Assessment of Novel Proteins Triggering Celiac Disease via Docking-Based Approach
Abstract
:1. Introduction
2. Results
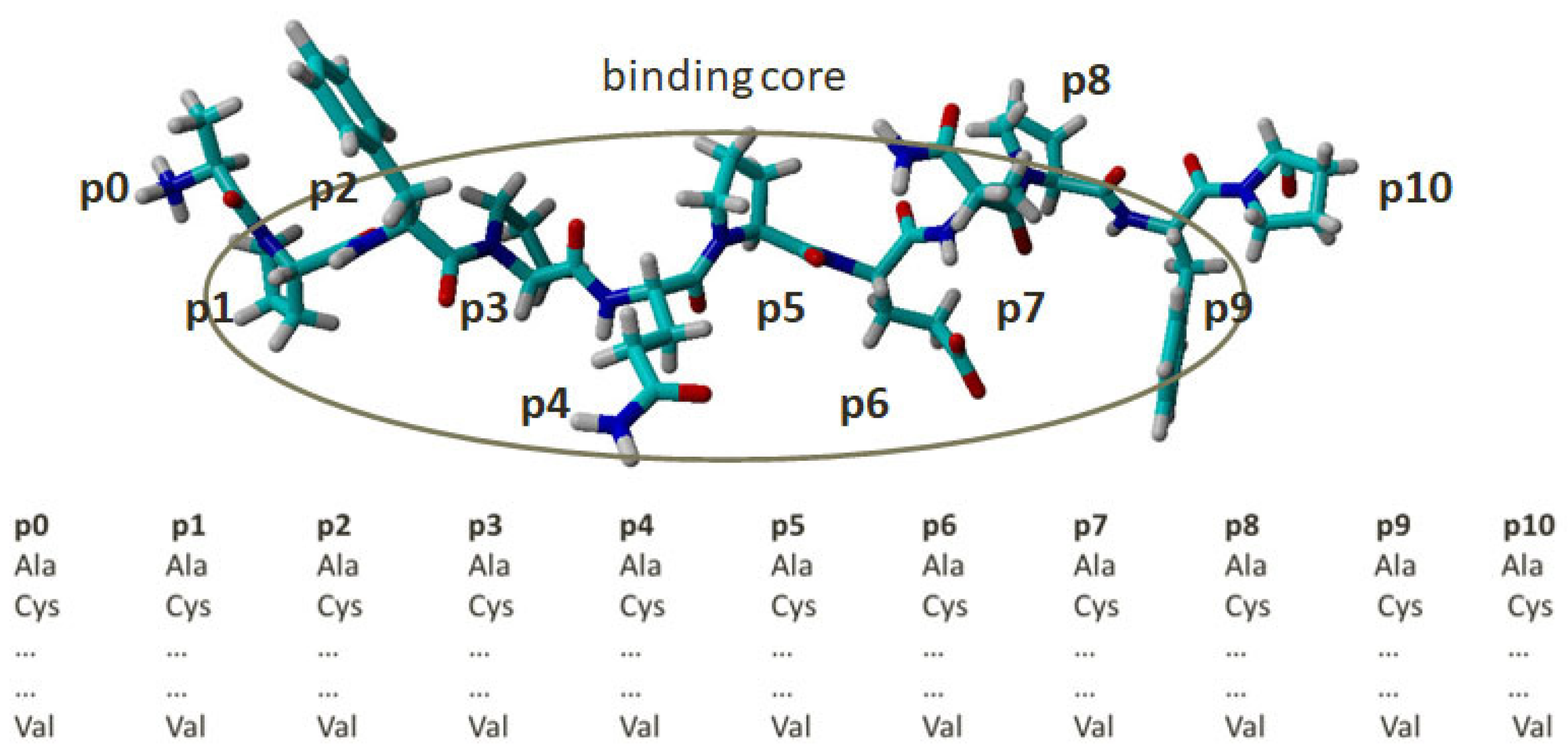
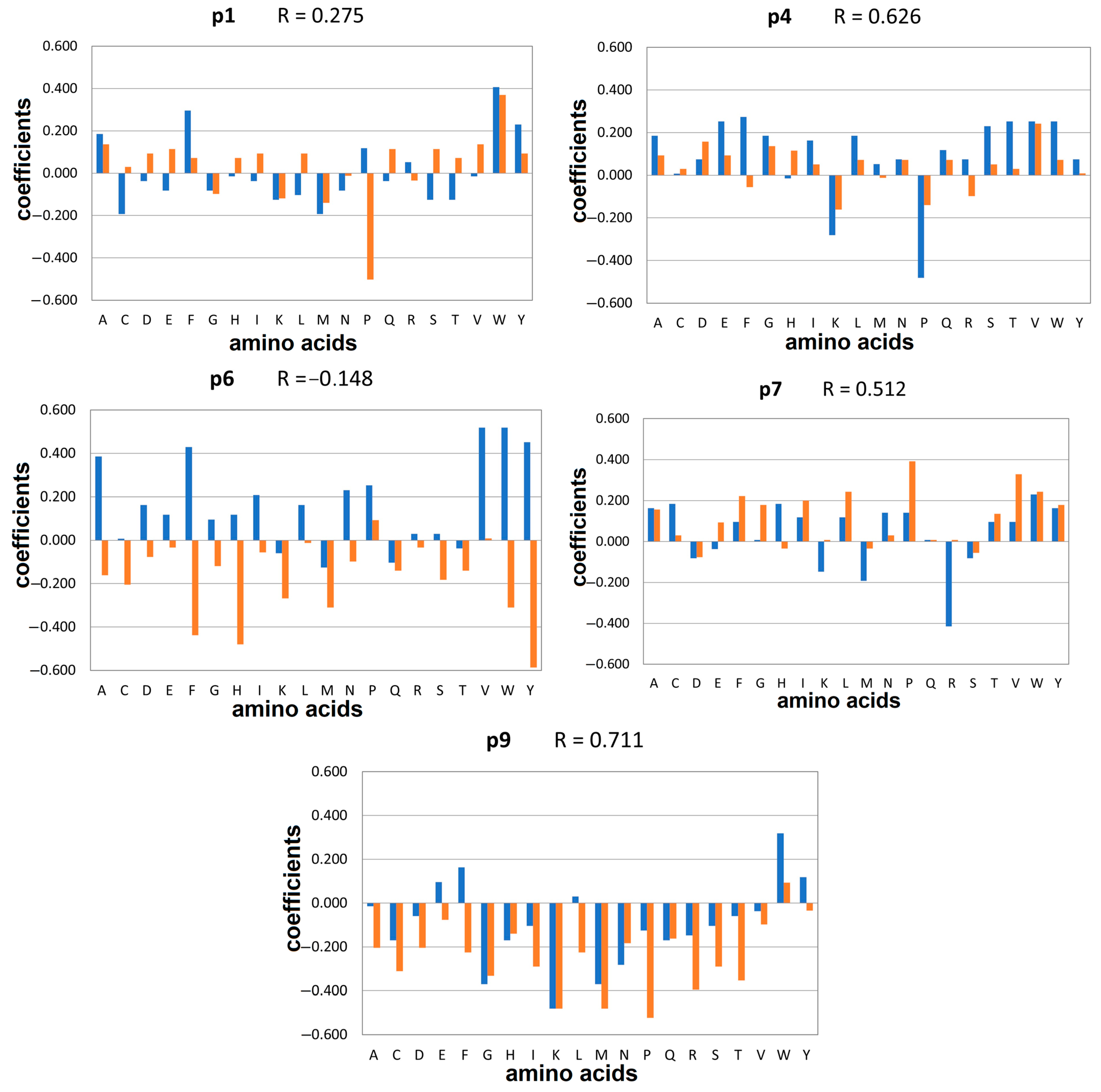
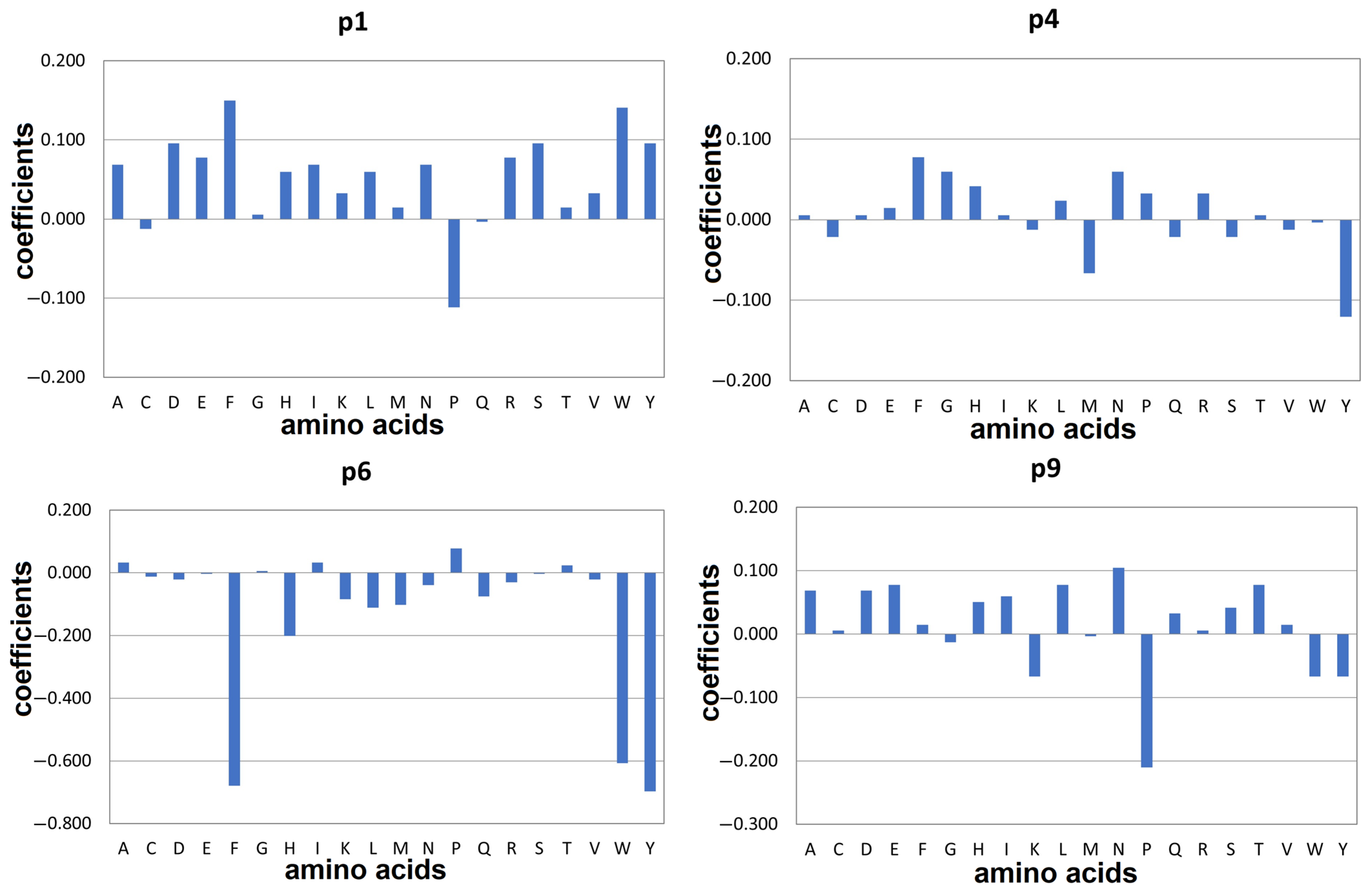
2.1. Docking-Based Quantitative Matrices (QMs) for Prediction of Peptide Binding to HLA-DQ2.5
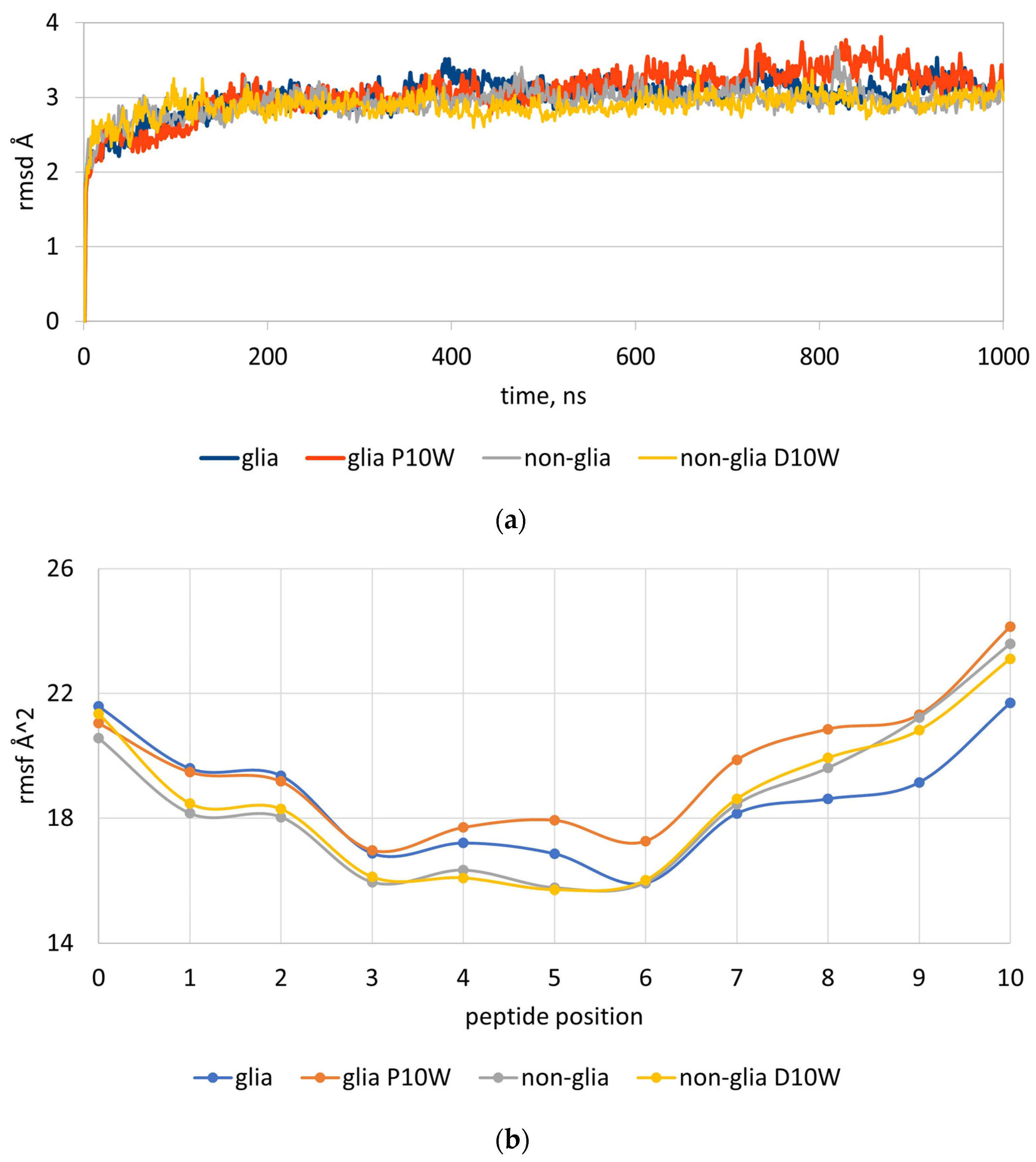
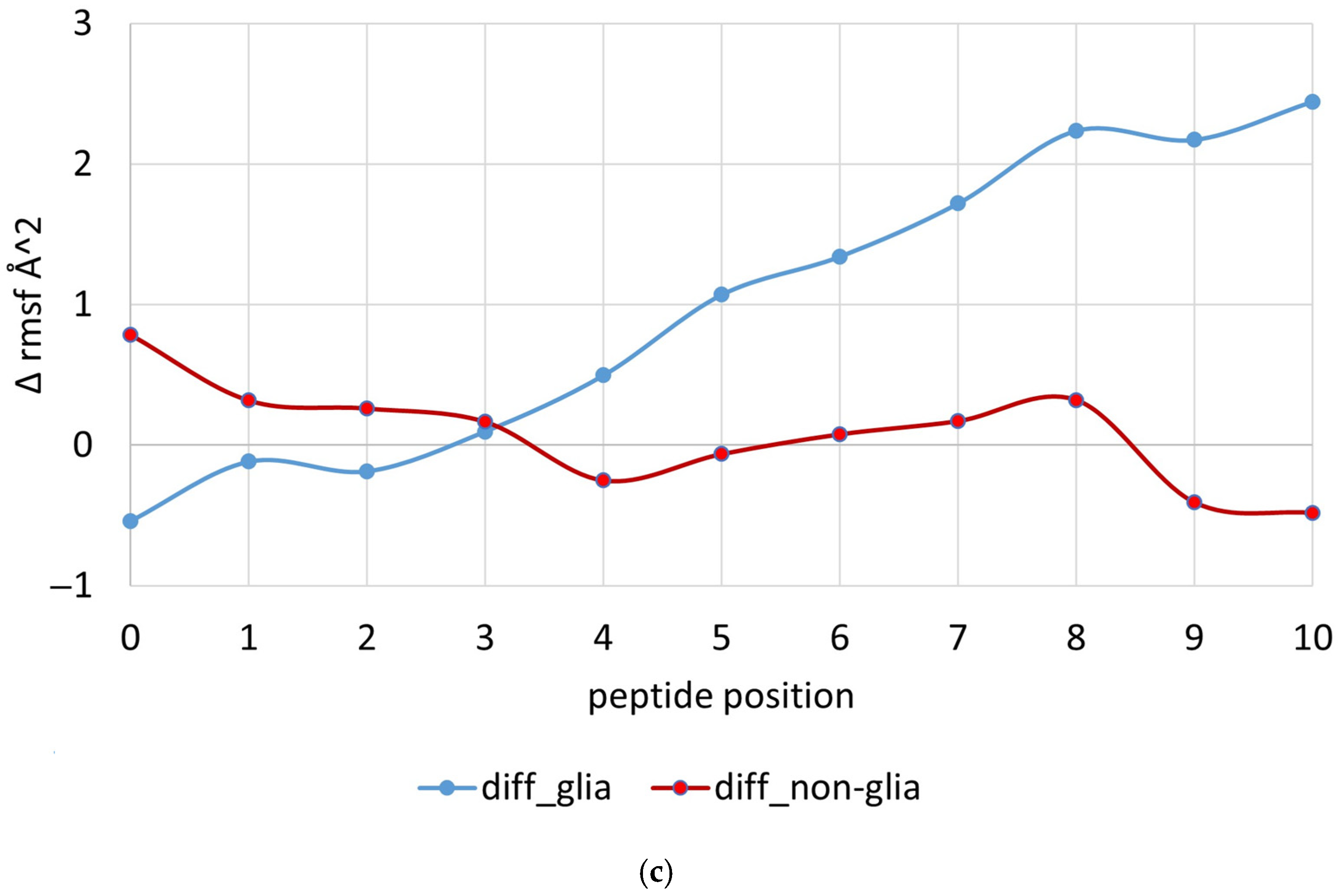
2.2. MD Simulations of Peptides Carrying Trp at p10
2.3. Docking-Based Quantitative Matrices (QMs) for Prediction of Peptide Binding to HLA-DQ8.1
3. Discussion
4. Materials and Methods
4.1. Structures and Combinatorial Libraries
4.2. Molecular Docking Protocol
4.3. MD Simulations Protocol
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Patronov, A.; Doytchinova, I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013, 3, 120139. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Barker, D.J.; Georgiou, X.; Cooper, M.A.; Flicek, P.; Marsh, S.G.E. IPD-IMGT/HLA database. Nucleic Acids Res. 2020, 48, D948–D955. [Google Scholar] [PubMed]
- Raychaudhuri, S.; Sandor, C.; Stahl, E.A.; Freudenberg, J.; Lee, H.S.; Jia, X.; Alfredsson, L.; Padyukov, L.; Klareskog, L.; Worthington, J.; et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 2012, 44, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Larid, G.; Pancarte, M.; Offer, G.; Clavel, C.; Martin, M.; Pradel, V.; Auger, I.; Lafforgue, P.; Roudier, J.; Serre, G.; et al. In rheumatoid arthritis patients, HLA-DRB1*04:01 and rheumatoid nodules are associated with ACPA to a particular fibrin epitope. Front. Immunol. 2021, 12, 692041. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.F.; Wu, C.J.; Chen, H.H.; Chang, F.M.; Liu, H.F.; Chu, C.C.; Lin, M.; Lee, Y.J. Molecular analysis of HLA-DRB1 allelic associations with systemic lupus erythematous and lupus nephritis in Taiwan. Lupus 2009, 18, 698–704. