Metabolic Engineering of Corynebacterium glutamicum for the Production of Flavonoids and Stilbenoids
Abstract
1. Introduction
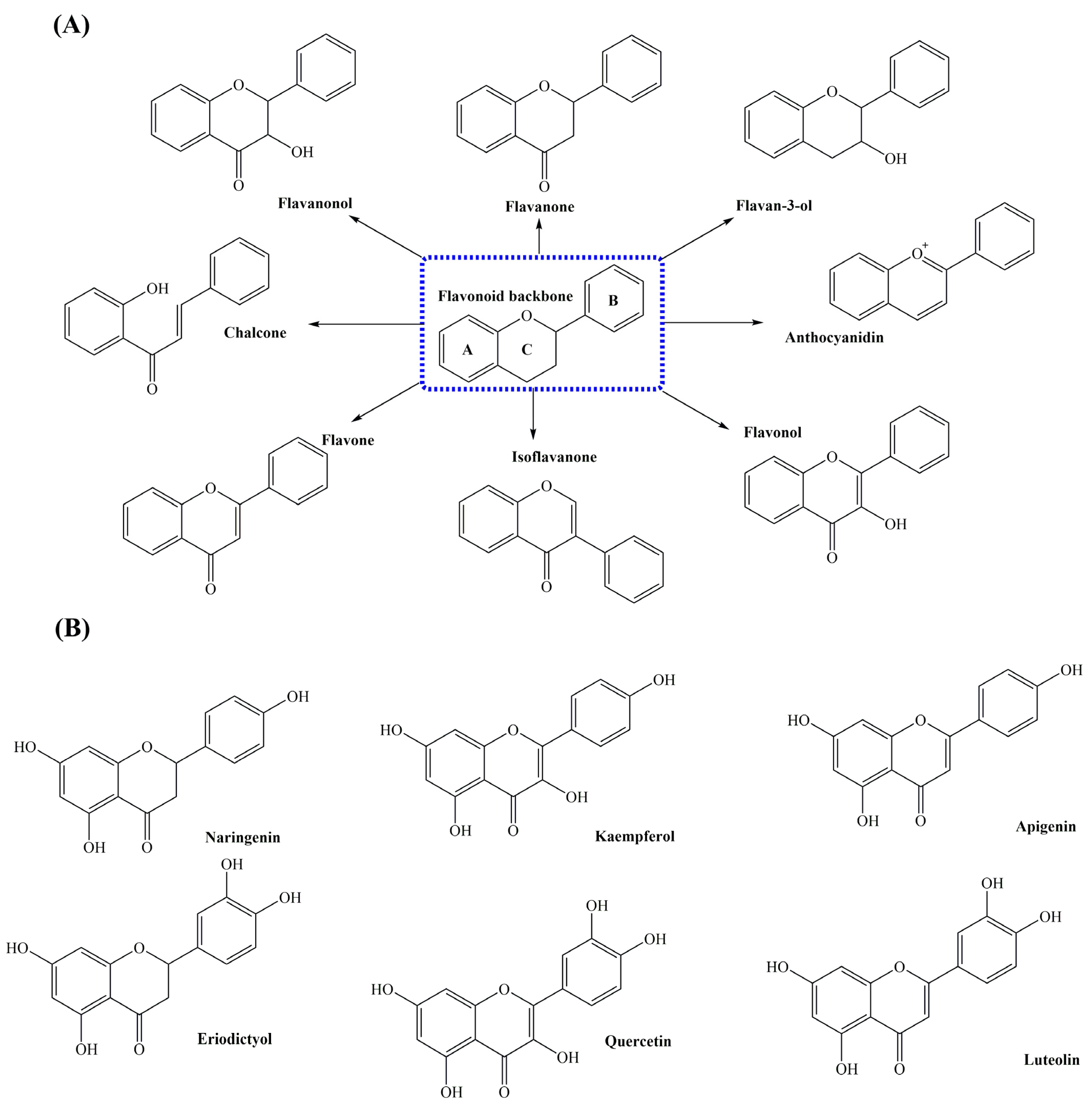
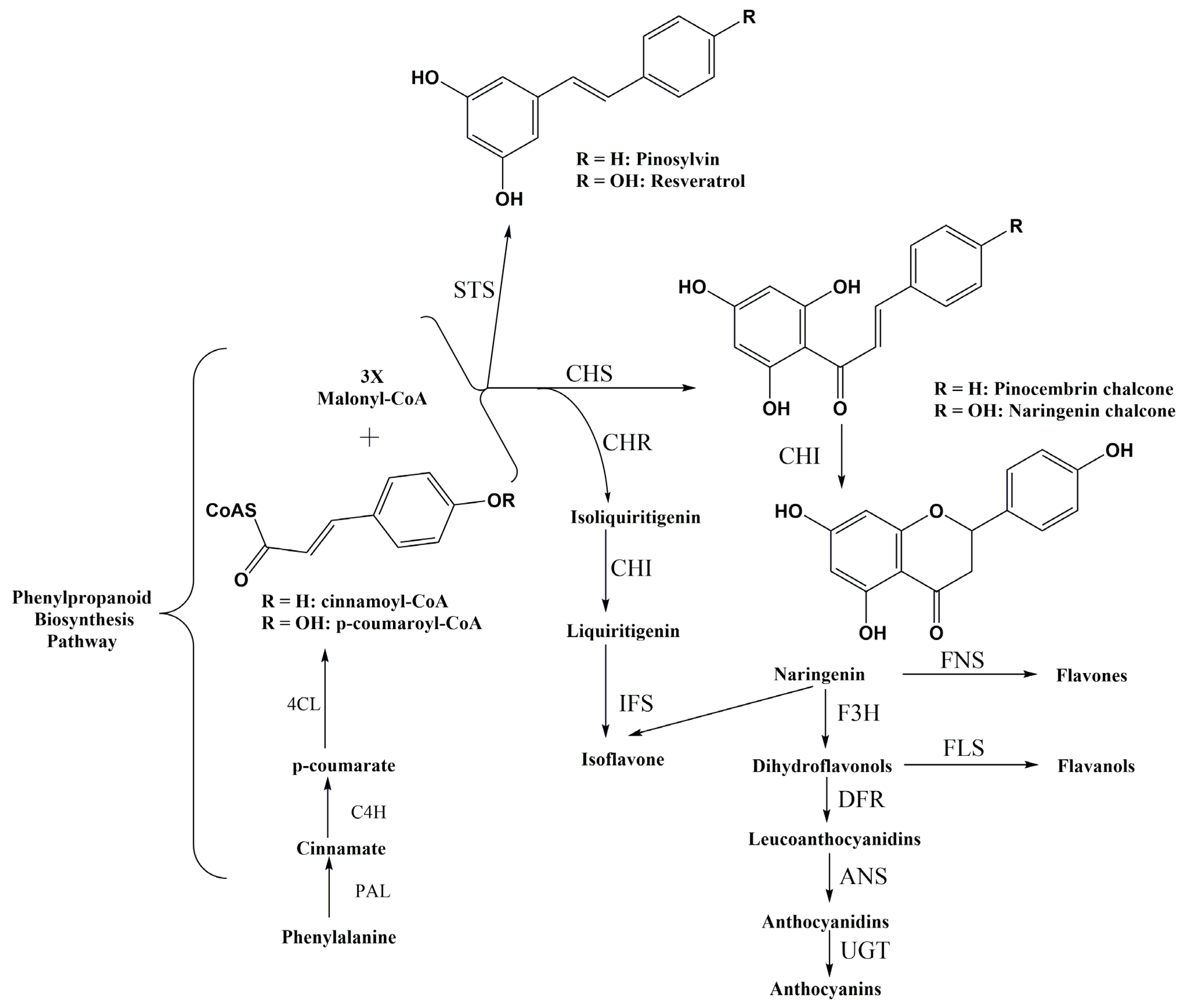
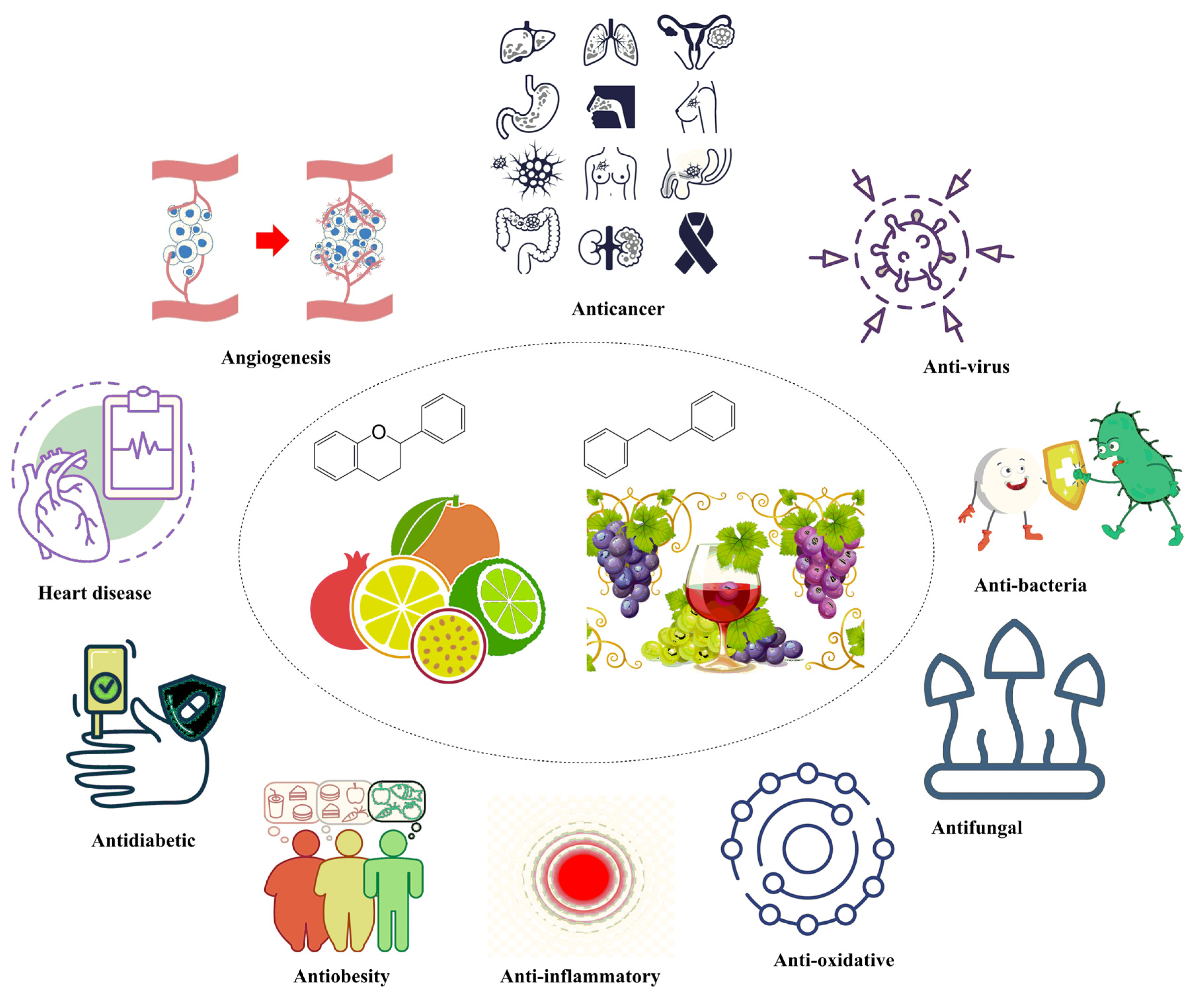
2. Flavonoids and Its Biological Activities
3. Stilbenoids and Their Biological Activities
4. Corynebacterium glutamicum as a Novel Platform to Produce Flavonoids and Stilbenoids
5. Metabolic Engineering of Corynebacterium glutamicum Cell Factories for Biosynthesis of Flavonoids and Stilbenoids
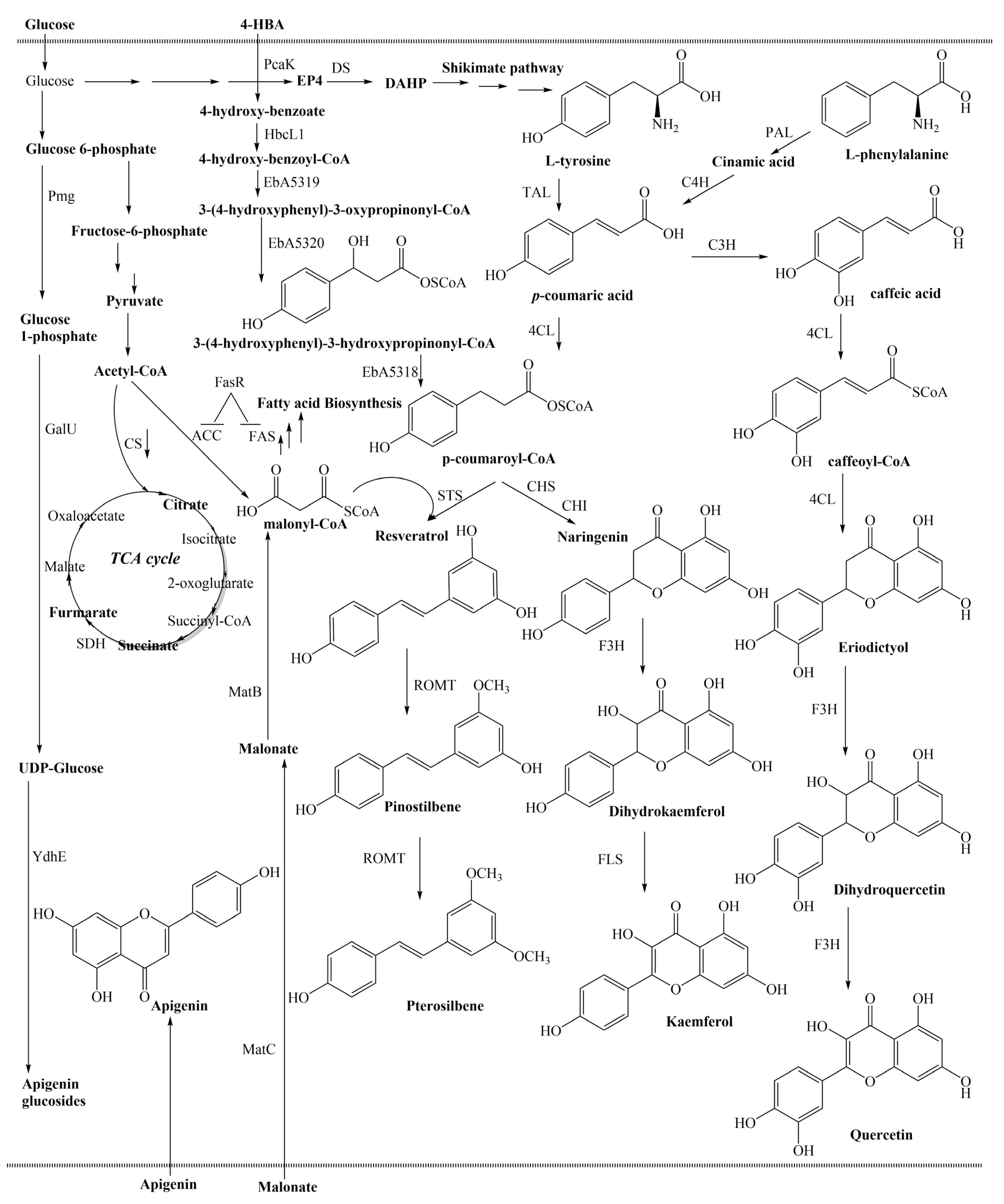
5.1. Establishing an Artificial Pathway
5.2. Enhancement of Intracellular Malonyl-CoA Level
5.3. Deletion and Downregulation of Competitive Pathway
5.4. Improving Intracellular UDP-Sugar Level for Biosynthesis of Diversified Flavonoids
6. Future Perspectives and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dias, M.C.; Pinto, D.C.G.A.; Silva, A.M.S. Plant flavonoids: Chemical characteristics and biological activity. Molecules 2021, 26, 5377. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef]
- Akinwumi, B.C.; Bordun, K.M.; Anderson, H.D. Biological activities of stilbenoids. Int. J. Mol. Sci. 2018, 19, 792. [Google Scholar] [CrossRef] [PubMed]
- Choy, K.W.; Murugan, D.; Leong, X.F.; Abas, R.; Alias, A.; Mustafa, M.R. Flavonoids as natural anti-inflammatory agents targeting nuclear factor-Kappa B (NFκB) signaling in cardiovascular diseases: A mini review. Front. Pharmacol. 2019, 10, 1295. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.; Lamb, A.J. Recent advances in understanding the antibacterial properties of flavonoids. Int. J. Antimicrob. Agents 2011, 38, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Tuli, H.S.; Garg, V.K.; Bhushan, S.; Uttam, V.; Sharma, U.; Jain, A.; Sak, K.; Yadav, V.; Lorenzo, J.M.; Dhama, K.; et al. Natural flavonoids exhibit potent anticancer activity by targeting microRNAs in cancer: A signature step hinting towards clinical perfection. Transl. Oncol. 2023, 27, 101596. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, N.; Singh, M.; Singh, S.R. Flavonoids: Food associations, therapeutic mechanisms, metabolism and nanoformulations. Food Res. Int. 2022, 157, 111442. [Google Scholar] [CrossRef] [PubMed]
- Gouveia, H.J.C.B.; Urquiza-Martínez, M.V.; Manhães-de-Castro, R.; Costa-de-Santana, B.J.R.; Villarreal, J.P.; Mercado-Camargo, R.; Torner, L.; de Souza Aquino, J.; Toscano, A.E.; Guzmán-Quevedo, O. Effects of the treatment with flavonoids on metabolic syndrome components in humans: A systematic review focusing on mechanisms of action. Int. J. Mol. Sci. 2022, 23, 8344. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.M.; Cidade, H.; Tiritan, M.E. Stereoselective synthesis of flavonoids: A brief overview. Molecules 2023, 28, 426. [Google Scholar] [CrossRef]
- Chaves, J.O.; de Souza, M.C.; da Silva, L.C.; Lachos-Perez, D.; Torres-Mayanga, P.C.; Machado, A.P.D.F.; Forster-Carneiro, T.; Vázquez-Espinosa, M.; González-de-Peredo, A.V.; Barbero, G.F.; et al. Extraction of flavonoids from natural sources using modern techniques. Front. Chem. 2020, 8, 507887. [Google Scholar] [CrossRef]
- Sheng, H.; Sun, X.; Yan, Y.; Yuan, Q.; Wang, J.; Shen, X. Metabolic engineering of microorganisms for the production of flavonoids. Front. Bioeng. Biotechnol. 2020, 8, 589069. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Lyu, Y.; Zeng, W.; Du, G.; Zhou, J.; Chen, J. Efficient biosynthesis of (2S)-Naringenin from p-coumaric acid in Saccharomyces cerevisiae. J. Agric. Food Chem. 2020, 68, 1015–1021. [Google Scholar] [CrossRef]
