The Isolation and Structure Elucidation of Spirotetronate Lobophorins A, B, and H8 from Streptomyces sp. CB09030 and Their Biosynthetic Gene Cluster
Abstract
1. Introduction
2. Results and Discussion
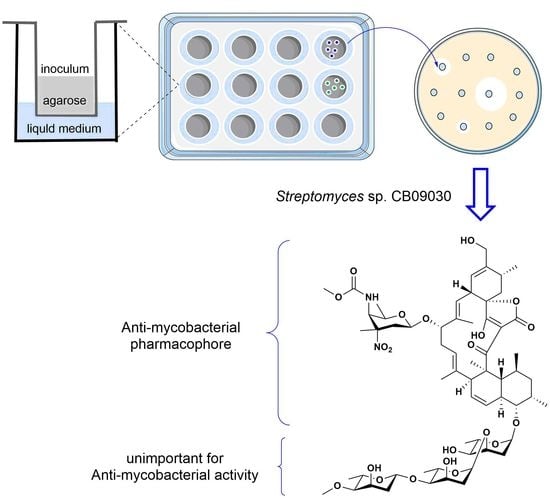
2.1. A Transwell-Based Culture Bioassay to Discover Streptomyces sp. CB09030 with Anti-Mycobacterial Potential
2.2. Isolation and Structure Elucidation of 1–3 from S. sp. CB09030
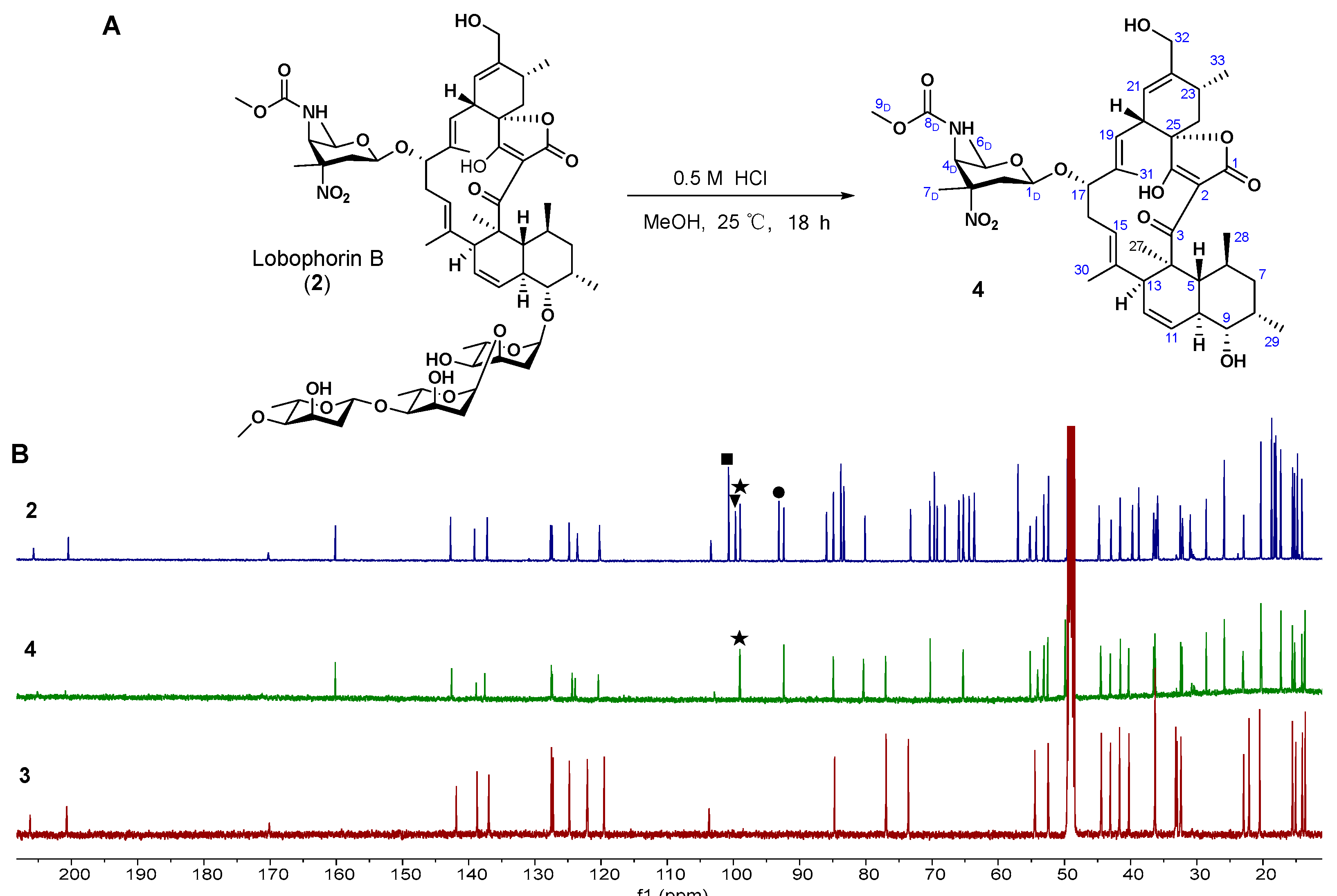
2.3. Acid Hydrolysis of 2 Leads to the Generation of 4
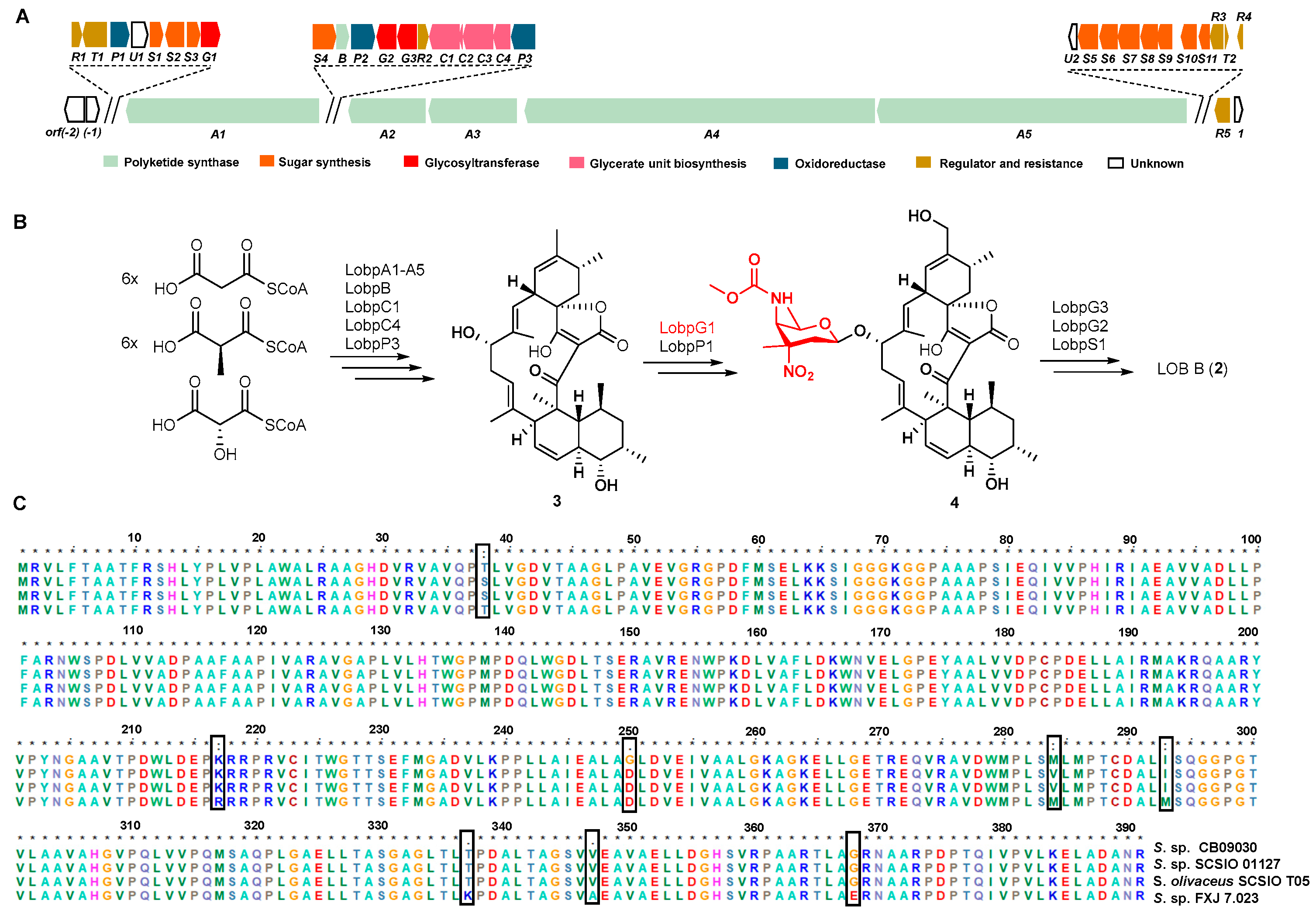
2.4. Bioinformatic Analysis of A Putative LOB BGC from S. sp. CB09030
2.5. The Antibacterial Activity of 1–4
3. Materials and Methods
3.1. A Transwell-Based Culture Bioassay
3.2. General Experimental Procedures
3.3. Streptomyces Fermentation in Shaking Flasks and Extraction
3.4. Isolation and Acid Hydrolysis of 2
3.5. Genome Sequencing and Analysis
3.6. Antibacterial Assay
3.7. In Vitro Cytotoxicity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kang, M.J.; Jones, B.D.; Mandel, A.L.; Hammons, J.C.; DiPasquale, A.G.; Rheingold, A.L.; La Clair, J.J.; Burkart, M.D. Isolation, structure elucidation, and antitumor activity of spirohexenolides A and B. J. Org. Chem. 2009, 74, 9054–9061. [Google Scholar] [CrossRef]
- Imai, H.S.; Suzuki, K.I.; Morioka, M.; Numasaki, Y.; Kadota, S.; Nagai, K.; Iwanami, M.; Saito, T. Okilactomycin, a novel antibiotic produced by a Streptomyces species I. Taxonomy, fermentation, isolation and characterization. J. Antibiot. 1987, 40, 1475–1482. [Google Scholar] [CrossRef]
- Nakai, R.; Kakita, S.; Asai, A.; Chiba, S.; Akinaga, S.; Mizukami, T.; Yamashita, Y. Chrolactomycin, a novel antitumor antibiotic produced by Streptomyces sp. J. Antibiot. 2001, 54, 836–839. [Google Scholar] [CrossRef]
