In Vivo Metabolite Profiling of DMU-212 in ApcMin/+ Mice Using UHPLC-Q/Orbitrap/LTQ MS
Abstract
1. Introduction
2. Results
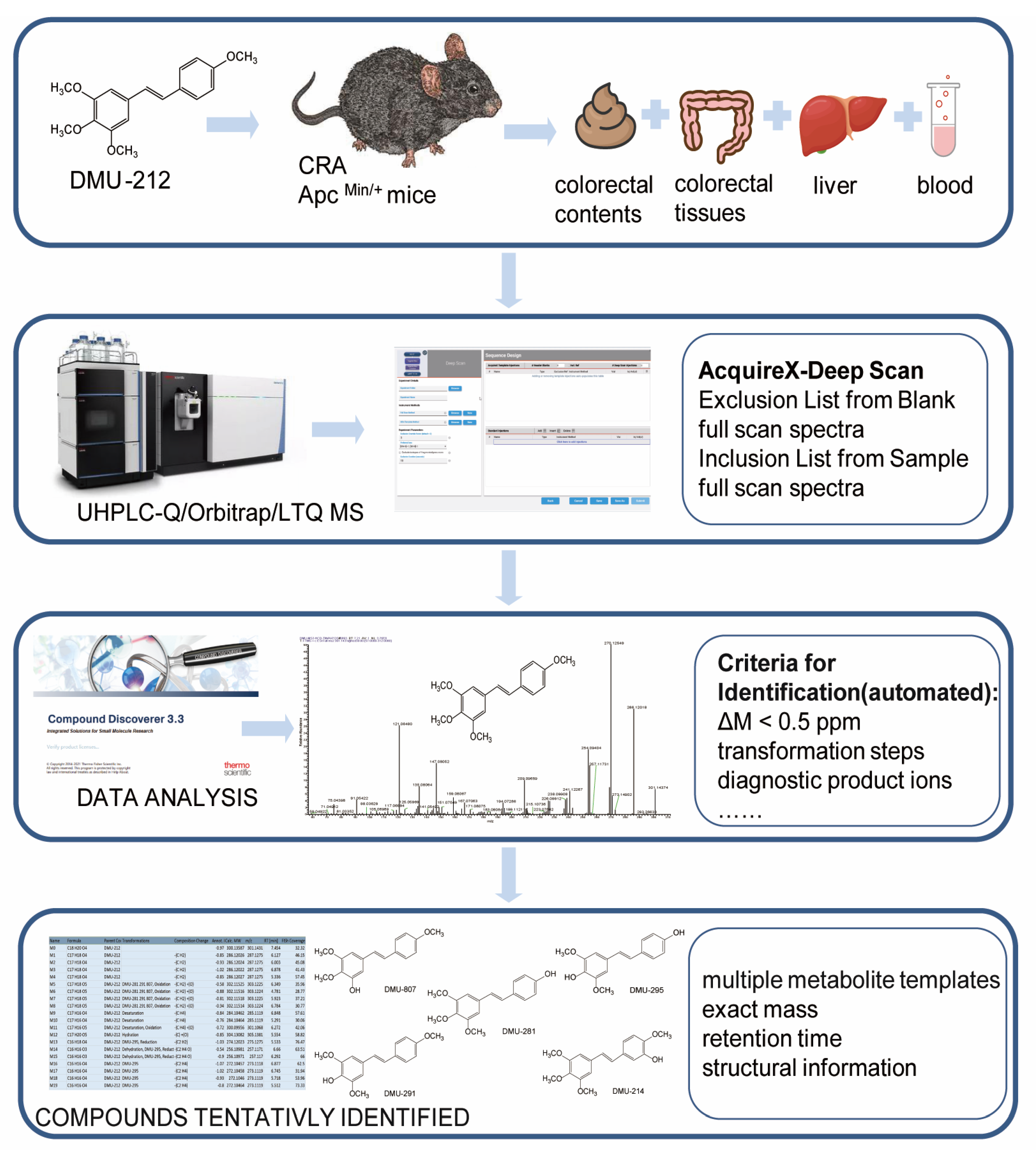
2.1. Establishment of the Analytical Strategy
2.2. Characteristic Fragments of DMU-212
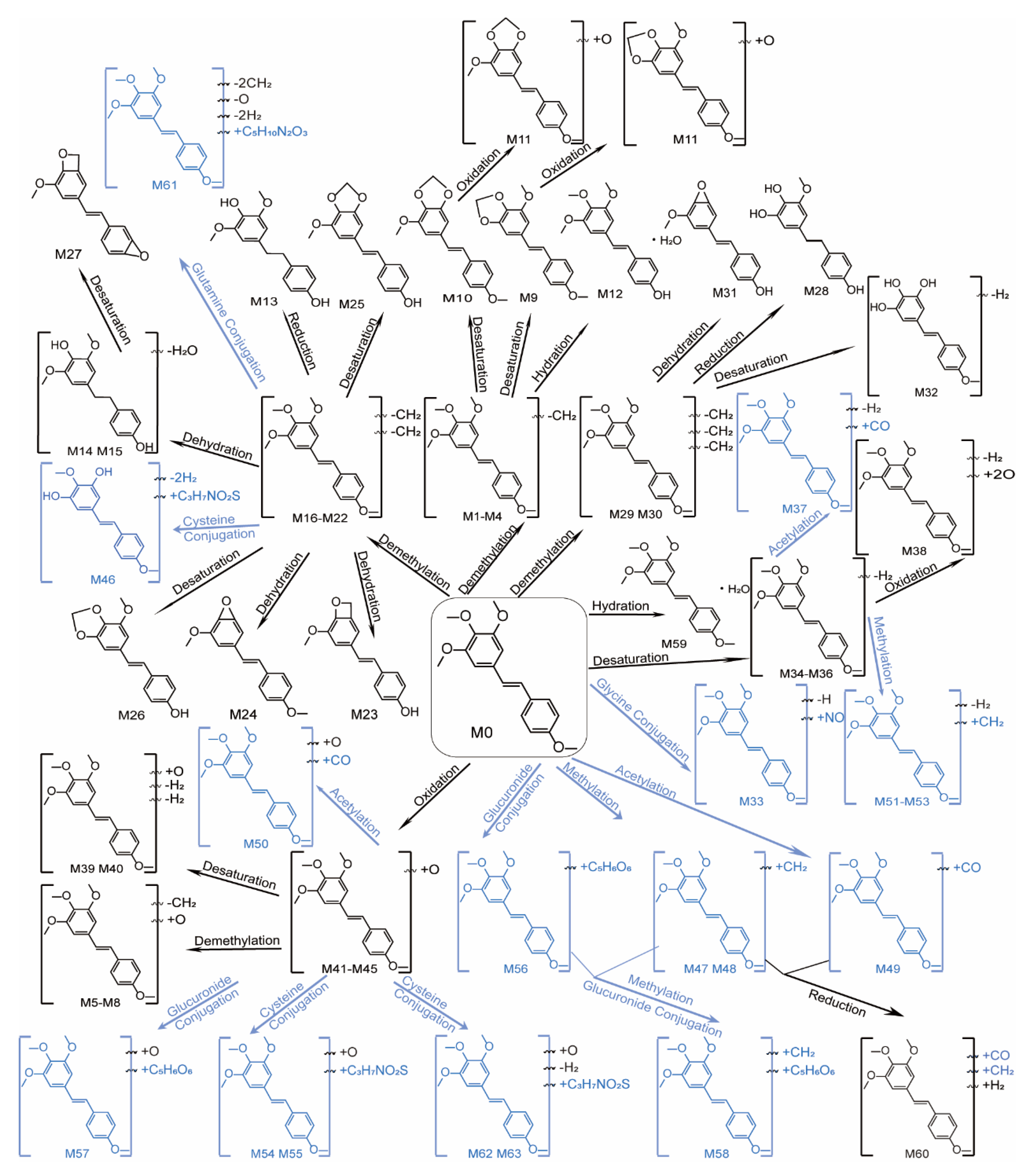
2.3. Comprehensive Characterization of Metabolites of DMU-212 In Vivo
2.3.1. Phase I Metabolites Identification
2.3.2. Phase II Metabolites Identification
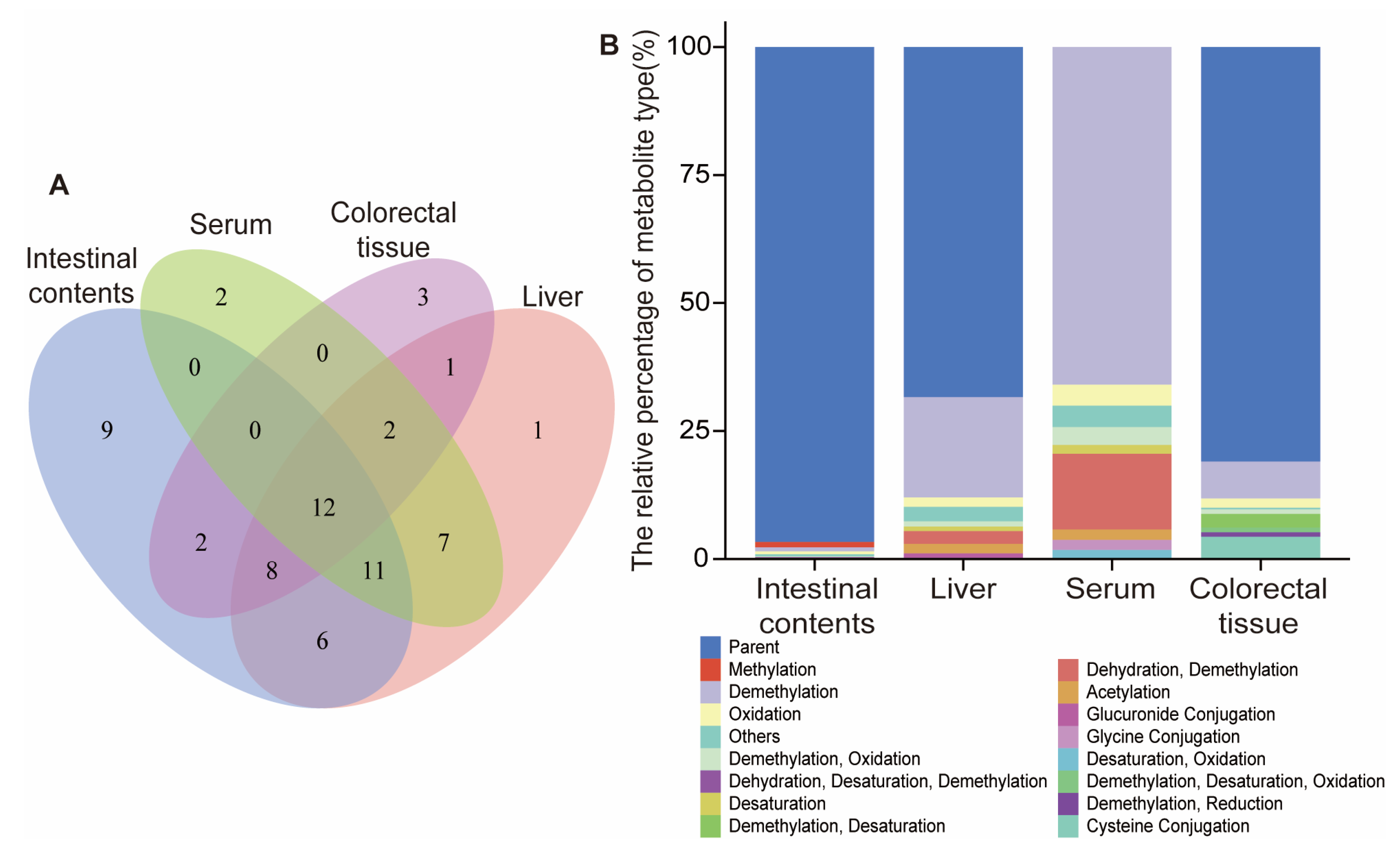
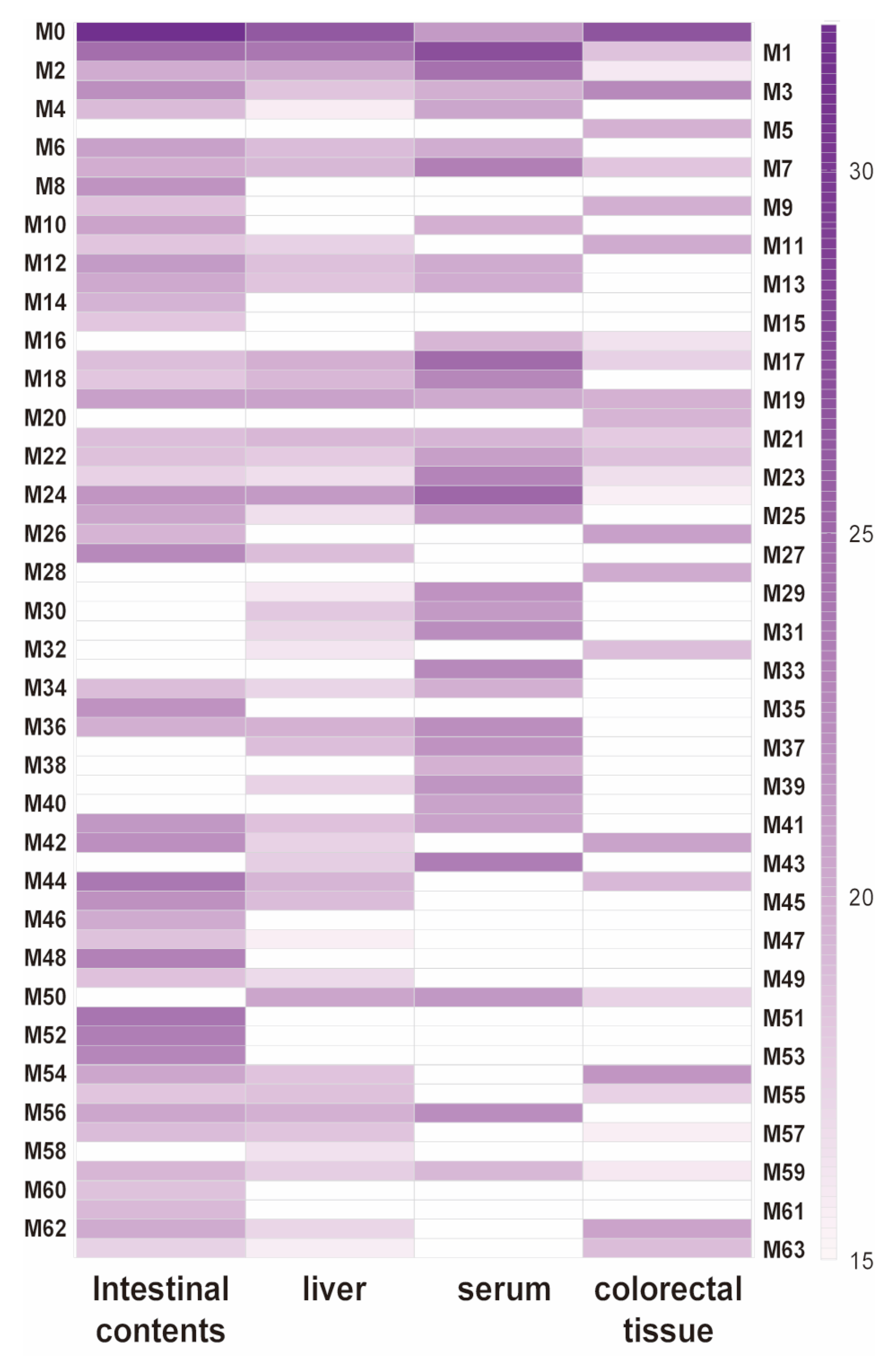
2.4. Metabolic Profiles of DMU-212
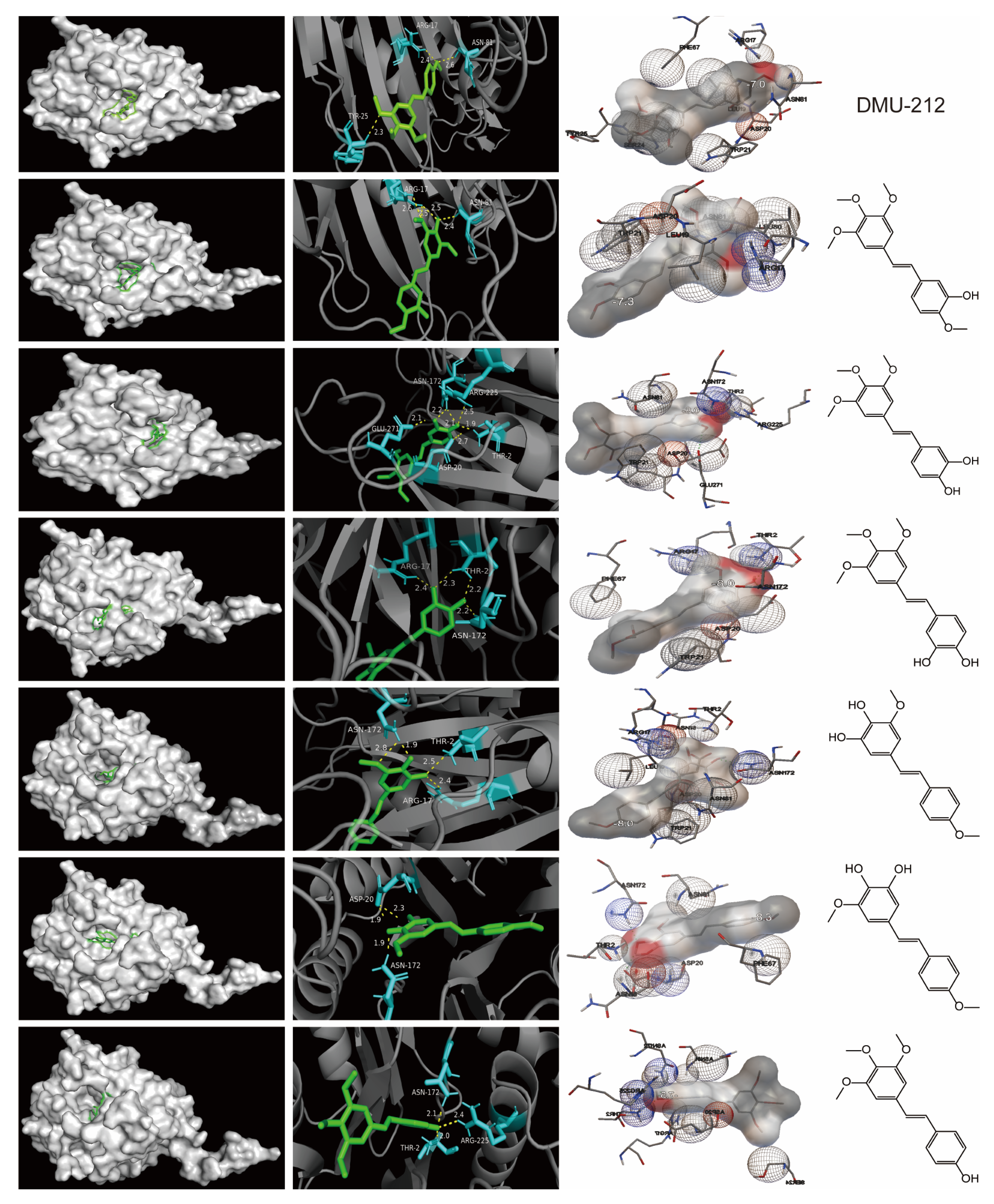
2.5. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Animal Experiments
4.3. Collection and Preparation of Biological Samples
4.4. UHPLC-Q/Orbitrap/LTQ MS Analysis
4.5. Data Processing Software
4.6. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Wu, K.; Ogino, S.; Giovannucci, E.L.; Chan, A.T.; Song, M. Association Between Risk Factors for Colorectal Cancer and Risk of Serrated Polyps and Conventional Adenomas. Gastroenterology 2018, 155, 355–373.e318. [Google Scholar] [CrossRef] [PubMed]