- Pei, J.; Sun, Q.; Gu, N.; Zhao, L.; Fang, X.; Tang, F.; Cao, F. Production of isoorientin and isovitexin from luteolin and apigenin using coupled catalysis of glycosyltransferase and sucrose synthase. Appl. Biochem. Biotechnol. 2020, 190, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Liga, S.; Paul, C.; Peter, F. Flavonoids: Overview of biosynthesis, biological activity, and current extraction techniques. Plants 2023, 12, 2732. [Google Scholar] [CrossRef] [PubMed]
- Okoye, C.O.; Jiang, H.; Wu, Y.; Li, X.; Gao, L.; Wang, Y.; Jiang, J. Bacterial biosynthesis of flavonoids: Overview, current biotechnology applications, challenges, and prospects. J. Cell. Physiol. 2023. [Google Scholar] [CrossRef]
- Xu, P.; Marsafari, M.; Zha, J.; Koffas, M. Microbial coculture for flavonoid synthesis. Trends Biotechnol. 2020, 38, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Tariq, H.; Asif, S.; Andleeb, A.; Hano, C.; Abbasi, B.H. Flavonoid production: Current trends in plant metabolic engineering and de novo microbial production. Metabolites 2023, 13, 124. [Google Scholar] [CrossRef]
- Ghiffary, M.R.; Prabowo, C.P.S.; Adidjaja, J.J.; Lee, S.Y.; Kim, H.U. Systems metabolic engineering of Corynebacterium glutamicum for the efficient production of β-alanine. Metab. Eng. 2022, 74, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Tachikawa, Y.; Okuno, M.; Itoh, T.; Hirasawa, T. Metabolic engineering with adaptive laboratory evolution for phenylalanine production by Corynebacterium glutamicum. J. Biosci. Bioeng. 2024, 137, 344–353. [Google Scholar] [CrossRef]
- Choi, J.W.; Jeon, E.J.; Jeong, K.J. Recent advances in engineering Corynebacterium glutamicum for utilization of hemicellulosic biomass. Curr. Opin. Biotechnol. 2019, 57, 17–24. [Google Scholar] [CrossRef]
- Zha, J.; Zang, Y.; Mattozzi, M.; Plassmeier, J.; Gupta, M.; Wu, X.; Clarkson, S.; Koffas, M.A.G. Metabolic engineering of Corynebacterium glutamicum for anthocyanin production. Microb. Cell Fact. 2018, 17, 143. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Han, S.; Zheng, S. Application of Corynebacterium glutamicum engineering display system in three generations of biorefinery. Microb. Cell Fact. 2022, 21, 14. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.; Anand, U.; Jha, N.K.; Korzeniewska, E.; Bontempi, E.; Proćków, J.; Dey, A. The soil bacterium, Corynebacterium glutamicum, from biosynthesis of value-added products to bioremediation: A master of many trades. Environ. Res. 2022, 213, 113622. [Google Scholar] [CrossRef]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The flavonoid biosynthesis network in plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Ma, Y.; Ma, X.; Gao, X.; Wu, W.; Zhou, B. Light induced regulation pathway of anthocyanin biosynthesis in plants. Int. J. Mol. Sci. 2021, 22, 11116. [Google Scholar] [CrossRef] [PubMed]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, C.; Di Ferdinando, M.; Fini, A.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants and developmental regulators: Relative significance in plants and humans. Int. J. Mol. Sci. 2013, 14, 3540–3555. [Google Scholar] [CrossRef] [PubMed]
- Shamsudin, N.F.; Ahmed, Q.U.; Mahmood, S.; Ali Shah, S.A.; Khatib, A.; Mukhtar, S.; Alsharif, M.A.; Parveen, H.; Zakaria, Z.A. Antibacterial effects of flavonoids and their structure-activity relationship study: A comparative interpretation. Molecules 2022, 27, 1149. [Google Scholar] [CrossRef]