- Nakai, R.; Ishida, H.; Asai, A.; Ogawa, H.; Yamamoto, Y.; Kawasaki, H.; Akinaga, S.; Mizukami, T.; Yamashita, Y. Telomerase inhibitors identified by a forward chemical genetics approach using a yeast strain with shortened telomere length. Cell Chem. Biol. 2006, 13, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Waitz, J.A.; Horan, A.C.; Kalyanpur, M.; Lee, B.K.; Loebenberg, D.; Marquez, J.A.; Miller, G.; Patel, M.G. Kijanimicin (sch 25663), a novel antibiotic produced by actinomad ura kijaniata scc 1256. J. Antibiot. 1981, 34, 1101–1106. [Google Scholar] [CrossRef]
- Ashton, R.J.; Kenig, M.D.; Luk, K.; Planterose, D.N.; Scott-Wood, G. MM 46115, a new antiviral antibiotic from Actinomadura pelletieri. J. Antibiot. 1990, 43, 1387–1393. [Google Scholar] [CrossRef]
- Bister, B.; Bischoff, D.; Strobele, M.; Riedlinger, J.; Reicke, A.; Wolter, F.; Bull, A.T.; Zahner, H.; Fiedler, H.P.; Sussmuth, R.D. Abyssomicin C—A polycyclic antibiotic from a marine Verrucosispora strain as an inhibitor of the p-aminobenzoic acid/tetrahydrofolate biosynthesis pathway. Angew. Chem. Int. Ed. 2004, 43, 2574–2576. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.D.; Jensen, P.R.; Fenical, W. Lobophorins A and B, new antiinflammatory macrolides produced by a tropical marine bacterium. Bioorg. Med. Chem. Lett. 1999, 9, 2003–2006. [Google Scholar] [CrossRef]
- Niu, S.W.; Li, S.M.; Chen, Y.C.; Tian, X.P.; Zhang, H.B.; Zhang, G.T.; Zhang, W.M.; Yang, X.H.; Zhang, S.; Ju, J.H. Lobophorins E and F, new spirotetronate antibiotics from a South China Sea-derived Streptomyces sp. SCSIO 01127. J. Antibiot. 2011, 64, 711–716. [Google Scholar] [CrossRef]
- Wei, R.B.; Xi, T.; Li, J.; Wang, P.; Li, F.C.; Lin, Y.C.; Qin, S. Lobophorin C and D, new kijanimicin derivatives from a marine sponge-associated actinomycetal strain AZS17. Mar. Drugs 2011, 9, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.X.; Wang, J.; Guo, H.; Hou, W.Y.; Yang, N.; Ren, B.; Liu, M.; Dai, H.Q.; Liu, X.T.; Song, F.H.; et al. Three antimycobacterial metabolites identified from a marine-derived Streptomyces sp. MS100061. Appl. Microbiol. Biotechnol. 2013, 97, 3885–3892. [Google Scholar] [CrossRef]
