; Lomax, M.E. Delayed repair of radiation induced clustered DNA damage: Friend or foe? Mutat. Res. /Fundam. Mol. Mech. Mutagen. 2011, 711, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Karwowski, B.T. Charge Transfer Depends on Its Diastereomeric Form: A Theoretical Study. Antioxidants 2023, 12, 881. [Google Scholar] [CrossRef] [PubMed]
- BIOVIA. BIOVIA. Discovery Studio Visualizer; v16.1.0.15350; BIOVIA: San Diego, CA, USA, 2015. [Google Scholar]
- Zhao, Y.; Pu, J.; Lynch, B.J.; Truhlar, D.G. Tests of second-generation and third-generation density functionals for thermochemical kinetics. Phys. Chem. Chem. Phys. 2004, 6, 673–676. [Google Scholar] [CrossRef]
- Lin, H.; Truhlar, D.G. Redistributed charge and dipole schemes for combined quantum mechanical and molecular mechanical calculations. J. Phys. Chem. A 2005, 109, 3991–4004. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Truhlar, D.G. QM/MM: What have we learned, where are we, and where do we go from here? Theor. Chem. Acc. 2007, 117, 185–199. [Google Scholar] [CrossRef]
- Mayhall, N.J.; Raghavachari, K. Charge transfer across ONIOM QM:QM boundaries: The impact of model system preparation. J. Chem. Theory Comput. 2010, 6, 3131–3136. [Google Scholar] [CrossRef] [PubMed]
- Hirshfeld, F.L. Bonded-atom fragments for describing molecular charge densities. Theor. Chim. Acta 1977, 44, 129–138. [Google Scholar] [CrossRef]
- Karwowski, B.T. Ionisation potential and electron affinity of free 5′,8-cyclopurine-2′-deoxynucleosides. DFT study in gaseous and aqueous phase. Cent. Eur. J. Chem. 2010, 8, 70–76. [Google Scholar] [CrossRef]
- Karwowski, B.T. The AT Interstrand Cross-Link: Structure, Electronic Properties, and Influence on Charge Transfer in dsDNA. Mol. Ther.—Nucleic Acids 2018, 13, 665–685. [Google Scholar] [CrossRef]
- Karwowski, B.T. The 2Ih and OXOG Proximity Consequences on Charge Transfer through ds -DNA: Theoretical Studies of Clustered DNA Damage. Molecules 2023, 28, 2180. [Google Scholar] [CrossRef]