- Heer, E.; Ruan, Y.; Mah, B.; Nguyen, T.; Lyons, H.; Poirier, A.; Boyne, D.J.; O’Sullivan, D.E.; Heitman, S.J.; Hilsden, R.J.; et al. The efficacy of chemopreventive agents on the incidence of colorectal adenomas: A systematic review and network meta-analysis. Prev. Med. 2022, 162, 107169. [Google Scholar] [CrossRef] [PubMed]
- Katona, B.W.; Weiss, J.M. Chemoprevention of Colorectal Cancer. Gastroenterology 2020, 158, 368–388. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Molavi, O.; Haddadi, A.; Lai, R.; Gossage, R.A.; Lavasanifar, A. Resveratrol analog trans 3,4,5,4’-tetramethoxystilbene (DMU-212) mediates anti-tumor effects via mechanism different from that of resveratrol. Cancer Chemother. Pharmacol. 2008, 63, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Sale, S.; Tunstall, R.G.; Ruparelia, K.C.; Potter, G.A.; Steward, W.P.; Gescher, A.J. Comparison of the effects of the chemopreventive agent resveratrol and its synthetic analog trans 3,4,5,4’-tetramethoxystilbene (DMU-212) on adenoma development in the Apc(Min+) mouse and cyclooxygenase-2 in human-derived colon cancer cells. Int. J. Cancer 2005, 115, 194–201. [Google Scholar] [CrossRef]
