Cell-Penetrating Peptides for Use in Development of Transgenic Plants
Abstract
1. Introduction
2. Cell-Penetrating Peptides (CPPs)
2.1. Categories of Cell-Penetrating Peptides
2.1.1. Cationic Type
2.1.2. Amphipathic Type
2.1.3. Hydrophobic Type
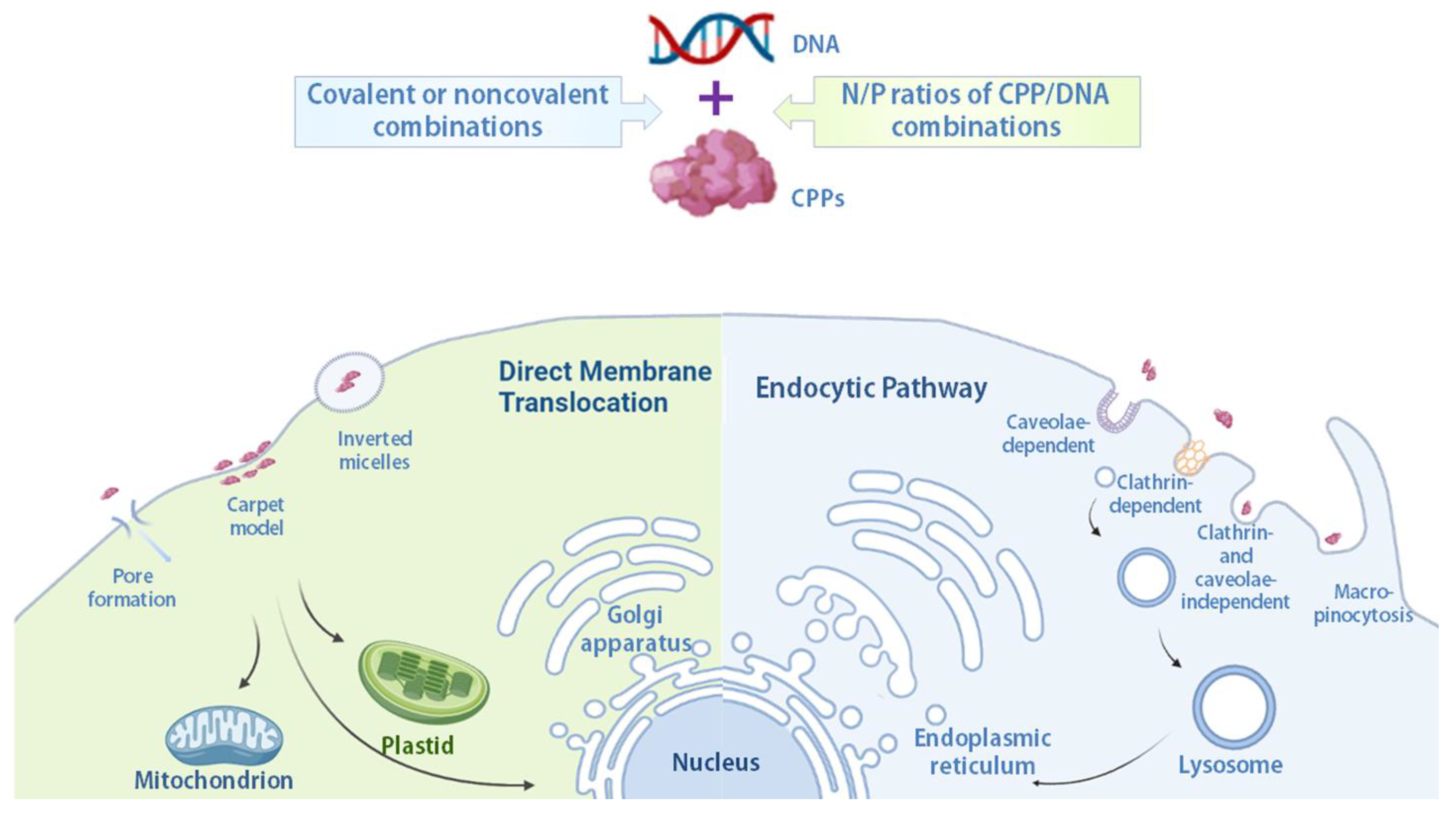
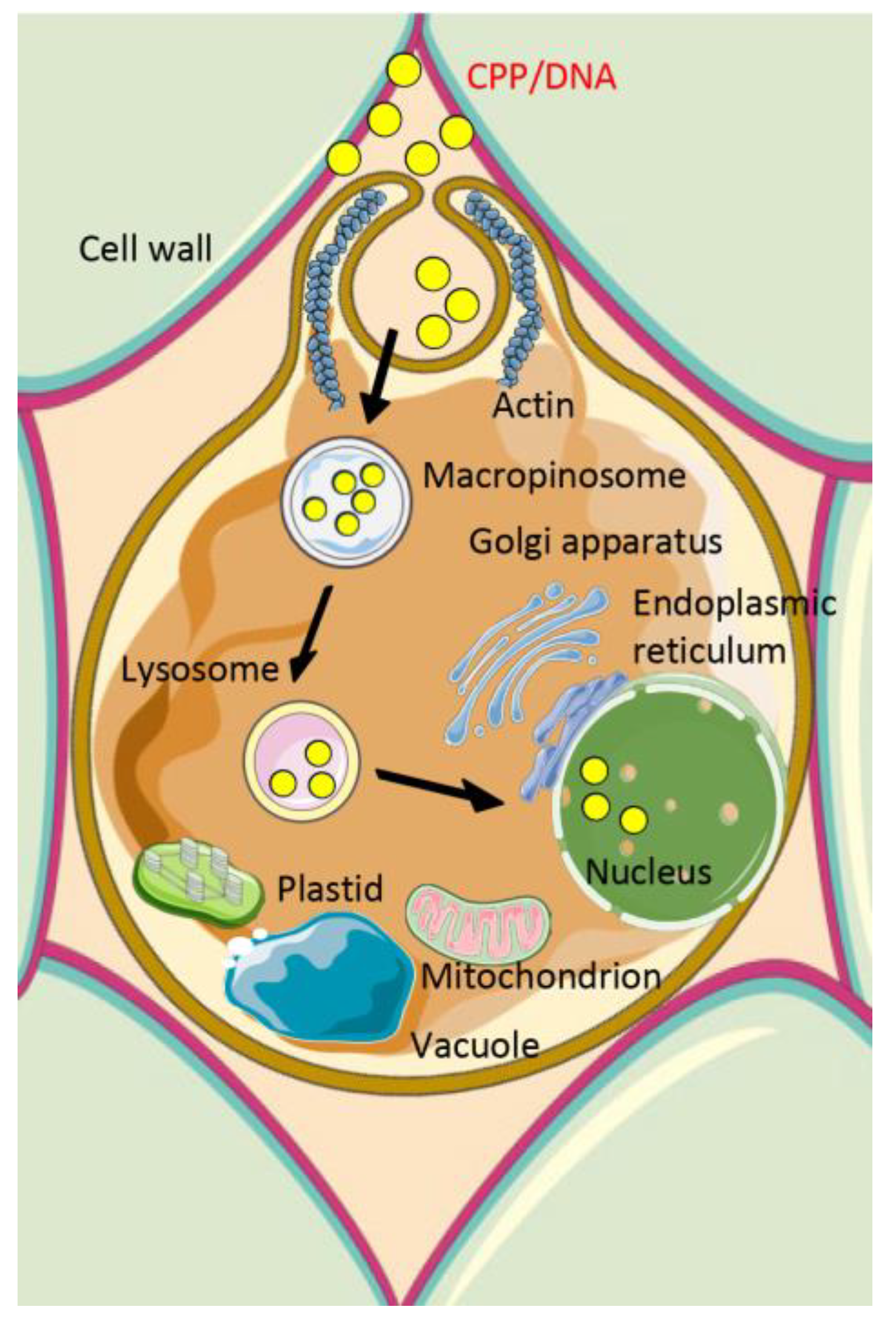
2.2. Mechanisms of Cellular Internalization of Cell-Penetrating Peptide/Cargo Complexes
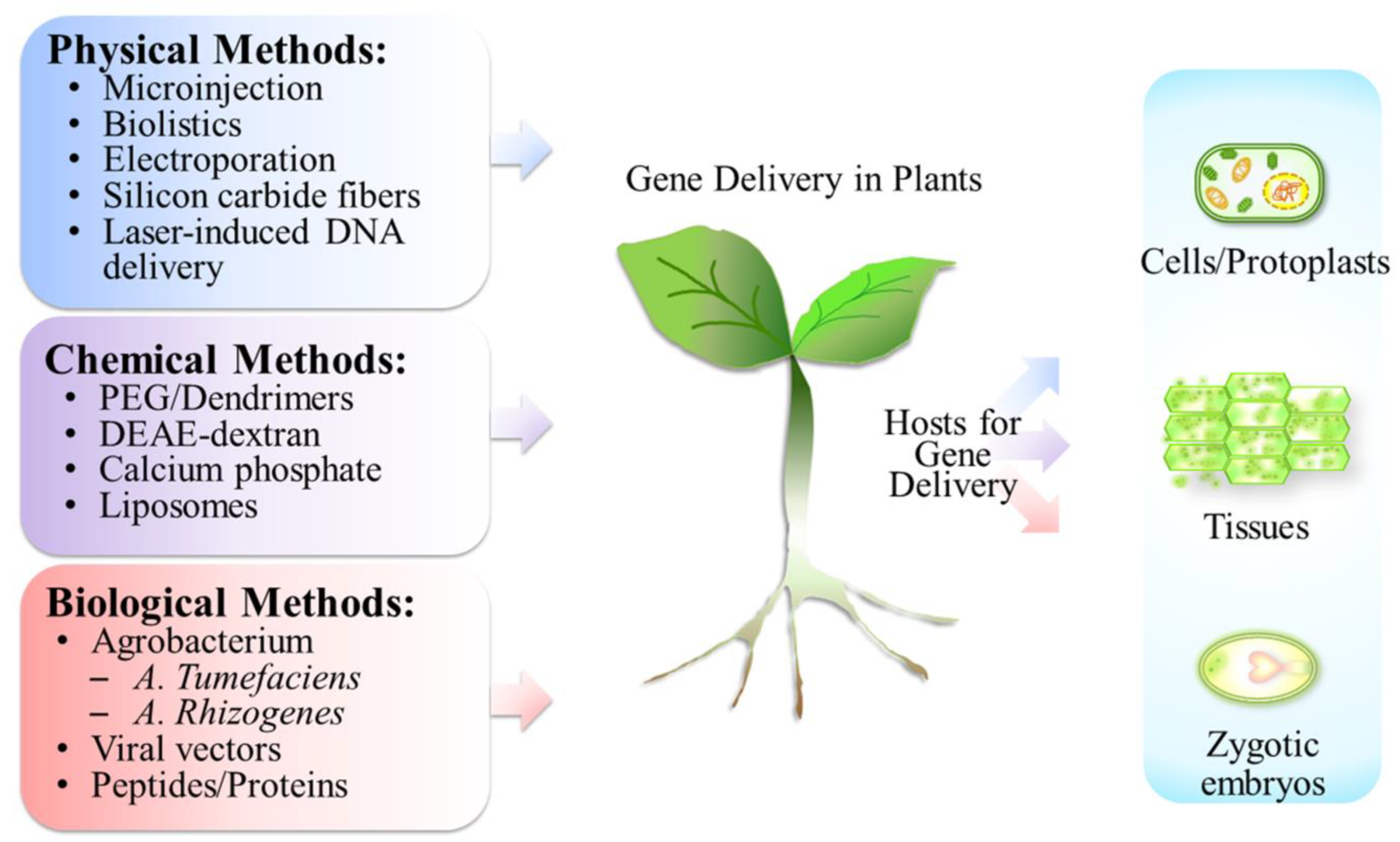
3. Subcellular Targets for Gene Delivery
3.1. Nucleus
3.2. Chloroplasts (Plastids)
3.3. Mitochondria
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed]
- Akanmu, A.O.; Babalola, O.O.; Venturi, V.; Ayilara, M.S.; Adeleke, B.S.; Amoo, A.E.; Sobowale, A.A.; Fadiji, A.E.; Glick, B.R. Plant disease management: Leveraging on the plant-microbe-soil interface in the biorational use of organic amendments. Front. Plant Sci. 2021, 12, 700507. [Google Scholar] [CrossRef] [PubMed]
- Sultana, M.S.; Millwood, R.J.; Mazarei, M.; Stewart, C.N., Jr. Proteinase inhibitors in legume herbivore defense: From natural to genetically engineered protectants. Plant Cell Rep. 2022, 41, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Then, C.; Miyazaki, J.; Bauer-Panskus, A. Deficiencies in the risk assessment of genetically engineered Bt cowpea approved for cultivation in Nigeria: A critical review. Plants 2022, 11, 380. [Google Scholar] [CrossRef]
- Haq, S.K.; Atif, S.M.; Khan, R.H. Protein proteinase inhibitor genes in combat against insects, pests, and pathogens: Natural and engineered phytoprotection. Arch. Biochem. Biophys. 2004, 431, 145–159. [Google Scholar] [CrossRef]
- Key, S.; Ma, J.K.; Drake, P.M. Genetically modified plants and human health. J. R. Soc. Med. 2008, 101, 290–298. [Google Scholar] [CrossRef]
- Nandy, D.; Maity, A.; Mitra, A.K. Target-specific gene delivery in plant systems and their expression: Insights into recent developments. J. Biosci. 2020, 45, 30. [Google Scholar] [CrossRef]
- Vain, P. Thirty years of plant transformation technology development. Plant Biotechnol. J. 2007, 5, 221–229. [Google Scholar] [CrossRef]
- Chen, Q.; Lai, H. Gene delivery into plant cells for recombinant protein production. Biomed. Res. Int. 2015, 2015, 932161. [Google Scholar] [CrossRef]
- Peng, L.H.; Gu, T.W.; Xu, Y.; Dad, H.A.; Liu, J.X.; Lian, J.Z.; Huang, L.Q. Gene delivery strategies for therapeutic proteins production in plants: Emerging opportunities and challenges. Biotechnol. Adv. 2022, 54, 107845. [Google Scholar] [CrossRef]
- Holl, N.J.; Lee, H.J.; Huang, Y.W. Evolutionary timeline of genetic delivery and gene therapy. Curr. Gene Ther. 2021, 21, 89–111. [Google Scholar] [CrossRef] [PubMed]
- Imani, J.; Kogel, K.H. Plant transformation techniques: Agrobacterium- and microparticle-mediated gene transfer in cereal plants. Methods Mol. Biol. 2020, 2124, 281–294. [Google Scholar]
- Ozyigit, I.I. Gene transfer to plants by electroporation: Methods and applications. Mol. Biol. Rep. 2020, 47, 3195–3210. [Google Scholar] [CrossRef] [PubMed]
- Hayashimoto, A.; Li, Z.; Murai, N. A polyethylene glycol-mediated protoplast transformation system for production of fertile transgenic rice plants. Plant Physiol. 1990, 93, 857–863. [Google Scholar] [CrossRef]
- Gad, A.E.; Rosenberg, N.; Altman, A. Liposome-mediated gene delivery into plant cells. Physiol. Plant. 1990, 79, 177–183. [Google Scholar] [CrossRef]