- Yang, J.Y.; Ma, Y.X.; Liu, Y.; Peng, X.J.; Chen, X.Z. A comprehensive review of natural flavonoids with anti-SARS-CoV-2 activity. Molecules 2023, 28, 2735. [Google Scholar] [CrossRef]
- Fusi, F.; Trezza, A.; Tramaglino, M.; Sgaragli, G.; Saponara, S.; Spiga, O. The beneficial health effects of flavonoids on the cardiovascular system: Focus on K+ channels. Pharmacol. Res. 2020, 152, 104625. [Google Scholar] [CrossRef] [PubMed]
- Duta-Bratu, C.G.; Nitulescu, G.M.; Mihai, D.P.; Olaru, O.T. Resveratrol and other natural oligomeric stilbenoid compounds and their therapeutic applications. Plants 2023, 12, 2935. [Google Scholar] [CrossRef] [PubMed]
- Esatbeyoglu, T.; Ewald, P.; Yasui, Y.; Yokokawa, H.; Wagner, A.E.; Matsugo, S.; Winterhalter, P.; Rimbach, G. Chemical characterization, free radical scavenging, and cellular antioxidant and anti-inflammatory properties of a stilbenoid-rich root extract of Vitis vinifera. Oxidative Med. Cell. Longev. 2016, 2016, 8591286. [Google Scholar] [CrossRef] [PubMed]
- Tisserant, L.P.; Aziz, A.; Jullian, N.; Jeandet, P.; Clement, C.; Courot, E.; Boitel-Conti, M. Enhanced stilbene production and excretion in Vitis vinifera cv Pinot Noir hairy root cultures. Molecules 2016, 21, 1703. [Google Scholar] [CrossRef] [PubMed]
- Mattio, L.M.; Catinella, G.; Dallavalle, S.; Pinto, A. Stilbenoids: A natural arsenal against bacterial pathogens. Antibiotics 2020, 9, 336. [Google Scholar] [CrossRef] [PubMed]
- Sen, K.; Khan, M.I.; Paul, R.; Ghoshal, U.; Asakawa, Y. Recent advances in the phytochemistry of Bryophytes: Distribution, structures and biological activity of Bibenzyl and Bisbibenzyl compounds. Plants 2023, 12, 4173. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.Y.; Chung, B.; Kwon, O.S.; Park, S.C.; Cho, E.; Oh, D.C.; Shin, J.; Oh, K.B. Inhibitory effects of Epipolythiodioxopiperazine fungal metabolites on isocitrate lyase in the glyoxylate cycle of Candida albicans. Mar. Drugs 2021, 19, 295. [Google Scholar] [CrossRef] [PubMed]
- Dubrovina, A.S.; Kiselev, K.V. Regulation of stilbene biosynthesis in plants. Planta 2017, 246, 597–623. [Google Scholar] [CrossRef] [PubMed]
- Valletta, A.; Iozia, L.M.; Leonelli, F. Impact of environmental factors on stilbene biosynthesis. Plants 2021, 10, 90. [Google Scholar] [CrossRef]
- Qayyum, Z.; Noureen, F.; Khan, M.; Khan, M.; Haider, G.; Munir, F.; Gul, A.; Amir, R. Identification and expression analysis of stilbene synthase genes in Arachis hypogaea in response to methyl jasmonate and salicylic acid induction. Plants 2022, 11, 1776. [Google Scholar] [CrossRef]
- Koo, H.B.; Hwang, H.S.; Han, J.Y.; Cheong, E.J.; Kwon, Y.S.; Choi, Y.E. Enhanced production of pinosylvin stilbene with aging of Pinus strobus callus and nematicidal activity of callus extracts against pinewood nematodes. Sci. Rep. 2022, 12, 770. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Becker, J.; Tsuge, Y.; Kawaguchi, H.; Kondo, A.; Marienhagen, J.; Bott, M.; Wendisch, V.F.; Wittmann, C. Advances in metabolic engineering of Corynebacterium glutamicum to produce high-value active ingredients for food, feed, human health, and well-being. Essays Biochem. 2021, 65, 197–212. [Google Scholar]
- Lee, M.J.; Kim, P. Recombinant protein expression system in Corynebacterium glutamicum and its application. Front. Microbiol. 2018, 9, 2523. [Google Scholar] [CrossRef] [PubMed]