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.L.; Taylor, K.E.; Fernando, M.M.A.; Nititham, J.; Alarcón-Riquelme, M.E.; Barcellos, L.F.; Behrens, T.W.; Cotsapas, C.; Gaffney, P.M.; Graham, R.R.; et al. Unraveling multiple MHC gene associations with systemic lupus erythematosus: Model choice indicates a role for HLA alleles and non-HLA genes in Europeans. Am. J. Hum. Genet. 2012, 91, 778–793. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.T.; Faridi, P.; Lim, J.J.; Ting, Y.T.; Onwukwe, G.; Bhattacharjee, P.; Jones, C.M.; Tresoldi, E.; Cameron, F.J.; La Gruta, N.L.; et al. T cell receptor recognition of hybrid insulin peptides bound to HLA-DQ8. Nat. Commun. 2021, 12, 5110. [Google Scholar] [CrossRef]
- Zhao, L.P.; Papadopoulos, G.K.; Moustakas, A.K.; Bondinas, G.P.; Carlsson, A.; Larsson, H.E.; Ludvigsson, J.; Marcus, C.; Persson, M.; Samuelsson, U.; et al. Nine residues in HLA-DQ molecules determine with susceptibility and resistance to type 1 diabetes among young children in Sweden. Sci. Rep. 2021, 11, 8821. [Google Scholar] [CrossRef]
- Fontenot, A.P.; Kotzin, B.L. Chronic beryllium disease: Immune-mediated destruction with implications for organ-specific autoimmunity. Tissue Antigens 2003, 62, 449–458. [Google Scholar] [CrossRef]
- Henderson, K.N.; Tye-Din, J.A.; Reid, H.H.; Chen, Z.; Borg, N.A.; Beissbarth, T.; Tatham, A.; Mannering, S.I.; Purcell, A.W.; Dudek, N.L.; et al. A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity 2007, 27, 23–34. [Google Scholar] [CrossRef]
- Cecilio, L.A.; Bonatto, M.W. The prevalence of HLA DQ2 and DQ8 in patients with celiac disease, in family and in general population. Arq. Bras. De Cir. Dig. 2015, 28, 183–185. [Google Scholar] [CrossRef] [PubMed]