- Alghuthaymi, M.A.; Ahmad, A.; Khan, Z.; Khan, S.H.; Ahmed, F.K.; Faiz, S.; Nepovimova, E.; Kuca, K.; Abd-Elsalam, K.A. Exosome/liposome-like nanoparticles: New carriers for CRISPR genome editing in plants. Int. J. Mol. Sci. 2021, 22, 7456. [Google Scholar] [CrossRef]
- McLenachan, S.; Sarsero, J.P.; Ioannou, P.A. Flow-cytometric analysis of mouse embryonic stem cell lipofection using small and large DNA constructs. Genomics 2007, 89, 708–720. [Google Scholar] [CrossRef]
- Davey, M.R.; Soneji, J.R.; Rao, M.N.; Kourmpetli, S.; Bhattacharya, A.; Kole, C. Generation and deployment of transgenic crop plants: An overview. In Transgenic Crop Plants: Principles and Development; Kole, C., Michler, C.H., Abbott, A.G., Hall, T.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 1, pp. 1–29. [Google Scholar]
- Hwang, H.H.; Yu, M.; Lai, E.M. Agrobacterium-mediated plant transformation: Biology and applications. Arab. Book 2017, 15, e0186. [Google Scholar] [CrossRef]
- Yusibov, V.; Rabindran, S.; Commandeur, U.; Twyman, R.M.; Fischer, R. The potential of plant virus vectors for vaccine production. Drugs R D 2006, 7, 203–217. [Google Scholar] [CrossRef]
- Liu, B.R.; Chiou, S.H.; Huang, Y.W.; Lee, H.J. Bio-membrane internalization mechanisms of arginine-rich cell-penetrating peptides in various species. Membranes 2022, 12, 88. [Google Scholar] [CrossRef]
- Taylor, R.E.; Zahid, M. Cell penetrating peptides, novel vectors for gene therapy. Pharmaceutics 2020, 12, 225. [Google Scholar] [CrossRef]
- Sokolova, V.; Epple, M. Inorganic nanoparticles are carriers of nucleic acids. Angew. Chem. Int. Ed. Engl. 2008, 47, 1382–1395. [Google Scholar] [CrossRef]
- Hingorani, D.V.; Camargo, M.F.; Quraishi, M.A.; Adams, S.R.; Advani, S.J. Tumor activated cell penetrating peptides to selectively deliver immune modulatory drugs. Pharmaceutics 2021, 13, 365. [Google Scholar] [CrossRef] [PubMed]
- Korivi, M.; Huang, Y.W.; Liu, B.R. Cell-penetrating peptides as a potential drug delivery system for effective treatment of diabetes. Curr. Pharm. Des. 2021, 27, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, B.R.; Lee, H.J.; Shannon, K.B.; Winiarz, J.G.; Wang, T.C.; Chiang, H.J.; Huang, Y.W. Nona-arginine facilitates delivery of quantum dots into cells via multiple pathways. J. Biomed. Biotechnol. 2010, 2010, 948543. [Google Scholar] [CrossRef] [PubMed]
- Habault, J.; Poyet, J.L. Recent advances in cell penetrating peptide-based anticancer therapies. Molecules 2019, 24, 927. [Google Scholar] [CrossRef]
- Xiao, Q.; Du, W.; Dong, X.; Du, S.; Ong, S.Y.; Tang, G.; Zhang, C.; Yang, F.; Li, L.; Gao, L.; et al. Cell-penetrating mitochondrion-targeting ligands for the universal delivery of small molecules, proteins and nanomaterials. Chemistry 2021, 27, 12207–12214. [Google Scholar] [CrossRef]
- Layek, B.; Lipp, L.; Singh, J. Cell penetrating peptide conjugated chitosan for enhanced delivery of nucleic acid. Int. J. Mol. Sci. 2015, 16, 28912–28930. [Google Scholar] [CrossRef]
- Kardani, K.; Milani, A.; Shabani, S.H.; Bolhassani, A. Cell penetrating peptides: The potent multi-cargo intracellular carriers. Expert Opin. Drug Deliv. 2019, 16, 1227–1258. [Google Scholar] [CrossRef]
- Chugh, A.; Eudes, F.; Shim, Y.S. Cell-penetrating peptides: Nanocarrier for macromolecule delivery in living cells. IUBMB Life 2010, 62, 183–193. [Google Scholar] [CrossRef]