- Li, X.; Cai, Z.; Sevilla, M.D. DFT calculations of the electron affinities of nucleic acid bases: Dealing with negative electron affinities. J. Phys. Chem. A 2002, 106, 1596–1603. [Google Scholar] [CrossRef]
- Kumar, A.; Sevilla, M.D. Photoexcitation of dinucleoside radical cations: A time-dependent density functional study. J. Phys. Chem. B 2006, 110, 24181–24188. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cave, R.J.; Newton, M.D. Generalization of the Mulliken-Hush treatment for the calculation of electron transfer matrix elements. Chem. Phys. Lett. 1996, 249, 15–19. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 16, Revision C.01; Gaussian Inc.: Wallingford, CT, USA, 2019. [Google Scholar]
 adiabatic radical anion,
adiabatic radical anion,  vertical radical anion, non-equilibrated solvent–solute interaction,
vertical radical anion, non-equilibrated solvent–solute interaction,  vertical radical cation/anion, equilibrated solvent–solute interaction, and
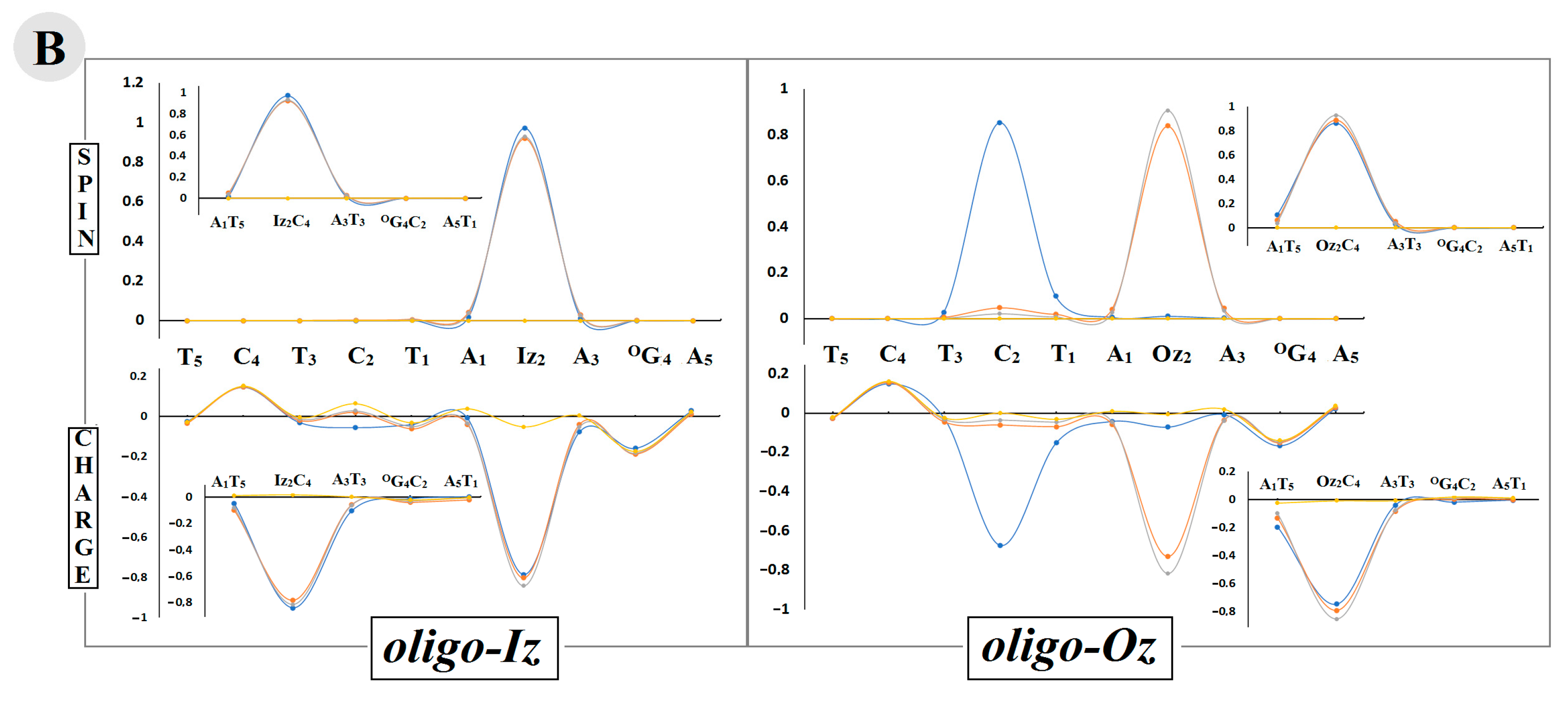
vertical radical cation/anion, equilibrated solvent–solute interaction, and  neutral form. The raw data of the charge and spin distribution have been given in Tables S3 and S4 (Supplementary Materials). (B) A graphical visualization of the spin distribution within oligo-Iz and oligo-Oz. The raw data have been given in Table S5.
neutral form. The raw data of the charge and spin distribution have been given in Tables S3 and S4 (Supplementary Materials). (B) A graphical visualization of the spin distribution within oligo-Iz and oligo-Oz. The raw data have been given in Table S5.
 adiabatic radical anion,
adiabatic radical anion,  vertical radical anion, non-equilibrated solvent–solute interaction,
vertical radical anion, non-equilibrated solvent–solute interaction,  vertical radical cation/anion, equilibrated solvent–solute interaction, and
vertical radical cation/anion, equilibrated solvent–solute interaction, and  neutral form. The raw data of the charge and spin distribution have been given in Tables S3 and S4 (Supplementary Materials). (B) A graphical visualization of the spin distribution within oligo-Iz and oligo-Oz. The raw data have been given in Table S5.