- Sale, S.; Verschoyle, R.D.; Boocock, D.; Jones, D.J.; Wilsher, N.; Ruparelia, K.C.; Potter, G.A.; Farmer, P.B.; Steward, W.P.; Gescher, A.J. Pharmacokinetics in mice and growth-inhibitory properties of the putative cancer chemopreventive agent resveratrol and the synthetic analogue trans 3,4,5,4’-tetramethoxystilbene. Br. J. Cancer 2004, 90, 736–744. [Google Scholar] [CrossRef]
- Ingle, R.G.; Zeng, S.; Jiang, H.; Fang, W.J. Current developments of bioanalytical sample preparation techniques in pharmaceuticals. J. Pharm. Anal. 2022, 12, 517–529. [Google Scholar] [CrossRef]
- Jia, B.; Park, D.; Hahn, Y.; Jeon, C.O. Metagenomic analysis of the human microbiome reveals the association between the abundance of gut bile salt hydrolases and host health. Gut Microbes 2020, 11, 1300–1313. [Google Scholar] [CrossRef]
- Yang, J.; Li, C.; Liu, Y.; Han, Y.; Zhao, H.; Luo, S.; Zhao, C.; Jiang, N.; Yang, M.; Sun, L. Using network pharmacology to explore the mechanism of Danggui-Shaoyao-San in the treatment of diabetic kidney disease. Front. Pharmacol. 2022, 13, 832299. [Google Scholar] [CrossRef]
- Obach, R.S. Pharmacologically active drug metabolites: Impact on drug discovery and pharmacotherapy. Pharmacol. Rev. 2013, 65, 578–640. [Google Scholar] [CrossRef]
- Wang, H.; Sun, Y.; Guo, W.; Wang, J.; Gao, J.; Peng, W.; Gu, J. Identification and high-throughput quantification of baicalein and its metabolites in plasma and urine. J. Ethnopharmacol. 2023, 301, 115853. [Google Scholar] [CrossRef]