- Chang, M.; Chou, J.C.; Lee, H.J. Cellular internalization of fluorescent proteins via arginine-rich intracellular delivery peptide in plant cells. Plant Cell Physiol. 2005, 46, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.W.; Hu, J.W.; Liu, B.R.; Lee, C.Y.; Li, J.F.; Chou, J.C.; Lee, H.J. Arginine-rich intracellular delivery peptides synchronously deliver covalently and noncovalently linked proteins into plant cells. J. Agric. Food Chem. 2010, 58, 2288–2294. [Google Scholar] [CrossRef] [PubMed]
- Chuah, J.A.; Numata, K. Stimulus-responsive peptide for effective delivery and release of DNA in plants. Biomacromolecules 2018, 19, 1154–1163. [Google Scholar] [CrossRef]
- Numata, K.; Horii, Y.; Oikawa, K.; Miyagi, Y.; Demura, T.; Ohtani, M. Library screening of cell-penetrating peptide for BY-2 cells, leaves of Arabidopsis, tobacco, tomato, poplar, and rice callus. Sci Rep 2018, 8, 10966. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Itami, J.; Oikawa, K.; Motoda, Y.; Kigawa, T.; Numata, K. Native protein delivery into rice callus using ionic complexes of protein and cell-penetrating peptides. PLoS ONE 2019, 14, e0214033. [Google Scholar] [CrossRef]
- Terada, K.; Gimenez-Dejoz, J.; Miyagi, Y.; Oikawa, K.; Tsuchiya, K.; Numata, K. Artificial cell-penetrating peptide containing periodic α-aminoisobutyric acid with long-term internalization efficiency in human and plant cells. ACS Biomater. Sci. Eng. 2020, 6, 3287–3298. [Google Scholar] [CrossRef]
- Miyamoto, T.; Tsuchiya, K.; Numata, K. Endosome-escaping micelle complexes dually equipped with cell-penetrating and endosome-disrupting peptides for efficient DNA delivery into intact plants. Nanoscale 2021, 13, 5679–5692. [Google Scholar] [CrossRef]
- Watanabe, K.; Odahara, M.; Miyamoto, T.; Numata, K. Fusion peptide-based biomacromolecule delivery system for plant cells. ACS Biomater. Sci. Eng. 2021, 7, 2246–2254. [Google Scholar] [CrossRef]
- Islam, M.M.; Odahara, M.; Yoshizumi, T.; Oikawa, K.; Kimura, M.; Su’etsugu, M.; Numata, K. Cell-penetrating peptide-mediated transformation of large plasmid DNA into Escherichia coli. ACS Synth. Biol. 2019, 8, 1215–1218. [Google Scholar] [CrossRef]
- Higuchi-Takeuchi, M.; Miyamoto, T.; Foong, C.P.; Goto, M.; Morisaki, K.; Numata, K. Peptide-mediated gene transfer into marine purple photosynthetic bacteria. Int. J. Mol. Sci. 2020, 21, 8625. [Google Scholar] [CrossRef]
- Gong, Z.; Ikonomova, S.P.; Karlsson, A.J. Secondary structure of cell-penetrating peptides during interaction with fungal cells. Protein Sci. 2018, 27, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Liu, B.R.; Dai, Y.H.; Lee, C.Y.; Chan, M.H.; Chen, H.H.; Chiang, H.J.; Lee, H.J. A gene delivery system for insect cells mediated by arginine-rich cell-penetrating peptides. Gene 2012, 493, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.H.; Liu, B.R.; Chiang, H.J.; Lee, H.J. Gene transport and expression by arginine-rich cell-penetrating peptides in Paramecium. Gene 2011, 489, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.R.; Liou, J.S.; Chen, Y.J.; Huang, Y.W.; Lee, H.J. Delivery of nucleic acids, proteins, and nanoparticles by arginine-rich cell-penetrating peptides in rotifers. Mar. Biotechnol. N. Y. 2013, 15, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, E.J.B.; Yoshida, S.; Kamei, N.; Iwamae, R.; Khafagy, E.S.; Olsen, J.; Rahbek, U.L.; Pedersen, B.L.; Takayama, K.; Takeda-Morishita, M. In vivo proof of concept of oral insulin delivery based on a co-administration strategy with the cell-penetrating peptide penetratin. J. Control. Release 2014, 189, 19–24. [Google Scholar] [CrossRef]