- Pan, H.Q.; Zhang, S.Y.; Wang, N.; Li, Z.L.; Hua, H.M.; Hu, J.C.; Wang, S.J. New spirotetronate antibiotics, lobophorins H and I, from a south China sea-derived Streptomyces sp. 12A35. Mar. Drugs 2013, 11, 3891–3901. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.J.; Koch, M.; Pond, C.D.; Mabeza, G.; Seronay, R.A.; Concepcion, G.P.; Barrows, L.R.; Olivera, B.M.; Schmidt, E.W. Structure and activity of lobophorins from a turrid mollusk-associated Streptomyces sp. J. Antibiot. 2014, 67, 121–126. [Google Scholar] [CrossRef]
- Song, C.F.; Pan, H.Q.; Hu, J.C. Isolation and identification of a new antibiotic, lobophorin J, from a deep sea-derived Streptomyces sp. 12A35. Chin. J. Antibiot. 2015, 40, 721–727. [Google Scholar]
- Brana, A.F.; Sarmiento-Vizcaino, A.; Osset, M.; Perez-Victoria, I.; Martin, J.; de Pedro, N.; de la Cruz, M.; Diaz, C.; Vicente, F.; Reyes, F.; et al. Lobophorin K, a new natural product with cytotoxic activity produced by Streptomyces sp. M-207 associated with the deep-sea coral lophelia pertusa. Mar. Drugs 2017, 15, 144. [Google Scholar] [CrossRef]
- Luo, M.H.; Tang, L.J.; Dong, Y.L.; Huang, H.B.; Deng, Z.X.; Sun, Y.H. Antibacterial natural products lobophorin L and M from the marine-derived Streptomyces sp. 4506. Nat. Prod. Res. 2020, 35, 5581–5587. [Google Scholar] [CrossRef] [PubMed]
- Cruz, P.G.; Fribley, A.M.; Miller, J.R.; Larsen, M.J.; Schultz, P.J.; Jacob, R.T.; Tamayo-Castillo, G.; Kaufman, R.J.; Sherman, D.H. Novel lobophorins inhibit oral cancer cell growth and induce Atf4- and chop-dependent cell death in murine fibroblasts. ACS Med. Chem. Lett. 2015, 6, 877–881. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Y.; Ding, W.J.; Qin, X.J.; Ju, J.H. Genome sequencing of Streptomyces olivaceus SCSIO T05 and activated production of lobophorin CR4 via metabolic engineering and genome mining. Mar. Drugs 2019, 17, 593. [Google Scholar] [CrossRef]