- Li, Y.; Sun, C.; Zhang, Y.; Chen, X.; Huang, H.; Han, L.; Xing, H.; Zhao, D.; Chen, X.; Zhang, Y. Phase I Metabolism of Pterostilbene, a Dietary Resveratrol Derivative: Metabolite Identification, Species Differences, Isozyme Contribution, and Further Bioactivation. J. Agric. Food Chem. 2023, 71, 331–346. [Google Scholar] [CrossRef]
- Han, Y.; Sun, H.; Zhang, A.; Yan, G.; Wang, X.J. Chinmedomics, a new strategy for evaluating the therapeutic efficacy of herbal medicines. Pharmacol. Ther. 2020, 216, 107680. [Google Scholar] [CrossRef]
- Prakash, C.; Shaffer, C.L.; Nedderman, A. Analytical strategies for identifying drug metabolites. Mass Spectrom. Rev. 2007, 26, 340–369. [Google Scholar] [CrossRef]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between drugs and the gut microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef]
- Javdan, B.; Lopez, J.G.; Chankhamjon, P.; Lee, Y.J.; Hull, R.; Wu, Q.; Wang, X.; Chatterjee, S.; Donia, M.S. Personalized Mapping of Drug Metabolism by the Human Gut Microbiome. Cell 2020, 181, 1661–1679.e1622. [Google Scholar] [CrossRef]