- Park, S.E.; Sajid, M.I.; Parang, K.; Tiwari, R.K. Cyclic cell-penetrating peptides as efficient intracellular drug delivery tools. Mol. Pharm. 2019, 16, 3727–3743. [Google Scholar] [CrossRef]
- Szabó, I.; Yousef, M.; Soltész, D.; Bató, C.; Mező, G.; Bánóczi, Z. Redesigning of cell-penetrating peptides to improve their efficacy as a drug delivery system. Pharmaceutics 2022, 14, 907. [Google Scholar] [CrossRef]
- Lehto, T.; Kurrikoff, K.; Langel, U. Cell-penetrating peptides for the delivery of nucleic acids. Expert Opin. Drug Deliv. 2012, 9, 823–836. [Google Scholar] [CrossRef]
- Ziemienowicz, A.; Pepper, J.; Eudes, F. Applications of CPPs in genome modulation of plants. Methods Mol. Biol. 2015, 1324, 417–434. [Google Scholar]
- Bilichak, A.; Sastry-Dent, L.; Sriram, S.; Simpson, M.; Samuel, P.; Webb, S.; Jiang, F.; Eudes, F. Genome editing in wheat microspores and haploid embryos mediated by delivery of ZFN proteins and cell-penetrating peptide complexes. Plant Biotechnol. J. 2020, 18, 1307–1316. [Google Scholar] [CrossRef]
- Kardani, K.; Bolhassani, A. Cppsite 2.0: An available database of experimentally validated cell-penetrating peptides predicting their secondary and tertiary structures. J. Mol. Biol. 2021, 433, 166703. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.R.; Huang, Y.W.; Aronstam, R.S.; Lee, H.J. Identification of a short cell-penetrating peptide from bovine lactoferricin for intracellular delivery of DNA in human A549 cells. PLoS ONE 2016, 11, e0150439. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Patel, V.; George, N.V.; Mallajosyula, S.S. KELM-CPPpred: Kernel extreme learning machine based prediction model for cell-penetrating peptides. J. Proteome Res. 2018, 17, 3214–3222. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-penetrating peptides: From basic research to clinics. Trends Pharmacol Sci 2017, 38, 406–424. [Google Scholar] [CrossRef]
- Vivès, E.; Brodin, P.; Lebleu, B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 1997, 272, 16010–16017. [Google Scholar] [CrossRef]
- Herce, H.D.; Garcia, A.E. Molecular dynamics simulations suggest a mechanism for translocation of the HIV-1 TAT peptide across lipid membranes. Proc. Natl. Acad. Sci. USA 2007, 104, 20805–20810. [Google Scholar] [CrossRef]
- Tietz, O.; Cortezon-Tamarit, F.; Chalk, R.; Able, S.; Vallis, K.A. Tricyclic cell-penetrating peptides for efficient delivery of functional antibodies into cancer cells. Nat. Chem. 2022, 14, 284–293. [Google Scholar] [CrossRef]
- Wender, P.A.; Mitchell, D.J.; Pattabiraman, K.; Pelkey, E.T.; Steinman, L.; Rothbard, J.B. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: Peptoid molecular transporters. Proc. Natl. Acad. Sci. USA 2000, 97, 13003–13008. [Google Scholar] [CrossRef]
- Derossi, D.; Calvet, S.; Trembleau, A.; Brunissen, A.; Chassaing, G.; Prochiantz, A. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J. Biol. Chem. 1996, 271, 18188–18193. [Google Scholar] [CrossRef]
- Drin, G.; Mazel, M.; Clair, P.; Mathieu, D.; Kaczorek, M.; Temsamani, J. Physico-chemical requirements for cellular uptake of pAntp peptide. Role of lipid-binding affinity. Eur. J. Biochem. 2001, 268, 1304–1314. [Google Scholar] [CrossRef]
- Lundin, P.; Johansson, H.; Guterstam, P.; Holm, T.; Hansen, M.; Langel, U.; Andaloussi, S.E.L. Distinct uptake routes of cell-penetrating peptide conjugates. Bioconjug. Chem. 2008, 19, 2535–2542. [Google Scholar] [CrossRef] [PubMed]
