Identification and Evaluation of Olive Phenolics in the Context of Amine Oxidase Enzyme Inhibition and Depression: In Silico Modelling and In Vitro Validation
Abstract
1. Introduction
2. Results and Discussion
2.1. In Silico Screen of Olea europaea Phenolics against FAD-Dependent Amine Oxidases
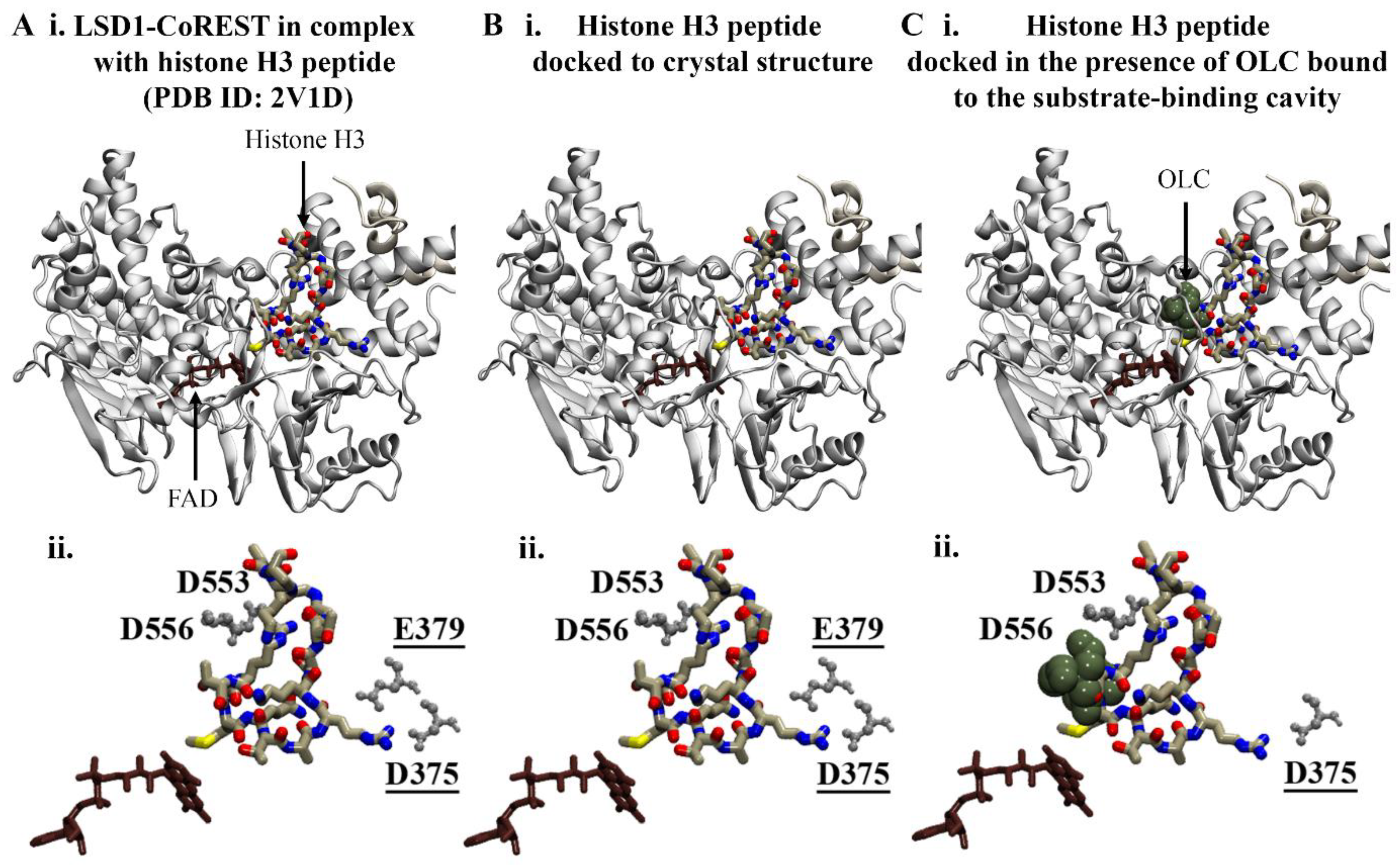
2.1.1. Substrate-Binding Cavity of LSD1
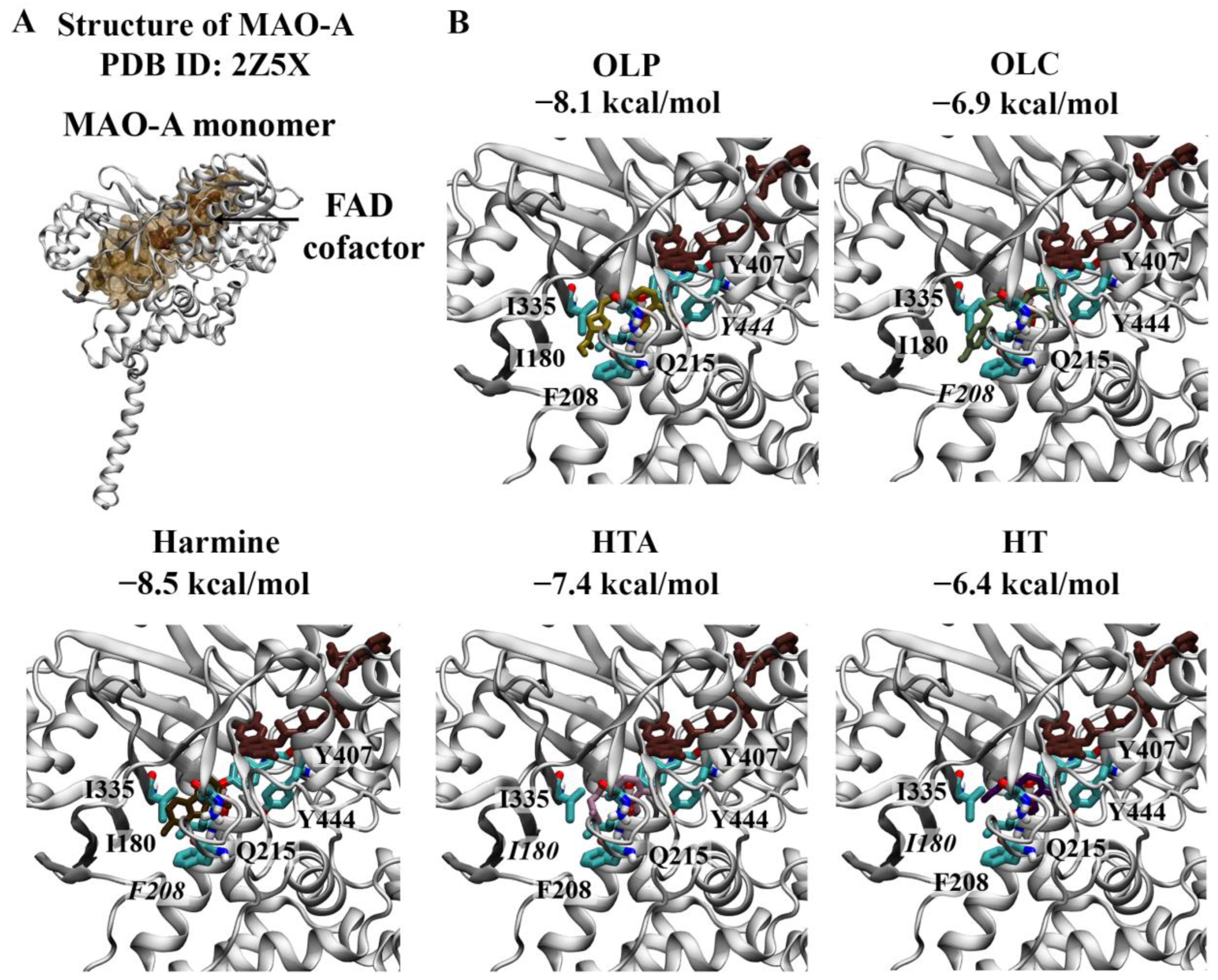
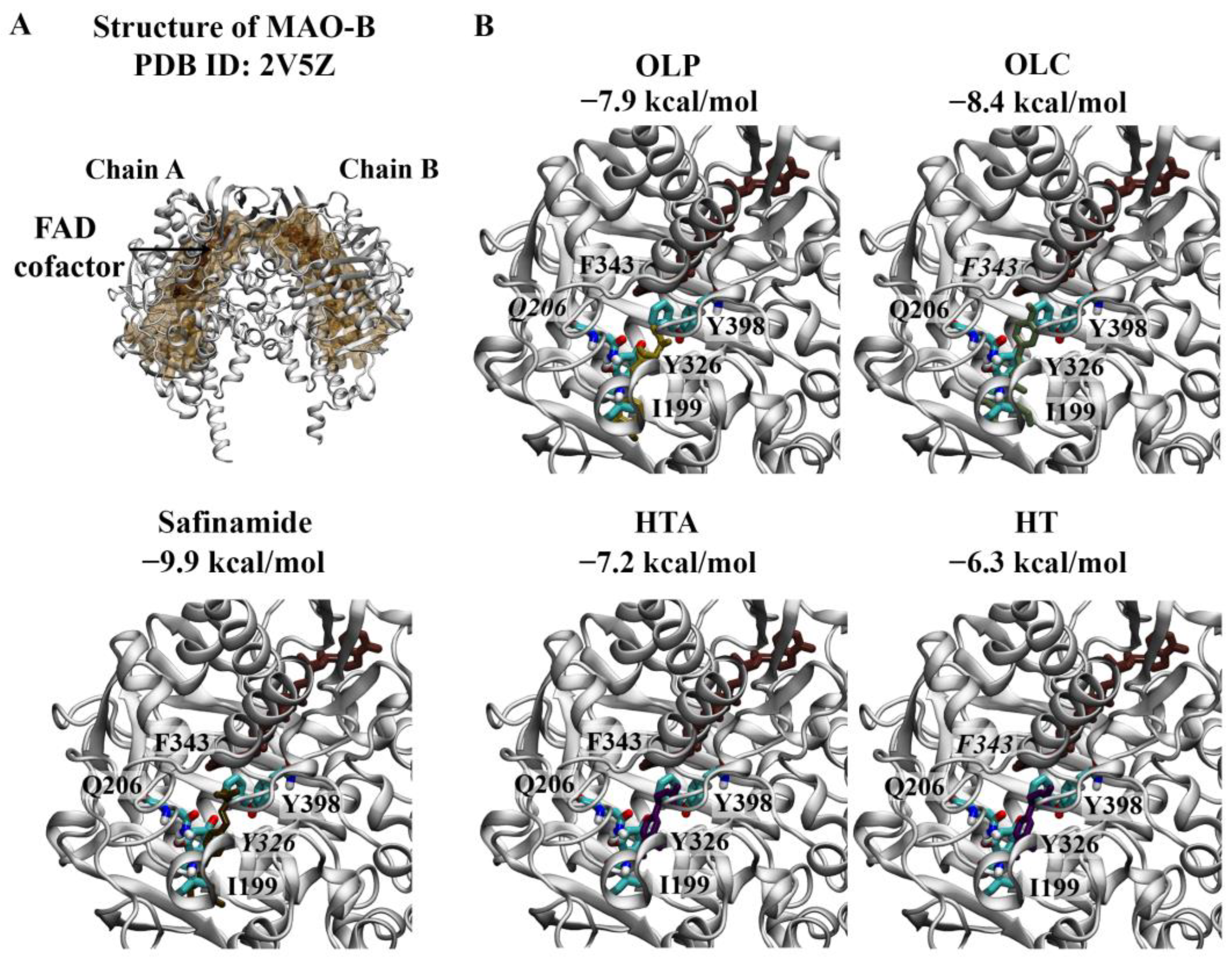
2.1.2. Monoamine Oxidases
2.2. Viability of BJ Cells
2.3. Inhibition of LSD1 Demethylase Activity