- Ludvigsson, J.F.; Leffler, D.A.; Bai, J.C.; Biagi, F.; Fasano, A.; Green, P.H.; Hadjivassiliou, M.; Kaukinen, K.; Kelly, C.P.; Leonard, J.N.; et al. The Oslo definitions for coeliac disease and related terms. Gut 2013, 62, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Kagnoff, M.F. Celiac Disease: Pathogenesis of a Model Immunogenetic Disease. J. Clin. Investig. 2007, 117, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Elli, L.; Branchi, F.; Tomba, C.; Villalta, D.; Norsa, L.; Ferretti, F.; Roncoroni, L.; Bardella, M.T. Diagnosis of gluten related disorders: Celiac disease, wheat allergy and non-celiac gluten sensitivity. World J. Gastroenterol. 2015, 21, 10331–10341. [Google Scholar] [CrossRef] [PubMed]
- Lebwohl, B.; Ludvigsson, J.F.; Green, P.H. Celiac disease and non-celiac gluten sensitivity. BMJ 2015, 351, h4347. [Google Scholar] [CrossRef] [PubMed]
- Ghazanfar, H.; Javed, N.; Lee, S.; Shaban, M.; Cordero, D.; Acherjee, T.; Hasan, K.Z.; Jyala, A.; Kandhi, S.; Hussain, A.N.; et al. Novel Therapies for Celiac Disease: A Clinical Review Article. Cureus 2023, 15, e39004. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Galarza, F.F.; McCabe, A.; Santos, E.J.; Jones, J.; Takeshita, L.Y.; Ortega-Rivera, N.D.; Del Cid-Pavon, G.M.; Ramsbottom, K.; Ghattaoraya, G.S.; Alfirevic, A.; et al. Allele frequency net database (AFND) 2020 update: Gold-standard data classification, open access genotype data and new query tools. Nucleic Acid Res. 2020, 48, D783–D788. [Google Scholar] [CrossRef]
- Liu, E.; Rewers, M.; Eisenbarth, G.S. Genetic Testing: Who Should Do the Testing and What Is the Role of Genetic Testing in the Setting of Celiac Disease? Gastroenterology 2005, 128, S33–S37. [Google Scholar] [CrossRef]