- Nishi, K.; Saigo, K. Cellular internalization of green fluorescent protein fused with herpes simplex virus protein VP22 via a lipid raft-mediated endocytic pathway independent of caveolae and Rho family GTPases but dependent on dynamin and Arf6. J. Biol. Chem. 2007, 282, 27503–27517. [Google Scholar] [CrossRef]
- Rhee, M.; Davis, P. Mechanism of uptake of C105Y, a novel cell-penetrating peptide. J. Biol. Chem. 2006, 281, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Oehlke, J.; Scheller, A.; Wiesner, B.; Krause, E.; Beyermann, M.; Klauschenz, E.; Melzig, M.; Bienert, M. Cellular uptake of an α-helical amphipathic model peptide with the potential to deliver polar compounds into the cell interior non-endocytically. Biochim. Biophys. Acta 1998, 1414, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.R.; Huang, Y.W.; Winiarz, J.G.; Chiang, H.J.; Lee, H.J. Intracellular delivery of quantum dots mediated by a histidine- and arginine-rich HR9 cell-penetrating peptide through the direct membrane translocation mechanism. Biomaterials 2011, 32, 3520–3537. [Google Scholar] [CrossRef]
- Chen, C.P.; Chou, J.C.; Liu, B.R.; Chang, M.; Lee, H.J. Transfection and expression of plasmid DNA in plant cells by an arginine-rich intracellular delivery peptide without protoplast preparation. FEBS Lett. 2007, 581, 1891–1897. [Google Scholar] [CrossRef]
- Liu, B.R.; Huang, Y.W.; Aronstam, R.S.; Lee, H.J. Comparative mechanisms of protein transduction mediated by cell-penetrating peptides in prokaryotes. J. Membr. Biol. 2015, 248, 355–368. [Google Scholar] [CrossRef]
- Liu, B.R.; Lo, S.Y.; Liu, C.C.; Chyan, C.L.; Huang, Y.W.; Aronstam, R.S.; Lee, H.J. Endocytic trafficking of nanoparticles delivered by cell-penetrating peptides comprised of nona-arginine and a penetration accelerating sequence. PLoS ONE 2013, 8, e67100. [Google Scholar] [CrossRef]
- Suresh, A.; Kim, Y.C. Translocation of cell penetrating peptides on Chlamydomonas reinhardtii. Biotechnol. Bioeng. 2013, 110, 2795–2801. [Google Scholar] [CrossRef]
- Simeoni, F.; Morris, M.C.; Heitz, F.; Divita, G. Insight into the mechanism of the peptide-based gene delivery system MPG: Implications for delivery of siRNA into mammalian cells. Nucleic Acids Res. 2003, 31, 2717–2724. [Google Scholar] [CrossRef]
- Wyman, T.B.; Nicol, F.; Zelphati, O.; Scaria, P.V.; Plank, C.; Szoka, F.C. Design, synthesis, and characterization of a cationic peptide that binds to nucleic acids and permeabilizes bilayers. Biochemistry 1997, 36, 3008–3017. [Google Scholar] [CrossRef] [PubMed]
- Henriques, S.T.; Costa, J.; Castanho, M.A.R.B. Translocation of β-galactosidase mediated by the cell-penetrating peptide pep-1 into lipid vesicles and human HeLa cells is driven by membrane electrostatic potential. Biochemistry 2005, 44, 10189–10198. [Google Scholar] [CrossRef] [PubMed]
- Henriques, S.T.; Quintas, A.; Bagatolli, L.A.; Homblé, F.; Castanho, M.A.R.B. Energy-independent translocation of cell-penetrating peptides occurs without formation of pores. A biophysical study with pep-1. Mol. Membr. Biol. 2007, 24, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Ragin, A.D.; Morgan, R.A.; Chmielewski, J. Cellular import mediated by nuclear localization signal peptide sequences. Chem. Biol. 2002, 9, 43–948. [Google Scholar] [CrossRef]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef]
- Shen, B.R.; Zhu, C.H.; Yao, Z.; Cui, L.L.; Zhang, J.J.; Yang, C.W.; He, Z.H.; Peng, X.X. An optimized transit peptide for effective targeting of diverse foreign proteins into chloroplasts in rice. Sci. Rep. 2017, 7, 46231. [Google Scholar] [CrossRef]