2.4. Inhibition of MAOs
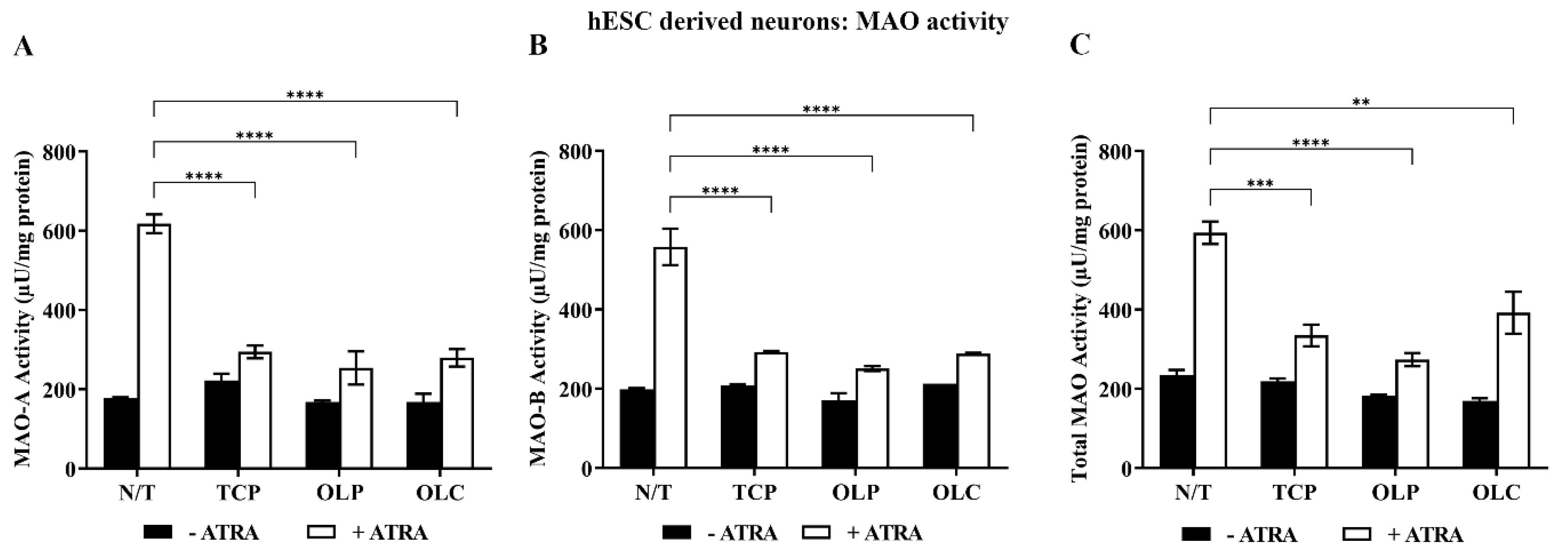
2.5. hESC-Derived Neurons: Inhibition of MAO Activity
3. Materials and Methods
3.1. Molecular Docking to FAD-Dependent Monoamine Oxidases
3.1.1. Preparation of Protein Structures and Ligands
3.1.2. Ligand-Binding Site Analysis and Molecular Docking
3.1.3. Protein–Peptide Docking
3.2. Cell Culture
3.2.1. BJ Cells
3.2.2. hESC-Derived Neurons
3.3. Cell Treatments and Analyses
3.3.1. Olea Europaea Phenolic Compounds
3.3.2. Western Blotting
3.3.3. Cell Viability
3.3.4. LSD1 Activity and H3K4 Mono-Methylation in BJ Cells
3.3.5. MAO Upregulation in BJ Cells
3.3.6. MAO Upregulation in hESC-Derived Neurons
3.3.7. MAO-A/B and Total MAO Activity Assay
3.4. Inhibitor Screening Assays
3.4.1. LSD1 Direct Screening Assay
3.4.2. MAO-A/B Direct Screening Assay
3.5. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- GBD 2019 Mental Disorders Collaborators. Global, regional, and national burden of 12 mental disorders in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet Psychiatry 2022, 9, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Penninx, B.W.J.H.; Lamers, F.; Milaneschi, Y.S. 03.02—Clinical heterogeneity in major depressive disorder. Eur. Neuropsychopharmacol. 2018, 28, S59–S60. [Google Scholar] [CrossRef]
- Goldberg, D. The heterogeneity of “major depression”. World Psychiatry 2011, 10, 226–228. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. International Classification of Diseases for Mortality and Morbidity Statistics (11th Revision); World Health Organization: Geneva, Switzerland, 2018.