- Tan, B.; Chen, S.; Zhang, Q.; Chen, Y.; Zhu, Y.; Khan, I.; Zhang, W.; Zhang, C. Heterologous expression leads to discovery of diversified lobophorin analogues and a flexible glycosyltransferase. Org. Lett. 2020, 22, 1062–1066. [Google Scholar] [CrossRef]
- Tan, B.; Zhang, Q.B.; Xiao, J.; Yuan, C.S.; Chen, Y.C.; Chen, S.Q.; Zhang, W.M.; Zhu, Y.G.; Zhang, C.S. Characterization of the P450 monooxygenase LobP1 as C-32 hydroxylase in lobophorin biosynthesis. Chin. J. Chem. 2022, 40. [Google Scholar] [CrossRef]
- Seyedsayamdost, M.R.; Stallforth, P. Special issue in honor of professor Jon Clardy. J. Nat. Prod. 2020, 83, 565–568. [Google Scholar] [CrossRef] [PubMed]
- Pishchany, G.; Mevers, E.; Ndousse-Fetter, S.; Horvath, D.J.; Paludo, C.R.; Silva-Junior, E.A.; Koren, S.; Skaar, E.P.; Clardy, J.; Kolter, R. Amycomicin is a potent and specific antibiotic discovered with a targeted interaction screen. Proc. Natl. Acad. Sci. USA 2018, 115, 10124–10129. [Google Scholar] [CrossRef] [PubMed]
- Bae, M.; Mevers, E.; Pishchany, G.; Whaley, S.G.; Charles, O.R.; Andes, D.R.; Currie, C.R.; Pupo, M.T.; Clardy, J. Chemical exchanges between multilateral symbionts. Org. Lett. 2021, 23, 1648–1652. [Google Scholar] [CrossRef] [PubMed]
- George, K.M.; Chatterjee, D.; Gunawardana, G.; Welty, D.; Hayman, J.; Lee, R.; Small, P.L.C. Mycolactone: A polyketide toxin from Mycobacterium ulcerans required for virulence. Science 1999, 283, 854–857. [Google Scholar] [CrossRef]
- Chany, A.C.; Tresse, C.; Casarotto, V.; Blanchard, N. History, biology and chemistry of Mycobacterium ulcerans infections (Buruli ulcer disease). Nat. Prod. Rep. 2013, 30, 1527–1567. [Google Scholar] [CrossRef]
- Silva, M.T.; Portaels, F.; Pedrosa, J. Pathogenetic mechanisms of the intracellular parasite Mycobacterium ulcerans leading to Buruli ulcer. Lancet 2009, 9, 696–710. [Google Scholar] [CrossRef]
- Vale, A.D.; Cabanes, D.; Sousa, S. Bacterial toxins as pathogen weapons against phagocytes. Front. Microbiol. 2016, 7, 42. [Google Scholar] [PubMed]