- Moustakas, A.K.; van de Wal, Y.; Routsias, J.; Kooy, Y.M.C.; van Veelen, P.; Drijfhout, J.W.; Koning, F.; Papadopoulos, G.K. Structure of Celiac Disease-Associated HLA-DQ8 and Non-Associated HLA-DQ9 Alleles in Complex with Two Disease-Specific Epitopes. Int. Immunol. 2000, 12, 1157–1166. [Google Scholar] [CrossRef]
- van de Wal, Y.; Kooy, Y.M.C.; Drijfhout, J.W.; Amons, R.; Papadopoulos, G.K.; Koning, F. Unique Peptide Binding Characteristics of the Disease-Associated DQ(α1*0501, β1*0201) vs the Non-Disease-Associated DQ(α1*0201, β1*0202) Molecule. Immunogenetics 1997, 46, 484–492. [Google Scholar] [CrossRef]
- Kumar, R.; Eastwood, A.L.; Brown, M.L.; Laurie, G.W. Human Genome Search in Celiac Disease: Mutated Gliadin T-Cell-like Epitope in Two Human Proteins Promotes T-Cell Activation. J. Mol. Biol. 2002, 319, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Costantini, S.; Rossi, M.; Colonna, G.; Facchiano, A.M. Modelling of HLA-DQ2 and Its Interaction with Gluten Peptides to Explain Molecular Recognition in Celiac Disease. J. Mol. Graph. Model. 2005, 23, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Gianfrani, C.; Siciliano, R.A.; Facchiano, A.M.; Camarca, A.; Mazzeo, M.F.; Costantini, S.; Salvati, V.M.; Maurano, F.; Mazzarella, G.; Iaquinto, G.; et al. Transamidation of Wheat Flour Inhibits the Response to Gliadin of Intestinal T Cells in Celiac Disease. Gastroenterology 2007, 133, 780–789. [Google Scholar] [CrossRef] [PubMed]
- Vartdal, F.; Johansen, B.H.; Friede, T.; Thorpe, C.J.; Stevanovic, S.; Eriksen, J.E.; Sletten, K.; Thorsby, E.; Rammensee, H.G.; Sollid, L.M. The peptide binding motif of the disease associated HLA-DQ (alpha 1* 0501, beta 1* 0201) molecule. Eur. J. Immunol. 1996, 26, 2764–2772. [Google Scholar] [CrossRef] [PubMed]