- Si, M.; Long, M.; Chaudhry, M.T.; Xu, Y.; Zhang, P.; Zhang, L.; Shen, X. Functional characterization of Corynebacterium glutamicum mycothiol S-conjugate amidase. PLoS ONE 2014, 9, e115075. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, J.; Bathe, B.; Bartels, D.; Bischoff, N.; Bott, M.; Burkovski, A.; Dusch, N.; Eggeling, L.; Eikmanns, B.J.; Gaigalat, L.; et al. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J. Biotechnol. 2003, 104, 5–25. [Google Scholar] [CrossRef]
- Kim, G.Y.; Kim, J.; Park, G.; Kim, H.J.; Yang, J.; Seo, S.W. Synthetic biology tools for engineering Corynebacterium glutamicum. Comput. Struct. Biotechnol. J. 2023, 21, 1955–1965. [Google Scholar] [CrossRef] [PubMed]
- Mhatre, A.; Shinde, S.; Jha, A.K.; Rodriguez, A.; Wardak, Z.; Jansen, A.; Gladden, J.M.; George, A.; Davis, R.W.; Varman, A.M. Corynebacterium glutamicum as an efficient omnivorous microbial host for the bioconversion of lignocellulosic biomass. Front. Bioeng. Biotechnol. 2022, 10, 827386. [Google Scholar]
- Kallscheuer, N.; Vogt, M.; Kappelmann, J.; Krumbach, K.; Noack, S.; Bott, M.; Marienhagen, J. Identification of the phd gene cluster responsible for phenylpropanoid utilization in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2016, 100, 1871–1881. [Google Scholar] [CrossRef] [PubMed]
- Kallscheuer, N.; Vogt, M.; Stenzel, A.; Gätgens, J.; Bott, M.; Marienhagen, J. Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones. Metab. Eng. 2016, 38, 47–55. [Google Scholar] [CrossRef]
- Kallscheuer, N.; Polen, T.; Bott, M.; Marienhagen, J. Reversal of β-oxidative pathways for the microbial production of chemicals and polymer building blocks. Metab. Eng. 2017, 42, 33–42. [Google Scholar] [CrossRef]
- Kallscheuer, N.; Vogt, M.; Bott, M.; Marienhagen, J. Functional expression of plant-derived O-methyltransferase, flavanone 3-hydroxylase, and flavonol synthase in Corynebacterium glutamicum for production of pterostilbene, kaempferol, and quercetin. J. Biotechnol. 2017, 258, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Kallscheuer, N.; Vogt, M.; Marienhagen, J. A novel synthetic pathway enables microbial production of polyphenols independent from the endogenous aromatic amino acid metabolism. ACS Synth. Biol. 2017, 6, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Braga, A.; Oliveira, J.; Silva, R.; Ferreira, P.; Rocha, I.; Kallscheuer, N.; Marienhagen, J.; Faria, N. Impact of the cultivation strategy on resveratrol production from glucose in engineered Corynebacterium glutamicum. J. Biotechnol. 2018, 265, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Milke, L.; Ferreira, P.; Kallscheuer, N.; Braga, A.; Vogt, M.; Kappelmann, J.; Oliveira, J.; Silva, A.R.; Rocha, I.; Bott, M.; et al. Modulation of the central carbon metabolism of Corynebacterium glutamicum improves malonyl-CoA availability and increases plant polyphenol synthesis. Biotechnol. Bioeng. 2019, 116, 1380–1391. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liu, J.; Liu, D.; Yuwen, M.; Koffas, M.A.G.; Zha, J. Biosynthesis of eriodictyol from tyrosine by Corynebacterium glutamicum. Microb. Cell Fact. 2022, 21, 86. [Google Scholar] [CrossRef] [PubMed]
- Milke, L.; Marienhagen, J. Engineering intracellular malonyl-CoA availability in microbial hosts and its impact on polyketide and fatty acid synthesis. Appl. Microbiol. Biotechnol. 2020, 104, 6057–6065. [Google Scholar] [CrossRef] [PubMed]