- Depressive Disorders. In Diagnostic and Statistical Manual of Mental Disorders; American Psychiatric Association Publishing: Washington, DC, USA, 2022. [CrossRef]
- Lee, S.; Jeong, J.; Kwak, Y.; Park, S.K. Depression research: Where are we now? Mol. Brain 2010, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Pitsillou, E.; Bresnehan, S.M.; Kagarakis, E.A.; Wijoyo, S.J.; Liang, J.; Hung, A.; Karagiannis, T.C. The cellular and molecular basis of major depressive disorder: Towards a unified model for understanding clinical depression. Mol. Biol. Rep. 2020, 47, 753–770. [Google Scholar] [CrossRef]
- Son, S.Y.; Ma, J.; Kondou, Y.; Yoshimura, M.; Yamashita, E.; Tsukihara, T. Structure of human monoamine oxidase A at 2.2-A resolution: The control of opening the entry for substrates/inhibitors. Proc. Natl. Acad. Sci. USA 2008, 105, 5739–5744. [Google Scholar] [CrossRef]
- Bach, A.W.; Lan, N.C.; Johnson, D.L.; Abell, C.W.; Bembenek, M.E.; Kwan, S.W.; Seeburg, P.H.; Shih, J.C. cDNA cloning of human liver monoamine oxidase A and B: Molecular basis of differences in enzymatic properties. Proc. Natl. Acad. Sci. USA 1988, 85, 4934–4938. [Google Scholar] [CrossRef]
- Binda, C.; Wang, J.; Pisani, L.; Caccia, C.; Carotti, A.; Salvati, P.; Edmondson, D.E.; Mattevi, A. Structures of human monoamine oxidase B complexes with selective noncovalent inhibitors: Safinamide and coumarin analogs. J. Med. Chem. 2007, 50, 5848–5852. [Google Scholar] [CrossRef] [PubMed]
- Thase, M.E. The role of monoamine oxidase inhibitors in depression treatment guidelines. J. Clin. Psychiatry 2012, 73 (Suppl. S1), 10–16. [Google Scholar] [CrossRef]
- Tian, H.; Hu, Z.; Xu, J.; Wang, C. The molecular pathophysiology of depression and the new therapeutics. MedComm 2022, 3, e156. [Google Scholar] [CrossRef]
- Liu, L.; Wang, H.; Chen, X.; Zhang, Y.; Zhang, H.; Xie, P. Gut microbiota and its metabolites in depression: From pathogenesis to treatment. eBioMedicine 2023, 90, 104527. [Google Scholar] [CrossRef] [PubMed]
- Leonard, B.E. Inflammation and depression: A causal or coincidental link to the pathophysiology? Acta Neuropsychiatr. 2018, 30, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Mitsis, T.; Papakonstantinou, E.; Vlachakis, D. Biological mechanisms underlying depression, epigenetics and their interplay (Review). Int. J. Epigen. 2023, 3, 3. [Google Scholar] [CrossRef]
- Wu, M.S.; Li, X.J.; Liu, C.Y.; Xu, Q.; Huang, J.Q.; Gu, S.; Chen, J.X. Effects of Histone Modification in Major Depressive Disorder. Curr. Neuropharmacol. 2022, 20, 1261–1277. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhou, C.; Yao, Y.; Wei, L.; Feng, Z.; Deng, L.; Song, Y. 3-(Piperidin-4-ylmethoxy)pyridine Containing Compounds Are Potent Inhibitors of Lysine Specific Demethylase 1. J. Med. Chem. 2016, 59, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Perillo, B.; Tramontano, A.; Pezone, A.; Migliaccio, A. LSD1: More than demethylation of histone lysine residues. Exp. Mol. Med. 2020, 52, 1936–1947. [Google Scholar] [CrossRef] [PubMed]
- Binda, C.; Li, M.; Hubalek, F.; Restelli, N.; Edmondson, D.E.; Mattevi, A. Insights into the mode of inhibition of human mitochondrial monoamine oxidase B from high-resolution crystal structures. Proc. Natl. Acad. Sci. USA 2003, 100, 9750–9755. [Google Scholar] [CrossRef] [PubMed]
- Allen, S.R.; Amrein, R.; Guentert, T.W.; Hartmann, D.; Lorscheid, T.; Schoerlin, M.P.; Vranesic, D. The Pharmacology of Reversible Monoamine Oxidase Inhibitors. Br. J. Psychiatry 1989, 155, 66–71. [Google Scholar] [CrossRef]