- Barre, A.; Simplicien, M.; Cassan, G.; Benoist, H.; Rougé, P. Docking of Peptide Candidates to HLA-DQ2 and HLA-DQ8 Basket as a Tool for Predicting Potential Immunotoxic Peptides toward Celiac Diseased People. Rev. Fr. Allergol. 2018, 58, 482–488. [Google Scholar] [CrossRef]
- Ting, Y.T.; Dahal-Koirala, S.; Kim, H.S.K.; Qiao, S.-W.; Neumann, R.S.; Lundin, K.E.A.; Petersen, J.; Reid, H.H.; Sollid, L.M.; Rossjohn, J. A Molecular Basis for the T Cell Response in HLA-DQ2.2 Mediated Celiac Disease. Proc. Natl. Acad. Sci. USA 2020, 117, 3063–3073. [Google Scholar] [CrossRef] [PubMed]
- Atanasova, M.; Patronov, A.; Dimitrov, I.; Flower, D.R.; Doytchinova, I. EpiDOCK: A Molecular Docking-Based Tool for MHC Class II Binding Prediction. Protein Eng. Des. Sel. 2013, 26, 631–634. [Google Scholar] [CrossRef]
- Dahal-Koirala, S.; Ciacchi, L.; Petersen, J.; Risnes, L.F.; Neumann, R.S.; Christophersen, A.; Lundin, K.; Reid, H.H.; Qiao, S.W.; Rossjohn, J.; et al. Discriminative T-cell receptor recognition of highly homologous HLA-DQ2-bound gluten epitopes. J. Biol. Chem. 2019, 294, 941–952. [Google Scholar] [CrossRef]
- van de Wal, Y.; Kooy, Y.M.; Drijfhout, J.W.; Amons, R.; Koning, F. Peptide binding characteristics of the coeliac disease-associated DQ (α1*0501, β1*0201) molecule. Immunogenetics 1996, 44, 246–253. [Google Scholar] [CrossRef]
- Koşaloğlu-Yalçın, Z.; Sidney, J.; Chronister, W.; Peters, B.; Sette, A. Comparison of HLA ligand elution data and binding predictions reveals varying prediction performance for the multiple motifs recognized by HLA-DQ2.5. Immunology 2021, 162, 235–247. [Google Scholar] [CrossRef]