- Amoah, O.J.; Ma, S.Y.; Thapa, S.B.; Nguyen, H.T.; Zaka, M.M.; Sohng, J.K. Biosynthesis of apigenin glucosides in engineered Corynebacterium glutamicum. J. Microbiol. Biotechnol. 2023, 34, 1–10. [Google Scholar] [CrossRef]
- Chin, Y.W.; Jang, S.W.; Shin, H.S.; Kim, T.W.; Kim, S.K.; Park, C.S.; Seo, D.H. Heterologous expression of Deinococcus geothermalis amylosucrase in Corynebacterium glutamicum for luteolin glucoside production. Enzyme Microb. Technol. 2020, 135, 109505. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, S.; Dietrich, D.; Becker, J.; Kohlstedt, M.; Wittmann, C. Microbial production of polyunsaturated fatty acids-high-value ingredients for aquafeed, superfoods, and pharmaceuticals. Curr. Opin. Biotechnol. 2021, 69, 199–211. [Google Scholar] [CrossRef]
- James, E.S.; Cronan, J.E. Expression of two Escherichia coli acetyl-CoA carboxylase subunits is autoregulated. J. Biol. Chem. 2004, 279, 2520–2527. [Google Scholar] [CrossRef]
- Brüsseler, C.; Radek, A.; Tenhaef, N.; Krumbach, K.; Noack, S.; Marienhagen, J. The myo-inositol/proton symporter IolT1 contributes to d-xylose uptake in Corynebacterium glutamicum. Bioresour. Technol. 2018, 249, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Ku, J.T.; Chen, A.Y.; Lan, E.I. Metabolic engineering design strategies for increasing acetyl-CoA flux. Metabolites 2020, 10, 166. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, D.; Qin, Z.; Zeng, W.; Zhou, J. Enhancing glycosylation of flavonoids by engineering the uridine diphosphate glucose supply in Escherichia coli. J. Agric. Food Chem. 2023, 71, 17842–17851. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhou, T.; Liu, H.; Wen, L.; Jiang, Y.; Yang, B. Heterologous biosynthesis of prenylated resveratrol through multiplex metabolic engineering in Escherichia coli. Green Chem. 2024, 26, 4792–4802. [Google Scholar] [CrossRef]
- Siedlecka-Kroplewska, K.; Wrońska, A.; Kmieć, Z. Piceatannol, a structural analog of resveratrol, is an apoptosis inducer and a nultidrug resistance modulator in HL-60 human acute myeloid leukemia cells. Int. J. Mol. Sci. 2021, 22, 10597. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Jiang, Y.; Zeng, B.; Yang, B. The cancer preventive activity and mechanisms of prenylated resveratrol and derivatives. Curr. Res. Toxicol. 2023, 5, 100113. [Google Scholar] [CrossRef] [PubMed]
- Sordon, S.; Popłoński, J.; Huszcza, E. Microbial glycosylation of flavonoids. Pol. J. Microbiol. 2016, 65, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Magar, R.T.; Sohng, J.K. A review on structure, modifications and structure-activity relation of quercetin and its derivatives. J. Microbiol. Biotechnol. 2020, 30, 11–20. [Google Scholar] [CrossRef]
- Tronina, T.; Łużny, M.; Dymarska, M.; Urbaniak, M.; Kozłowska, E.; Piegza, M.; Stępień, Ł.; Janeczko, T. Glycosylation of quercetin by selected entomopathogenic filamentous fungi and prediction of its products’ bioactivity. Int. J. Mol. Sci. 2023, 24, 11857. [Google Scholar] [CrossRef]
- Duan, H.Y.; Wang, J.; Zha, L.P.; Peng, H.S.; Zhao, Y.P.; Yuan, Y.; Huang, L.Q. Molecular cloning and functional characterization of an isoflavone glucosyltransferase from Pueraria thomsonii. Chin. J. Nat. Med. 2022, 20, 133–138. [Google Scholar] [CrossRef]
- Wang, Q.; Zhu, J.; Li, B.; Li, S.; Yang, Y.; Wang, Q.; Xu, W.; Wang, L. Functional identification of anthocyanin glucosyltransferase genes: A Ps3GT catalyzes pelargonidin to pelargonidin 3-O-glucoside painting the vivid red flower color of Paeonia. Planta 2023, 257, 65. [Google Scholar] [CrossRef] [PubMed]