neutral form. The raw data of the charge and spin distribution have been given in Tables S3 and S4 (Supplementary Materials). (B) A graphical visualization of the spin distribution within oligo-Iz and oligo-Oz. The raw data have been given in Table S5.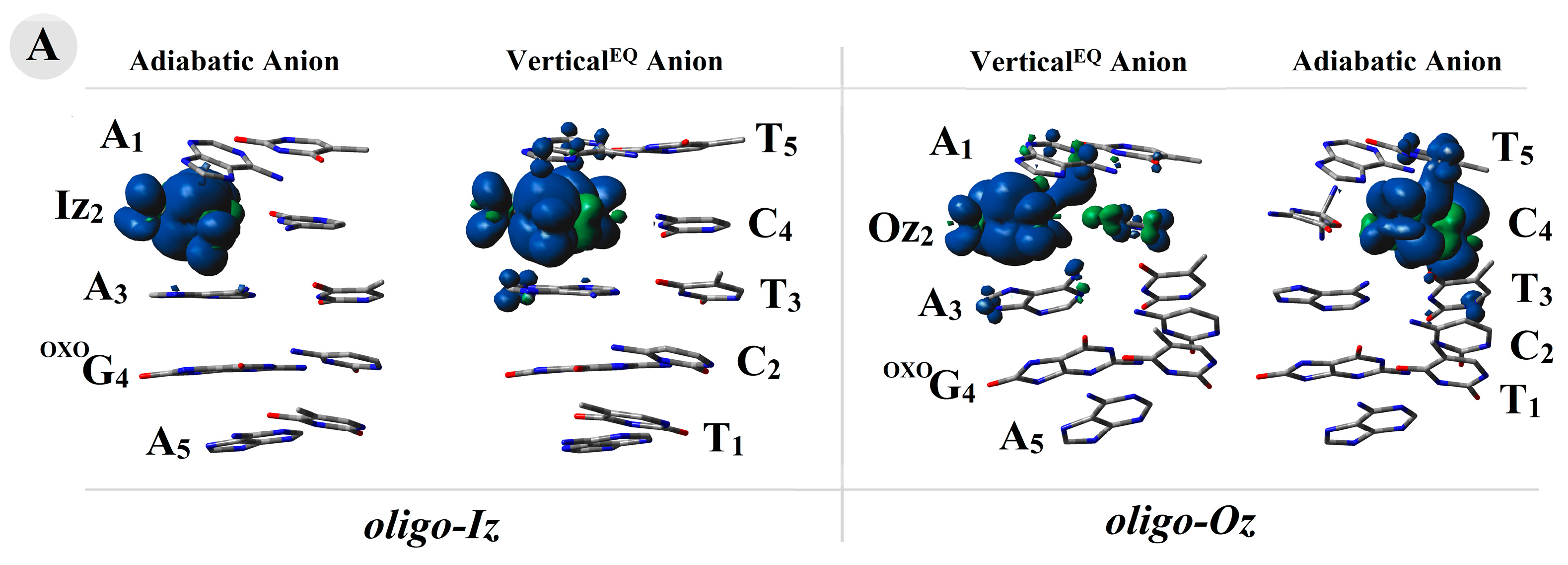
| VIPNE | VIPEQ | AIP | VEANE | VEAEQ | AEA | |
|---|---|---|---|---|---|---|
| oligo-Iz | (a) 6.47 | 5.85 | 5.47 | 1.53 | 2.22 | 2.83 |
| (b) 6.25 | 5.78 | 5.37 | 1.08 | 2.14 | 2.81 | |
| oligo-Oz | (a) 6.52 | 5.89 | 5.48 | 0.95 | 1.49 | 2.06 |
| (b) 6.36 | 5.82 | 5.40 | 0.48 | 1.58 | 1.78 | |
| oligo-N * | *(a) 6.72 | 6.08 | 5.65 | 0.84 | 1.58 | 2.09 |
| *(b) 6.48 | 5.98 | 5.58 | 0.60 | 1.34 | 1.90 | |
| RMSD: Anion versus Neutral | RMSD: Cation versus Neutral | |||||
| ds-DNA | BP | PS-Frame | ds-DNA | BP | PS-Frame | |
| oligo-Iz | 0.72 | 0.49 | 0.87 | 0.18 | 0.15 | 0.21 |
| oligo-Oz | 0.46 | 0.38 | 0.52 | 0.59 | 0.46 | 0.69 |
| oligo-N * | 0.17 | 0.16 | 0.17 | 0.36 | 0.29 | 0.42 |
| oligo-Iz | oligo-Oz | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Base Pair | VIP | AIP | VEA | AEA | Base Pair | VIP | AIP | VEA | AEA |
| A1T5 | 6.63 | 6.65 | 1.43 | 1.41 | A1T5 | 6.65 | 6.72 | 1.39 | 1.40 |
| Iz2C4 | 7.04 | 7.03 | 2.45 | 2.97 | Oz2C4 | 7.01 | 7.03 | 1.82 | 1.89 |
| A3T3 | 6.65 | 6.66 | 1.43 | 1.31 | A3T3 | 6.62 | 6.63 | 1.42 | 1.44 |
| OXOG4C2 | 5.94 | 5.51 | 1.51 | 1.53 | OXOG4C2 | 5.91 | 5.56 | 1.52 | 1.48 |
| A5T1 | 6.73 | 6.69 | 1.43 | 1.43 | A5T1 | 6.70 | 6.68 | 1.43 | 1.43 |
| GC [39] | 6.13 | 5.83 | 1.52 | 1.95 | AT [39] | 6.65 | 6.60 | 1.40 | 1.40 |
| System | Electron-Hole Transfer | Graphics | ||||
|---|---|---|---|---|---|---|
| oligo-Iz | λ | ΔG | Ea | V12 | kHT [s−1] |  |
| A1T5 ← Iz2C4 | −0.02 | −0.38 | −2.04 | 0.22 | 0.00 | |
| Iz2C4 → A3T3 | −0.02 | −0.37 | −2.28 | 0.23 | 0.00 | |
| A3T3 → OXOG4C2 | 0.44 | −1.15 | 0.28 | 0.43 | 8.49 × 1010 | |
| OXOG4C2 ← A5T1 | 0.34 | −1.18 | 0.52 | 0.39 | 7.22 × 106 | |
| A1T5 ← A3T3 | −0.03 | −0.01 | −0.01 | 0.01 | 0.00 | |
| Iz2C4 → OXOG4C2 | 0.43 | −1.52 | 0.70 | 0.59 | 1.23 × 104 | |
| A3T3 ← A5T1 | −0.01 | −0.03 | −0.04 | 0.04 | 0.00 | |
| oligo-Iz | Excess Electron Transfer | |||||
| A1T5 → Iz2C4 | 0.52 | −1.56 | 0.52 | 0.37 | 5.73 × 106 |  |
| Iz2C4 ← A3T3 | 0.61 | −1.66 | 0.45 | 0.41 | 8.86 × 107 | |