- Commission Implementing Regulation (EU) No 503/2013 of 3 April 2013 on applications for authorisation of genetically modified food and feed in accordance with Regulation (EC) No 1829/2003 of the European Parliament and of the Council and amending Commission Regulations (EC) No 641/2004 and (EC) No 1981/2006. OJ L157, 8.6.2013. pp. 1–48. Available online: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2013:157:0001:0048:EN:PDF (accessed on 28 November 2023).
- EFSA Panel on Genetically Modified Organisms (GMO); Naegeli, H.; Birch, A.N.; Casacuberta, J.; De Schrijver, A.; Gralak, M.A.; Guerche, P.; Jones, H.; Manachini, B.; Messéan, A.; et al. Guidance on allergenicity assessment of genetically modified plants. EFSA J. 2017, 15, e04862. [Google Scholar]
- Sidney, J.; Steen, A.; Moore, C.; Ngo, S.; Chung, J.; Peters, B.; Sette, A. Divergent motifs but overlapping binding repertoires of six HLA-DQ molecules frequently expressed in the worldwide human population. J. Immunol. 2010, 185, 4189–4198. [Google Scholar] [CrossRef]
- Stepniak, D.; Wiesner, M.; de Ru, A.H.; Moustakas, A.K.; Drijfhout, J.W.; Papadopoulos, G.K.; van Veelen, P.A.; Koning, F. Large-scale characterization of natural ligands explains the unique gluten-binding properties of HLA-DQ2. J. Immunol. 2008, 180, 3268–3278. [Google Scholar] [CrossRef] [PubMed]
- Kwok, W.W.; Nepom, G.T.; Raymond, F.C. HLA-DQ polymorphisms are highly selective for peptide binding interactions. J. Immunol. 1995, 155, 2468–2476. [Google Scholar] [CrossRef] [PubMed]
- Kwok, W.W.; Domeier, M.E.; Johnson, M.L.; Nepom, G.T.; Koelle, D.M. HLA-DQB1 codon 57 is critical for peptide binding and recognition. J. Exp. Med. 1996, 183, 1253–1258. [Google Scholar] [CrossRef] [PubMed]