- Yoshizumi, T.; Oikawa, K.; Chuah, J.A.; Kodama, Y.; Numata, K. Selective gene delivery for integrating exogenous DNA into plastid and mitochondrial genomes using peptide-DNA complexes. Biomacromolecules 2018, 19, 1582–1591. [Google Scholar] [CrossRef]
- Oikawa, K.; Tateishi, A.; Odahara, M.; Kodama, Y.; Numata, K. Imaging of the entry pathway of a cell-penetrating peptide-DNA complex from the extracellular space to chloroplast nucleoids across multiple membranes in Arabidopsis leaves. Front. Plant Sci. 2021, 12, 759871. [Google Scholar] [CrossRef]
- Eggenberger, K.; Mink, C.; Wadhwani, P.; Ulrich, A.S.; Nick, P. Using the peptide BP100 as a cell-penetrating tool for the chemical engineering of actin filaments within living plant cells. Chembiochem 2011, 12, 132–137. [Google Scholar] [CrossRef]
- Chuah, J.A.; Yoshizumi, T.; Kodama, Y.; Numata, K. Gene introduction into the mitochondria of Arabidopsis thaliana via peptide-based carriers. Sci. Rep. 2015, 5, 7751. [Google Scholar] [CrossRef]
- Lakshmanan, M.; Kodama, Y.; Yoshizumi, T.; Sudesh, K.; Numata, K. Rapid and efficient gene delivery into plant cells using designed peptide carriers. Biomacromolecules 2013, 14, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Terada, K.; Gimenez-Dejoz, J.; Kurita, T.; Oikawa, K.; Uji, H.; Tsuchiya, K.; Numata, K. Synthetic mitochondria-targeting peptides incorporating α-aminoisobutyric acid with a stable amphiphilic helix conformation in plant cells. ACS Biomater. Sci. Eng. 2021, 7, 1475–1484. [Google Scholar] [CrossRef] [PubMed]
- Green, M.; Loewenstein, P.M. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell 1988, 55, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Frankle, A.D.; Pabo, C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, X.; Wang, F. Intracellular transduction and potential of Tat PTD and its analogs: From basic drug delivery mechanism to application. Expert Opin. Drug Deliv. 2012, 9, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Zahid, M.; Robbins, P.D. Cell-type specific penetrating peptides: Therapeutic promises and challenges. Molecules 2015, 20, 13055–13070. [Google Scholar] [CrossRef]
- Xie, J.; Bi, Y.; Zhang, H.; Dong, S.; Teng, L.; Lee, R.J.; Yang, Z. Cell-penetrating peptides in diagnosis and treatment of human diseases: From preclinical research to clinical application. Front. Pharmacol. 2020, 11, 697. [Google Scholar] [CrossRef]
- Pouny, Y.; Rapaport, D.; Mor, A.; Nicolas, P.; Shai, Y. Interaction of antimicrobial dermaseptin and its fluorescently labeled analogues with phospholipid membranes. Biochemistry 1992, 31, 12416–12423. [Google Scholar] [CrossRef]
- Gautam, A.; Chaudhary, K.; Kumar, R.; Sharma, A.; Kapoor, P.; Tyagi, A.; Open Source Drug Discovery Consortium; Raghava, G.P.S. In silico approaches for designing highly effective cell penetrating peptides. J. Transl. Med. 2013, 11, 74. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Yoneyama, S.; Murase, O.; Miyajima, K. Transbilayer transport of ions and lipids coupled with mastoparan X translocation. Biochemistry 1996, 35, 8450–8456. [Google Scholar] [CrossRef]
- Chang, M.; Chou, J.C.; Chen, C.P.; Liu, B.R.; Lee, H.J. Noncovalent protein transduction in plant cells by macropinocytosis. New Phytol. 2007, 174, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Chen, C.W.; Buj, R.; Tangudu, N.K.; Fang, R.S.; Leon, K.E.; Dahl, E.S.; Varner, E.L.; von Krusenstiern, E.; Cole, A.R.; et al. ATM inhibition drives metabolic adaptation via induction of macropinocytosis. J. Cell Biol. 2023, 222, e202007026. [Google Scholar] [CrossRef] [PubMed]