- Corona, G.; Spencer, J.P.; Dessì, M.A. Extra virgin olive oil phenolics: Absorption, metabolism, and biological activities in the GI tract. Toxicol. Ind. Health 2009, 25, 285–293. [Google Scholar] [CrossRef]
- Estruch, R.; Ros, E.; Salas-Salvadó, J.; Covas, M.-I.; Corella, D.; Arós, F.; Gómez-Gracia, E.; Ruiz-Gutiérrez, V.; Fiol, M.; Lapetra, J.; et al. Primary Prevention of Cardiovascular Disease with a Mediterranean Diet Supplemented with Extra-Virgin Olive Oil or Nuts. N. Engl. J. Med. 2018, 378, e34. [Google Scholar] [CrossRef]
- Keys, A.; Keys, M. How to Eat Well and Stay Well the Mediterranean Way; Doubleday: New York, NY, USA, 1975. [Google Scholar]
- Menotti, A.; Puddu, P.E. How the Seven Countries Study contributed to the definition and development of the Mediterranean diet concept: A 50-year journey. Nutr. Metab. Cardiovasc. Dis. 2015, 25, 245–252. [Google Scholar] [CrossRef]
- Pett, K.D.; Willett, W.C.; Vartiainen, E.; Katz, D.L. The Seven Countries Study. Eur. Heart J. 2017, 38, 3119–3121. [Google Scholar] [CrossRef]
- Psaltopoulou, T.; Sergentanis, T.N.; Panagiotakos, D.B.; Sergentanis, I.N.; Kosti, R.; Scarmeas, N. Mediterranean diet, stroke, cognitive impairment, and depression: A meta-analysis. Ann. Neurol. 2013, 74, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Jacka, F.N.; O’Neil, A.; Opie, R.; Itsiopoulos, C.; Cotton, S.; Mohebbi, M.; Castle, D.; Dash, S.; Mihalopoulos, C.; Chatterton, M.L.; et al. A randomised controlled trial of dietary improvement for adults with major depression (the ‘SMILES’ trial). BMC Med. 2017, 15, 23. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, G.K.; Keast, R.S.J.; Morel, D.; Lin, J.; Pika, J.; Han, Q.; Lee, C.-H.; Smith, A.B.; Breslin, P.A.S. Ibuprofen-like activity in extra-virgin olive oil. Nature 2005, 437, 45–46. [Google Scholar] [CrossRef]
- Bonvino, N.P.; Liang, J.; McCord, E.D.; Zafiris, E.; Benetti, N.; Ray, N.B.; Hung, A.; Boskou, D.; Karagiannis, T.C. OliveNet™: A comprehensive library of compounds from Olea europaea. Database 2018, 2018, bay016. [Google Scholar] [CrossRef]
- Nan, J.N.; Ververis, K.; Bollu, S.; Rodd, A.L.; Swarup, O.; Karagiannis, T.C. Biological effects of the olive polyphenol, hydroxytyrosol: An extra view from genome-wide transcriptome analysis. Hell. J. Nucl. Med. 2014, 17 (Suppl. 1), 62–69. [Google Scholar] [PubMed]
- Gorzynik-Debicka, M.; Przychodzen, P.; Cappello, F.; Kuban-Jankowska, A.; Marino Gammazza, A.; Knap, N.; Wozniak, M.; Gorska-Ponikowska, M. Potential Health Benefits of Olive Oil and Plant Polyphenols. Int. J. Mol. Sci. 2018, 19, 686. [Google Scholar] [CrossRef] [PubMed]
- Serreli, G.; Deiana, M. Biological Relevance of Extra Virgin Olive Oil Polyphenols Metabolites. Antioxidants 2018, 7, 170. [Google Scholar] [CrossRef]
- Bianco, A.; Chiacchio, M.A.; Grassi, G.; Iannazzo, D.; Piperno, A.; Romeo, R. Phenolic components of Olea europea: Isolation of new tyrosol and hydroxytyrosol derivatives. Food Chem. 2006, 95, 562–565. [Google Scholar] [CrossRef]
- Yin, W.; Löf, M.; Chen, R.; Hultman, C.M.; Fang, F.; Sandin, S. Mediterranean diet and depression: A population-based cohort study. Int. J. Behav. Nut. Phys. Act. 2021, 18, 153. [Google Scholar] [CrossRef] [PubMed]
- Bizzozero-Peroni, B.; Martínez-Vizcaíno, V.; Fernández-Rodríguez, R.; Jiménez-López, E.; Núñez de Arenas-Arroyo, S.; Saz-Lara, A.; Díaz-Goñi, V.; Mesas, A.E. The impact of the Mediterranean diet on alleviating depressive symptoms in adults: A systematic review and meta-analysis of randomized controlled trials. Nut. Rev. 2024, nuad176. [Google Scholar] [CrossRef]