- Zádori, Z.S.; Király, K.; Al-Khrasani, M.; Gyires, K. Interactions between NSAIDs, opioids and the gut microbiota—Future perspectives in the management of inflammation and pain. Pharmacol. Ther. 2023, 241, 108327. [Google Scholar] [CrossRef]
- Dikeocha, I.J.; Al-Kabsi, A.M.; Miftahussurur, M.; Alshawsh, M.A. Pharmacomicrobiomics: Influence of gut microbiota on drug and xenobiotic metabolism. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2022, 36, e22350. [Google Scholar] [CrossRef]
- McCoubrey, L.E.; Gaisford, S.; Orlu, M.; Basit, A.W. Predicting drug-microbiome interactions with machine learning. Biotechnol. Adv. 2022, 54, 107797. [Google Scholar] [CrossRef]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A.L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019, 570, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Maini Rekdal, V.; Bess, E.N.; Bisanz, J.E.; Turnbaugh, P.J.; Balskus, E.P. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science 2019, 364, eaau6323. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.S.; Mayers, J.R.; Zhang, Y.; Bhosle, A.; Glasser, N.R.; Nguyen, L.H.; Ma, W.; Bae, S.; Branck, T.; Song, K.; et al. Gut microbial metabolism of 5-ASA diminishes its clinical efficacy in inflammatory bowel