- Mallams, A.K.; Puar, M.S.; Rossman, R.R.; McPhail, A.T.; Macfarlane, R.D.; Stephens, R.L. Kijanimicin. Part 3. structure and absolute stereochemistry of 1983. J. Chem. Soc. 1983, 1, 1497–1534. [Google Scholar]
- Li, S.M.; Xiao, J.; Zhu, Y.G.; Zhang, G.T.; Yang, C.F.; Zhang, H.B.; Ma, L.; Zhang, C.S. Dissecting glycosylation steps in lobophorin biosynthesis implies an iterative glycosyltransferase. Org. Lett. 2013, 15, 1374–1377. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]


| Compounds | M. smegmatis MC2 155 | B. subtilis 62305 |
|---|---|---|
| 1 | 8 | 8 |
| 2 | 4 | 1 |
| 3 | 8 | 32 |
| 4 | 8 | 32 |
| ampicillin | - | 0.5 |
| rifampicin | 2 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, J.; Peng, D.; Peng, F.-F.; Zhang, Q.-B.; Duan, Y.-W.; Huang, Y. The Isolation and Structure Elucidation of Spirotetronate Lobophorins A, B, and H8 from Streptomyces sp. CB09030 and Their Biosynthetic Gene Cluster. Molecules 2023, 28, 3597. https://doi.org/10.3390/molecules28083597
Shi J, Peng D, Peng F-F, Zhang Q-B, Duan Y-W, Huang Y. The Isolation and Structure Elucidation of Spirotetronate Lobophorins A, B, and H8 from Streptomyces sp. CB09030 and Their Biosynthetic Gene Cluster. Molecules. 2023; 28(8):3597. https://doi.org/10.3390/molecules28083597
Chicago/Turabian StyleShi, Jie, Dian Peng, Fei-Fei Peng, Qing-Bo Zhang, Yan-Wen Duan, and Yong Huang. 2023. "The Isolation and Structure Elucidation of Spirotetronate Lobophorins A, B, and H8 from Streptomyces sp. CB09030 and Their Biosynthetic Gene Cluster" Molecules 28, no. 8: 3597. https://doi.org/10.3390/molecules28083597
APA StyleShi, J., Peng, D., Peng, F.-F., Zhang, Q.-B., Duan, Y.-W., & Huang, Y. (2023). The Isolation and Structure Elucidation of Spirotetronate Lobophorins A, B, and H8 from Streptomyces sp. CB09030 and Their Biosynthetic Gene Cluster. Molecules, 28(8), 3597. https://doi.org/10.3390/molecules28083597