- Amoah, O.J.; Nguyen, H.T.; Sohng, J.K. N-Glucosylation in Corynebacterium glutamicum with YdhE from Bacillus lichenformis. Molecules 2022, 27, 3405. [Google Scholar] [CrossRef] [PubMed]
- Wendisch, V.F.; Bott, M.; Eikmanns, B.J. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr. Opin. Microbiol. 2006, 9, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Tao, D.; Ren, M.; Cheng, L. A combinational optimization method for efficient production of indigo by the recombinant Escherichia coli with expression of monooxygenase and malate dehydrogenase. Foods 2023, 12, 502. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chu, R.; Wei, W.; Song, W.; Ye, C.; Chen, X.; Wu, J.; Liu, L.; Gao, C. Systems engineering of Escherichia coli for high-level glutarate production from glucose. Nat. Commun. 2024, 15, 1032. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.P.; Pan, Y.; Niu, F.X.; Liao, Y.L.; Huang, M.; Liu, J.Z. Biosensor-assisted evolution for high-level production of 4-hydroxyphenylacetic acid in Escherichia coli. Metab. Eng. 2022, 70, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Du, G.; Chen, J.; Zhou, J. Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Sci. Rep. 2015, 5, 13477. [Google Scholar] [CrossRef] [PubMed]
- Koju, N.; Qin, Z.H.; Sheng, R. Reduced nicotinamide adenine dinucleotide phosphate in redox balance and diseases: A friend or foe? Acta Pharmacol. Sin. 2022, 43, 1889–1904. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Mu, X.; Nie, Y.; Xu, Y. Improving the production of NAD+ via multi-strategy metabolic engineering in Escherichia coli. Metab. Eng. 2021, 64, 122–133. [Google Scholar] [CrossRef]
- Tong, Y.; Lyu, Y.; Xu, S.; Zhang, L.; Zhou, J. Optimum chalcone synthase for flavonoid biosynthesis in microorganisms. Crit. Rev. Biotechnol. 2021, 41, 1194–1208. [Google Scholar] [CrossRef]
- Noguchi, A.; Horikawa, M.; Fukui, Y.; Fukuchi-Mizutani, M.; Iuchi-Okada, A.; Ishiguro, M.; Kiso, Y.; Nakayama, T.; Ono, E. Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in Lamiales. Plant Cell 2009, 21, 1556–1572. [Google Scholar] [CrossRef]
- Wang, X.; Li, C.; Zhou, Z.; Zhang, Y. Identification of three (iso)flavonoid glucosyltransferases from Pueraria lobata. Front. Plant Sci. 2019, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Nabavi, S.M.; Samec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Chen, J.; Solem, C.; Jensen, P.R.; Liu, J.M. Disruption of the oxidative pentose phosphate pathway stimulates high-yield production using resting Corynebacterium glutamicum in the absence of external electron acceptors. App. Environ. Microbiol. 2020, 86, e02114-20. [Google Scholar] [CrossRef]
- Son, J.; Choi, I.H.; Lim, C.G.; Jang, J.H.; Bang, H.B.; Cha, J.W.; Jeon, E.J.; Sohn, M.G.; Yun, H.J.; Kim, S.C.; et al. Production of cinnamaldehyde through whole-cell bioconversion from trans-cinnamic acid using engineered Corynebacterium glutamicum. J. Agric. Food Chem. 2022, 70, 2656–2663. [Google Scholar] [CrossRef]
- Heinisch, J.J.; Murra, A.; Fernández Murillo, L.; Schmitz, H.P. The role of glucose-6-phosphate dehydrogenase in the wine yeast Hanseniaspora uvarum. Int. J. Mol. Sci. 2024, 25, 2395. [Google Scholar] [CrossRef]
- Snoek, T.; Chaberski, E.K.; Ambri, F.; Kol, S.; Bjørn, S.P.; Pang, B.; Barajas, J.F.; Welner, D.H.; Jensen, M.K.; Keasling, J.D. Evolution-guided engineering of small-molecule biosensors. Nucleic Acids Res. 2020, 48, e3. [Google Scholar] [CrossRef]