- Brown, E.S.; Chandler, P.A. Mood and Cognitive Changes During Systemic Corticosteroid Therapy. Prim. Care Companion J. Clin. Psychiatry 2001, 3, 17–21. [Google Scholar] [CrossRef]
- Warrington, T.P.; Bostwick, J.M. Psychiatric Adverse Effects of Corticosteroids. Mayo Clin. Proc. 2006, 81, 1361–1367. [Google Scholar] [CrossRef]
- Pitsillou, E.; Liang, J.; Hung, A.; Karagiannis, T.C. Chromatin modification by olive phenolics: In silico molecular docking studies utilising the phenolic groups categorised in the OliveNet™ database against lysine specific demethylase enzymes. J. Mol. Graph. Model. 2020, 97, 107575. [Google Scholar] [CrossRef] [PubMed]
- Pitsillou, E.; Liang, J.; Hung, A.; Karagiannis, T.C. Molecular docking utilising the OliveNet™ library reveals novel phenolic compounds which may potentially target key proteins associated with major depressive disorder. Comput. Biol. Chem. 2020, 86, 107234. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H.; Sato, S.; Hashimoto, T.; Matsuno, K.; Umehara, T. Crystal Structure of LSD1 in Complex with 4-[5-(Piperidin-4-ylmethoxy)-2-(p-tolyl)pyridin-3-yl]benzonitrile. Molecules 2018, 23, 1538. [Google Scholar] [CrossRef]
- Speranzini, V.; Rotili, D.; Ciossani, G.; Pilotto, S.; Marrocco, B.; Forgione, M.; Lucidi, A.; Forneris, F.; Mehdipour, P.; Velankar, S.; et al. Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features. Sci. Adv. 2016, 2, e1601017. [Google Scholar] [CrossRef]
- Maiques-Diaz, A.; Somervaille, T.C. LSD1: Biologic roles and therapeutic targeting. Epigenomics 2016, 8, 1103–1116. [Google Scholar] [CrossRef]
- Forneris, F.; Binda, C.; Adamo, A.; Battaglioli, E.; Mattevi, A. Structural Basis of LSD1-CoREST Selectivity in Histone H3 Recognition*. J. Biol. Chem. 2007, 282, 20070–20074. [Google Scholar] [CrossRef]
- Edelstein, S.B.; Castiglione, C.M.; Breakfield, X.O. Monoamine oxidase activity in normal and Lesch-Nyhan fibroblasts. J. Neurochem. 1978, 31, 1247–1254. [Google Scholar] [CrossRef] [PubMed]
- González-Acedo, A.; Ramos-Torrecillas, J.; Illescas-Montes, R.; Costela-Ruiz, V.J.; Ruiz, C.; Melguizo-Rodríguez, L.; García-Martínez, O. The Benefits of Olive Oil for Skin Health: Study on the Effect of Hydroxytyrosol, Tyrosol, and Oleocanthal on Human Fibroblasts. Nutrients 2023, 15, 2077. [Google Scholar] [CrossRef] [PubMed]
- Cuyàs, E.; Castillo, D.; Llorach-Parés, L.; Lozano-Sánchez, J.; Verdura, S.; Nonell-Canals, A.; Brunet, J.; Bosch-Barrera, J.; Joven, J.; Valdés, R.; et al. Computational de-orphanization of the olive oil biophenol oleacein: Discovery of new metabolic and epigenetic targets. Food Chem. Toxicol. 2019, 131, 110529. [Google Scholar] [CrossRef]
- Cuyàs, E.; Gumuzio, J.; Lozano-Sánchez, J.; Carreras, D.; Verdura, S.; Llorach-Parés, L.; Sanchez-Martinez, M.; Selga, E.; Pérez, G.J.; Scornik, F.S.; et al. Extra Virgin Olive Oil Contains a Phenolic Inhibitor of the Histone Demethylase LSD1/KDM1A. Nutrients 2019, 11, 1656. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.-C.; Shen, D.-D.; Ren, M.; Liu, X.-Q.; Wang, Z.-R.; Liu, Y.; Zhang, Q.-N.; Zhao, L.-J.; Zhao, L.-J.; Ma, J.-L.; et al. Baicalin, a natural LSD1 inhibitor. Bioorganic Chem. 2016, 69, 129–131. [Google Scholar] [CrossRef] [PubMed]
- Abdulla, A.; Zhao, X.; Yang, F. Natural Polyphenols Inhibit Lysine-Specific Demethylase-1 in vitro. J. Biochem. Pharmacol. Res. 2013, 1, 56–63. [Google Scholar] [PubMed]