disease. Nat. Med. 2023, 29, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Koppel, N.; Maini Rekdal, V.; Balskus, E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356, eaag2770. [Google Scholar] [CrossRef]
- Xu, J.; Chen, H.B.; Li, S.L. Understanding the Molecular Mechanisms of the Interplay Between Herbal Medicines and Gut Microbiota. Med. Res. Rev. 2017, 37, 1140–1185. [Google Scholar] [CrossRef]
- Zhao, M.; Ma, J.; Li, M.; Zhang, Y.; Jiang, B.; Zhao, X.; Huai, C.; Shen, L.; Zhang, N.; He, L.; et al. Cytochrome P450 Enzymes and Drug Metabolism in Humans. Int. J. Mol. Sci. 2021, 22, 12808. [Google Scholar] [CrossRef]
- Piotrowska-Kempisty, H.; Klupczyńska, A.; Trzybulska, D.; Kulcenty, K.; Sulej-Suchomska, A.M.; Kucińska, M.; Mikstacka, R.; Wierzchowski, M.; Murias, M.; Baer-Dubowska, W.; et al. Role of CYP1A1 in the biological activity of methylated resveratrol analogue, 3,4,5,4’-tetramethoxystilbene (DMU-212) in ovarian cancer A-2780 and non-cancerous HOSE cells. Toxicol. Lett. 2017, 267, 59–66. [Google Scholar] [CrossRef]
- Hoffmann, M.F.; Preissner, S.C.; Nickel, J.; Dunkel, M.; Preissner, R.; Preissner, S. The Transformer database: Biotransformation of xenobiotics. Nucleic Acids Res. 2014, 42, D1113–D1117. [Google Scholar] [CrossRef]