- Cazier, A.P.; Blazeck, J. Advances in promoter engineering: Novel applications and predefined transcriptional control. Biotechnol. J. 2021, 16, e2100239. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, M.; Nozaki, K.; Umeno, D.; Ishii, J.; Kondo, A. Robust and flexible platform for directed evolution of yeast genetic switches. Nat. Commun. 2021, 12, 1846. [Google Scholar] [CrossRef]
- Rao, X.; Li, D.; Su, Z.; Nomura, C.T.; Chen, S.; Wang, Q. A smart RBS library and its prediction model for robust and accurate fine-tuning of gene expression in Bacillus species. Metab. Eng. 2024, 81, 1–9. [Google Scholar] [CrossRef]
- Yang, D.; Kim, W.J.; Yoo, S.M.; Choi, J.H.; Ha, S.H.; Lee, M.H.; Lee, S.Y. Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, 9835–9844. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.N.; Shin, W.S.; Seo, S.Y.; Choi, S.S.; Song, J.S.; Kim, J.Y.; Park, J.H.; Lee, D.; Kim, S.Y.; Lee, S.J.; et al. Corynebacterium cell factory design and culture process optimization for muconic acid biosynthesis. Sci. Rep. 2018, 8, 18041. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, S.; Zhao, S.; Sun, P.; Zhang, Z.; Xu, Q. Productivity enhancement in L-lysine fermentation using oxygen-enhanced bioreactor and oxygen vector. Front. Bioeng. Biotechnol. 2023, 11, 1181963. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Chang, J.; Kim, M.; Lee, M.E.; Shin, H.Y.; Ok Han, S. Isopropanol production using engineered Corynebacterium glutamicum from waste rice straw biomass. Bioresour. Technol. 2024, 396, 130416. [Google Scholar] [CrossRef]
- Egbune, E.O.; Ezedom, T.; Odeghe, O.B.; Orororo, O.C.; Egbune, O.U.; Ehwarieme, A.D.; Aganbi, E.; Ebuloku, C.S.; Chukwuegbo, A.O.; Bogard, E.; et al. Solid-state fermentation production of L-lysine by Corynebacterium glutamicum (ATCC 13032) using agricultural by-products as substrate. World J. Microbiol. Biotechnol. 2023, 40, 20. [Google Scholar] [CrossRef]

| Strains | Genes or Related Gene Cassettes | Products | Substrate/Precusor | Titer (mg/L) | Major Media | Carbon Source | Ref. |
|---|---|---|---|---|---|---|---|
| C. glutamicum ATCC 13032 | Anthocyanidin synthase (ANS) from Petunia hybrida, and 3-O-glucosyltransferase (3GT) from Arabidopsis thaliana; glucokinase (GLK), phosphoglucomutase (PGM), UTP–glucose-1-phosphate uridylyltransferase (GalU1) | Cyanidin 3-O-glucoside (C3G) | catechin | 40 | AMM medium | glucose, fructose, or sucrose | [21] |
| MB001DelAro3 | STS from Arachis hypogaea, 4CL from Petroselinum crispum | Pinosylvin | cinnamic acid, | 121 | CGXII medium | glucose | [48] |
| Resveratrol | p-coumaric acid | 158 | |||||
| Piceatannol | caffeic acid | 56 | |||||
| CHS, CHI from Petunia hybrida | Naringenin | p-coumaric acid | 35 | ||||
| Eriodictyol | caffeic acid | 37 | |||||
| MB001DelAro4 | STS from Arachis hypogaea, 4CL from Petroselinum crispum, TAL from Flavobacterium johnsoniae (TALFj), AroH of E. coli | Resveratrol | 59 | CGXII medium | glucose | [49] | |
| Naringenin | 32 | ||||||
| MB001DelAro4 | 4CL from Petroselinum crispum, STS from Arachis hypogaea, resveratrol-di-Omethyltransferase (OMT) from Vitis vinifera, maltose-binding protein MalE of E. coli | di-O-Methylated pterostilbene | p-coumaric acid | 42 | CGXII medium | glucose | [51] |
| MB001DelAro4 | 4CL from Petroselinum crispum, CHS and CHI from P. hybrida, F3H from Petunia x hybrida, FLS from P. deltoides | Kaempferol | p-coumaric acid | 23 | |||