- Kwok, W.W.; Domeier, M.E.; Raymond, F.C.; Byers, P.; Nepom, G.T. Allele-specific motifs characterize HLA-DQ interactions with a diabetes-associated peptide derived from glutamic acid decarboxylase. J. Immunol. 1996, 156, 2171–2177. [Google Scholar] [CrossRef] [PubMed]
- Andreatta, M.; Nielsen, M. Characterizing the binding motifs of 11 common human HLA-DP and HLA-DQ molecules using NNAlign. Immunology 2012, 136, 306–311. [Google Scholar] [CrossRef]
- Bergseng, E.; Dørum, S.; Arntzen, M.Ø.; Nielsen, M.; Nygård, S.; Buus, S.; de Souza, G.A.; Sollid, L.M. Different binding motifs of the celiac disease-associated HLA molecules DQ2.5, DQ2.2, and DQ7.5 revealed by relative quantitative proteomics of endogenous peptide repertoires. Immunogenetics 2015, 67, 73–84. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E., 3rd; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Sofware for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]

| QM | Sensitivity | Specificity | Accuracy | Cutoff |
|---|---|---|---|---|
| HLA-DQ2.5 | ||||
| α-gliadin peptide library | 95 | 83 | 89 | 0.1 |
| non-gliadin peptide library | 93 | 89 | 91 | 0.2 |
| HLA-DQ8.1 | ||||
| α-gliadin peptide library | 94 | 94 | 94 | 0.3 |
| Contribution | DQ2.5 | DQ8.1 α-Glia aa | ||
|---|---|---|---|---|
| α-Glia aa | non-Glia aa | |||
| P1 | positive | W: π-π—F53α; hpho—W43α, F51α, F54α, N82β, L85β | W: hb—R53α; π-π—F33α; F52α, F54α; hpho—F33α, F52α, F54α, N82β, E86β | F: hb—R53α; hpho—Y9α, F54α, L85β |
| negative | M: hpho—W43α, L85β | P: hpho—F54α | P: hpho—F54α | |
| P4 | positive | F: hb—Y9α, N62α; hpho—F11β, L26β, K71β | V: hb—Y9α; hpho—L26β, K71β, V78β | F: hb—Y9α, N62α; hpho—F11β, L26β, T28β, K71β, E74β |
| negative | P: hpho—V78β | K: hb—Y9α, G10α; intramolecular hb with G5; hpho—L26β, K71β | Y: hb—Y9α, N62α; hpho—F11β, T28β, E74β, V78β | |