- Shi, M.; Xu, B. VO(acac)(2)-catalyzed oxidative coupling reactions of phosphonium salts. J. Org. Chem. 2002, 67, 294–297. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Li, X.; Zhou, X.; Yang, L.; Sun, H.; Kong, L.; Yan, G.; Han, Y.; Wang, X. In Vivo Metabolite Profiling of DMU-212 in ApcMin/+ Mice Using UHPLC-Q/Orbitrap/LTQ MS. Molecules 2023, 28, 3828. https://doi.org/10.3390/molecules28093828
Li J, Li X, Zhou X, Yang L, Sun H, Kong L, Yan G, Han Y, Wang X. In Vivo Metabolite Profiling of DMU-212 in ApcMin/+ Mice Using UHPLC-Q/Orbitrap/LTQ MS. Molecules. 2023; 28(9):3828. https://doi.org/10.3390/molecules28093828
Chicago/Turabian StyleLi, Jing, Xinghua Li, Xiaohang Zhou, Le Yang, Hui Sun, Ling Kong, Guangli Yan, Ying Han, and Xijun Wang. 2023. "In Vivo Metabolite Profiling of DMU-212 in ApcMin/+ Mice Using UHPLC-Q/Orbitrap/LTQ MS" Molecules 28, no. 9: 3828. https://doi.org/10.3390/molecules28093828
APA StyleLi, J., Li, X., Zhou, X., Yang, L., Sun, H., Kong, L., Yan, G., Han, Y., & Wang, X. (2023). In Vivo Metabolite Profiling of DMU-212 in ApcMin/+ Mice Using UHPLC-Q/Orbitrap/LTQ MS. Molecules, 28(9), 3828. https://doi.org/10.3390/molecules28093828