- Derakhshankhah, H.; Jafari, S. Cell penetrating peptides: A concise review with emphasis on biomedical applications. Biomed. Pharmacother. 2018, 108, 1090–1096. [Google Scholar] [CrossRef]
- Deshayes, S.; Morris, M.C.; Divita, G.; Heitz, F. Interactions of amphipathic CPPs with model membranes. Biochim. Biophys. Acta 2006, 1758, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, W.B.; Guha, S.; Wimley, W.C. Synthetic molecular evolution of hybrid cell penetrating peptides. Nat. Commun. 2018, 9, 2568. [Google Scholar] [CrossRef] [PubMed]
- Chugh, A.; Eudes, F. Study of uptake of cell penetrating peptides and their cargoes in permeabilized wheat immature embryos. FEBS J. 2008, 275, 2403–2414. [Google Scholar] [CrossRef]
- Holl, N.J.; Dey, M.; Huang, Y.W.; Chiou, S.H.; Lee, H.J. Lactoferricin-derived L5a cell-penetrating peptide for delivery of DNA into cells. Methods Mol. Biol. 2021, 2211, 113–121. [Google Scholar]
- Thagun, C.; Chuah, J.A.; Numata, K. Targeted gene delivery into various plastids mediated by clustered cell-penetrating and chloroplast-targeting peptides. Adv. Sci. Weinh. 2019, 6, 1902064. [Google Scholar] [CrossRef]
- Thagun, C.; Motoda, Y.; Kigawa, T.; Kodama, Y.; Numata, K. Simultaneous introduction of multiple biomacromolecules into plant cells using a cell-penetrating peptide nanocarrier. Nanoscale 2020, 12, 18844–18856. [Google Scholar] [CrossRef]
- Thagun, C.; Horii, Y.; Mori, M.; Fujita, S.; Ohtani, M.; Tsuchiya, K.; Kodama, Y.; Odahara, M.; Numata, K. Non-transgenic gene modulation via spray delivery of nucleic acid/peptide complexes into plant nuclei and chloroplasts. ACS Nano. 2022, 16, 3506–3521. [Google Scholar] [CrossRef]
- Fagerlund, R.; Mélen, K.; Kinnunen, L.; Julkunen, I. Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and importin alpha 5. J. Biol. Chem. 2002, 277, 30072–30078. [Google Scholar] [CrossRef] [PubMed]
- Kurnaeva, M.A.; Zalevsky, A.O.; Arifulin, E.A.; Lisitsyna, O.M.; Tvorogova, A.V.; Shubina, M.Y.; Bourenkov, G.; Tikhomirova, M.A.; Potashnikova, D.M.; Kachalova, A.I.; et al. Molecular coevolution of nuclear and nucleolar localization signals inside the basic domain of HIV-1 Tat. J. Virol. 2022, 96, e0150521. [Google Scholar] [CrossRef] [PubMed]
- Kroth, P.G. Protein transport into secondary plastids and the evolution of primary and secondary plastids. Int. Rev. Cytol. 2002, 221, 191–255. [Google Scholar] [PubMed]
- Larkin, R.M. Influence of plastids on light signalling and development. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130232. [Google Scholar] [CrossRef]
- Maliga, P. Transplastomic technology for safer and better transgenic crops. Nat. Biotechnol. 1999, 17, 28. [Google Scholar] [CrossRef]
- Waheed, M.T.; Ismail, H.; Gottschamel, J.; Mirza, B.; Lössl, A.G. Plastids: The green frontiers for vaccine production. Front. Plant Sci. 2015, 6, 1005. [Google Scholar] [CrossRef]
- De Marchis, F.; Bellucci, M. Plastid transformation in sugar beet: An important industrial crop. Methods Mol. Biol. 2021, 2317, 283–290. [Google Scholar]
- Bhushan, S.; Kuhn, C.; Berglund, A.K.; Roth, C.; Glaser, E. The role of the N-terminal domain of chloroplast targeting peptides in organellar protein import and miss-sorting. FEBS Lett. 2006, 580, 3966–3972. [Google Scholar] [CrossRef]