| A3T3 → OXOG4C2 | 0.10 | −0.22 | 0.03 | 0.01 | 7.10 × 1011 | |
| OXOG4C2 ← A5T1 | −0.02 | −0.09 | −0.16 | 0.05 | 0.00 | |
| A1T5 ← A3T3 | −0.11 | −0.10 | −0.10 | 0.07 | 0.00 | |
| Iz2C4 ← OXOG4C2 | 0.54 | −1.44 | 0.37 | 0.34 | 1.32 × 109 | |
| A3T3 → A5T1 | 0.10 | −0.12 | 0.002 | 0.07 | 2.74 × 1014 | |
| oligo-Oz | Electron-Hole Transfer | |||||
| A1T5 ← Oz2C4 | −0.03 | −0.31 | −0.88 | 0.21 | 0.00 |  |
| Oz2C4 → A3T3 | 0.02 | −0.40 | 1.57 | 0.32 | 3.43 × 10−11 | |
| A3T3→OXOG4C2 | 0.37 | −1.07 | 0.33 | 0.39 | 9.95 × 109 | |
| OXOG4C2 ← A5T1 | 0.36 | −1.12 | 0.39 | 0.39 | 1.04 × 109 | |
| A1T5 → A3T3 | 0.00 | −0.09 | 0.49 | 0.02 | 4.54 × 105 | |
| Oz2C4 → OXOG4C2 | 0.38 | −1.47 | 0.76 | 0.59 | 1.22 × 103 | |
| A3T3 ← A5T1 | −0.01 | −0.05 | −0.09 | 0.04 | 0.00 | |
| oligo-Oz | Excess Electron Transfer | |||||
| A1T5 → Oz2C4 | 0.13 | −0.49 | 0.25 | 0.10 | 2.38 × 1010 | 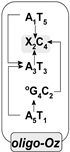 |
| Oz2C4 ← A3T3 | 0.06 | −0.46 | 0.60 | 0.07 | 1.95 × 104 | |
| A3T3 → OXOG4C2 | −0.04 | −0.04 | −0.04 | 0.08 | 0.00 | |
| OXOG4C2 ← A5T1 | 0.03 | −0.05 | 0.00 | 0.05 | 2.19 × 1014 | |
| A1T5 → A3T3 | 0.07 | −0.04 | 0.004 | 0.05 | 1.44 × 1014 | |
| Oz2C4 ← OXOG4C2 | 0.04 | −0.41 | 0.78 | 0.09 | 4.50 × 101 | |
| A3T3 ← A5T1 | 0.01 | −0.01 | 0.001 | 0.07 | 7.02 × 1014 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karwowski, B.T. The Influence of Clustered DNA Damage Containing Iz/Oz and OXOdG on the Charge Transfer through the Double Helix: A Theoretical Study. Molecules 2024, 29, 2754. https://doi.org/10.3390/molecules29122754
Karwowski BT. The Influence of Clustered DNA Damage Containing Iz/Oz and OXOdG on the Charge Transfer through the Double Helix: A Theoretical Study. Molecules. 2024; 29(12):2754. https://doi.org/10.3390/molecules29122754
Chicago/Turabian StyleKarwowski, Bolesław T. 2024. "The Influence of Clustered DNA Damage Containing Iz/Oz and OXOdG on the Charge Transfer through the Double Helix: A Theoretical Study" Molecules 29, no. 12: 2754. https://doi.org/10.3390/molecules29122754
APA StyleKarwowski, B. T. (2024). The Influence of Clustered DNA Damage Containing Iz/Oz and OXOdG on the Charge Transfer through the Double Helix: A Theoretical Study. Molecules, 29(12), 2754. https://doi.org/10.3390/molecules29122754