- Wang, J.; Zhang, X.; Yan, J.; Li, W.; Jiang, Q.; Wang, X.; Zhao, D.; Cheng, M. Design, synthesis and biological evaluation of curcumin analogues as novel LSD1 inhibitors. Bioorganic Med. Chem. Lett. 2019, 29, 126683. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.-C.; Guan, Y.-Y.; Zhai, X.-Y.; Ding, L.-N.; Qin, W.-P.; Shen, D.-D.; Liu, X.-Q.; Sun, X.-D.; Zheng, Y.-C.; Liu, H.-M. Discovery of resveratrol derivatives as novel LSD1 inhibitors: Design, synthesis and their biological evaluation. Eur. J. Med. Chem. 2017, 126, 246–258. [Google Scholar] [CrossRef]
- Jia, G.; Cang, S.; Ma, P.; Song, Z. Capsaicin: A “hot” KDM1A/LSD1 inhibitor from peppers. Bioorganic Chem. 2020, 103, 104161. [Google Scholar] [CrossRef]
- Melfi, F.; Carradori, S.; Angeli, A.; D’Agostino, I. Nature as a source and inspiration for human monoamine oxidase B (hMAO-B) inhibition: A review of the recent advances in chemical modification of natural compounds. Expert Opin. Drug Discov. 2023, 18, 851–879. [Google Scholar] [CrossRef]
- Bester, E.; Petzer, A.; Petzer, J.P. Coumarin derivatives as inhibitors of d-amino acid oxidase and monoamine oxidase. Bioorganic Chem. 2022, 123, 105791. [Google Scholar] [CrossRef]
- Myburg, T.; Petzer, A.; Petzer, J.P. The inhibition of monoamine oxidase by harmine derivatives. Results Chem. 2022, 4, 100607. [Google Scholar] [CrossRef]
- Zhang, Z.; Hamada, H.; Gerk, P.M. Selectivity of Dietary Phenolics for Inhibition of Human Monoamine Oxidases A and B. Biomed. Res. Int. 2019, 2019, 8361858. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System; Version 1.8; Schrödinger LLC: New York, NY, USA, 2015.
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminformatics. 2011, 3, 33. [Google Scholar] [CrossRef]
- Jendele, L.; Krivak, R.; Skoda, P.; Novotny, M.; Hoksza, D. PrankWeb: A web server for ligand binding site prediction and visualization. Nucleic Acids Res. 2019, 47, W345–W349. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Schrödinger Release 2022-2: Maestro; Schrödinger LLC: New York, NY, USA, 2022.
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Zhou, P.; Jin, B.; Li, H.; Huang, S.Y. HPEPDOCK: A web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018, 46, W443–W450. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karagiannis, T.C.; Ververis, K.; Liang, J.J.; Pitsillou, E.; Liu, S.; Bresnehan, S.M.; Xu, V.; Wijoyo, S.J.; Duan, X.; Ng, K.; et al. Identification and Evaluation of Olive Phenolics in the Context of Amine Oxidase Enzyme Inhibition and Depression: In Silico Modelling and In Vitro Validation. Molecules 2024, 29, 2446. https://doi.org/10.3390/molecules29112446
Karagiannis TC, Ververis K, Liang JJ, Pitsillou E, Liu S, Bresnehan SM, Xu V, Wijoyo SJ, Duan X, Ng K, et al. Identification and Evaluation of Olive Phenolics in the Context of Amine Oxidase Enzyme Inhibition and Depression: In Silico Modelling and In Vitro Validation. Molecules. 2024; 29(11):2446. https://doi.org/10.3390/molecules29112446
Chicago/Turabian StyleKaragiannis, Tom C., Katherine Ververis, Julia J. Liang, Eleni Pitsillou, Siyao Liu, Sarah M. Bresnehan, Vivian Xu, Stevano J. Wijoyo, Xiaofei Duan, Ken Ng, and et al. 2024. "Identification and Evaluation of Olive Phenolics in the Context of Amine Oxidase Enzyme Inhibition and Depression: In Silico Modelling and In Vitro Validation" Molecules 29, no. 11: 2446. https://doi.org/10.3390/molecules29112446
APA StyleKaragiannis, T. C., Ververis, K., Liang, J. J., Pitsillou, E., Liu, S., Bresnehan, S. M., Xu, V., Wijoyo, S. J., Duan, X., Ng, K., Hung, A., Goebel, E., & El-Osta, A. (2024). Identification and Evaluation of Olive Phenolics in the Context of Amine Oxidase Enzyme Inhibition and Depression: In Silico Modelling and In Vitro Validation. Molecules, 29(11), 2446. https://doi.org/10.3390/molecules29112446