| P6 | positive | W: hb—N62α; π-π—F11β, W61β hpho—V65α, L66α, F11β, S30β, W61β | P: hpho—V65α, L66α, F11β | P: hpho—V65α, F11β, Y30β |
| negative | M: hb—N62α; hpho—N62α,V65α, F11β | Y: hb—N62α; π-π—F11β, W61β hpho—V65α, Y9β, F11β | Y: π-π—F11β hpho—N62α,V65α, F11β | |
| P7 | positive | W: hb—N69α; π-π—F11β, F47β, W61β; hpho—F11β, S28β, F47β, W61β, K71β | P: hb—N69α; hpho—W61β, K71β | - |
| negative | R: hb—N69α; intramolecular hb with Q6; hpho—F47β, W61β | D: hb—N69α, K71β; hpho—W61β | - | |
| P9 | positive | W: hb—N69α; hpho—S72α, L73α, I37β | W: hb—N69α; hpho—L73α, I37β, W61β | N: hb—H68α, N69α |
| negative | K: hb—N69α; hpho—S72α | P: no contacts | P: hb—H68α; hpho—W61β | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atanasova, M.; Dimitrov, I.; Fernandez, A.; Moreno, J.; Koning, F.; Doytchinova, I. Assessment of Novel Proteins Triggering Celiac Disease via Docking-Based Approach. Molecules 2024, 29, 138. https://doi.org/10.3390/molecules29010138
Atanasova M, Dimitrov I, Fernandez A, Moreno J, Koning F, Doytchinova I. Assessment of Novel Proteins Triggering Celiac Disease via Docking-Based Approach. Molecules. 2024; 29(1):138. https://doi.org/10.3390/molecules29010138
Chicago/Turabian StyleAtanasova, Mariyana, Ivan Dimitrov, Antonio Fernandez, Javier Moreno, Frits Koning, and Irini Doytchinova. 2024. "Assessment of Novel Proteins Triggering Celiac Disease via Docking-Based Approach" Molecules 29, no. 1: 138. https://doi.org/10.3390/molecules29010138
APA StyleAtanasova, M., Dimitrov, I., Fernandez, A., Moreno, J., Koning, F., & Doytchinova, I. (2024). Assessment of Novel Proteins Triggering Celiac Disease via Docking-Based Approach. Molecules, 29(1), 138. https://doi.org/10.3390/molecules29010138