| Quercetin | caffeic acid | 10 | |||||
| MB001DelAro4 | 4CL from Petroselinum crispum, STS from Arachis hypogaea, 4-hydroxybenzoate: CoA ligase (HbcL1), enoyl-CoA hydratase (EbA5318), a 3-hydroxyacyl-CoA dehydrogenase (EbA5320) and a β-ketothiolase (EbA5319) from “Aromatoleum aromaticum” EbN1; | Resveratrol | 4-hydroxybenzoic acid | 5 | CGXII medium | glucose | [52] |
| MB001DelAro4 | STS Arachis hypogaea, 4CL from Petroselinum crispum, TAL from Flavobacterium johnsoniae (TALFj), AroH of E. coli | Resveratrol | p-coumaric acid | 12 | CGXII medium | glucose | [45] |
| C. glutamicum Nar1_C7 | 4CL from Petroselinum crispum, CHS and CHI from Petunia x hybrida, accBC (cg0802) and accD1 (dtsR1, cg0812) of C. glutamicum, ΔfasR, gltA can be downregulated by exchanging its native promoter to weaker promoter variants citrate synthase (CS) coding gltA (cg0949), ∆sdhCAB (cg0445-47) operon encoding the succinate dehydrogenase complex (SDH) | Naringenin | 24 | CGXII medium | glucose | [54] | |
| C. glutamicum Res1_C7 | STS from Arachis hypogaea, 4CL from Petroselinum crispum, TAL from Flavobacterium johnsoniae (TALFj), AroH of E. coli | Resveratrol | 112 | ||||
| C. glutamicum ATCC 13032 | TAL from Rhodotorula glutinis. 4CL from Arabidopsis thaliana, Petroselium crispum, and Vitis vinifera; CHS from Petunia hybrida, or Citrus maxima, CHI of Citrus maxima or Medicago sativa | Eriodictyol | Tyrosine | 14.10 | AMM medium | Glucose | [55] |
| C. glutamicum Nor2 C5 mufasOBCD1 PO6-iolT1 ∆pyc | ∆cg0344-47, cg0502, cg1226 and cg2625-40, 4CL from Petroselinum crispum and replacement of the native gltA promoter with the dapA promoter variant C7 (PgltA::PdapA-C7), pentaketide chromone synthase from Aloe arborescens | Noreugenin | casamino acids | 53.32 | CGXII medium | Glucose | [56] |
| C. glutamicum ATCC 13032 | YdhE from Bacillus licheniformis, galU1 (UDP-glucose pyrophosphorylase), and pgm (phosphoglucomutase) | apigenin glucosides | Sorbito | BHI medium | Glucose | [57] | |
| C. glutamicum ATCC 13032 | amylosucrase from Deinococcus geothermalis | Luteolin glucoside | BHI medium | Sucrose | [58] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, L.L.; Tran, C.T.B.; Pham, D.T.K.; Nguyen, H.T.A.; Nguyen, M.H.; Pham, N.M.; Nguyen, A.T.V.; Phan, D.T.; Do, H.M.; Nguyen, Q.H. Metabolic Engineering of Corynebacterium glutamicum for the Production of Flavonoids and Stilbenoids. Molecules 2024, 29, 2252. https://doi.org/10.3390/molecules29102252
Chu LL, Tran CTB, Pham DTK, Nguyen HTA, Nguyen MH, Pham NM, Nguyen ATV, Phan DT, Do HM, Nguyen QH. Metabolic Engineering of Corynebacterium glutamicum for the Production of Flavonoids and Stilbenoids. Molecules. 2024; 29(10):2252. https://doi.org/10.3390/molecules29102252
Chicago/Turabian StyleChu, Luan Luong, Chau T. Bang Tran, Duyen T. Kieu Pham, Hoa T. An Nguyen, Mi Ha Nguyen, Nhung Mai Pham, Anh T. Van Nguyen, Dung T. Phan, Ha Minh Do, and Quang Huy Nguyen. 2024. "Metabolic Engineering of Corynebacterium glutamicum for the Production of Flavonoids and Stilbenoids" Molecules 29, no. 10: 2252. https://doi.org/10.3390/molecules29102252
APA StyleChu, L. L., Tran, C. T. B., Pham, D. T. K., Nguyen, H. T. A., Nguyen, M. H., Pham, N. M., Nguyen, A. T. V., Phan, D. T., Do, H. M., & Nguyen, Q. H. (2024). Metabolic Engineering of Corynebacterium glutamicum for the Production of Flavonoids and Stilbenoids. Molecules, 29(10), 2252. https://doi.org/10.3390/molecules29102252