- Scarpulla, R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008, 88, 611–638. [Google Scholar] [CrossRef]
- Omura, T. Mitochondria-targeting sequence, a multi-role sorting sequence recognized at all steps of protein import into mitochondria. J. Biochem. 1998, 123, 1010–1016. [Google Scholar] [CrossRef]
| CPP/Others | Primary Sequence | Categories | Target Cells/Tissues/Species | Entry Mechanisms | Cytotoxicity | References |
|---|---|---|---|---|---|---|
| L5a | RRWQW | Amphipathic | A549 cells | Direct membrane translocation | Up to 10 μM is not toxic | [53] |
| Tat PTD (48–60) | GRKKRRQRRRPPQ | Cationic | HeLa, HL116, CCL39 cells | Direct membrane translocation | Up to 100 μM is not toxic | [56] |
| HeLa, CHO cells | Direct membrane translocation (pore formation) | 1 μM, 3-day treatment without cytotoxicity | [57,58] | |||
| R9 | RRRRRRRRR * | Cationic | Jurkat, murine B, human PBL cells | Endocytosis | – | [59] |
| Plant tissues | Energy-independent pathway (link GFP with covalent manner) | Up to 2 μg is not toxic | [32] | |||
| pAntp | RQIKIWFQNRRMKWKK | Cationic | K562, HeLa cells | Direct membrane translocation (inverted micelle) | – | [60,61] |
| CHO cells | Endocytosis (macropinocytosis) | Up to 10 μM is not toxic | [62] | |||
| VP22 | DAATATRGRSAASRPTERPRAPARSASRPRRPVD | Amphipathic | CHO-K1, HeLa cells | Endocytosis (macropinocytosis) | – | [29,63] |
| C105Y | CSIPPEVKFNKPFVYLI | Hydrophobic | HuH7 cells | Energy-independent pathway (cell entry) | – | [55,64] |
| Energy-dependent pathway (nucleolar entry) | ||||||
| MAP | KLALKLALKALKAALKLA | Amphipathic | HeLa, endothelial cells | Energy-dependent pathway Energy-independent pathway | Up to 10 μM is not toxic – | [29,62,65] |
| HR9 | CHHHHHRRRRRRRRRHHHHHC | Cationic | A549, Sf9, plant cells, paramecia, rotifers, prokaryotes | Direct membrane translocation | Up to 60 μM is not toxic | [21,66] |
| SR9 | RRRRRRRRR | Cationic | A549 cells | – (Link protein with noncovalent manners) | Up to 16 μM is not toxic | [21] |
| A549 cells | Multiple energy-dependent pathways (Link nanoparticles with noncovalent manners) | Up to 60 μM is not toxic | [26] | |||
| Plant tissues | Macropinocytosis (link GFP or DNA with noncovalent manners) | Up to 16.6 μM is not toxic | [32,67] | |||
| Prokaryotes | Macropinocytosis (Link nanoparticles with noncovalent manners) | Up to 48 μM is not toxic | [68] | |||
| PR9 | FFLIPKGRRRRRRRRR | Cationic | A549 cells | Endocytosis (Link nanoparticles with noncovalent manners) | Up to 60 μM is not toxic | [69] |
| pVEC | LLIILRRRIRKQAHAHSK | Amphipathic | HeLa cells | Endocytosis | Up to 10 μM is not toxic | [62] |
| Green alga | Direct membrane translocation | No toxicity | [70] | |||
| MPG | GALFLGFLGAAGSTMGAWSQPKKKRKV | Amphipathic | HS-68, Cos-7, HeLa cells | Direct membrane translocation | – | [29,71] |
| KALA | WEAKLAKALAKALAKHLAKALAKALKACEA | Amphipathic | CV-1, Hep G2, C2C12, K562, CaCo2 cells | Endocytosis | Toxic at the concentration ≥ 25 μM | [29,72] |
| GALA | WEAALAEALAAEALAEHLAEALAEALEALAA | Amphipathic | CV-1, Hep G2, C2C12, K562, CaCo2 cells | Endocytosis | – | [29,72] |
| Pep-1 | KETWWETWWTEWSQPKKKRKV | Amphipathic | HeLa cells | Direct membrane translocation | – | [73,74] |
| NLS | CGYGPKKKRKVGG | Cationic (or amphipathic) | MCF-7, KB, HT29, MIAPACA2, PC3 cells | Energy-independent pathway | – | [75,76] |
| RC2 | MQVWPIEGIKKFETLSYLPPL | Chloroplast transit peptide (CTP; not a CPP) | Rice chloroplasts | – | – | [77] |
| KH9-AtOEP34 | KHKHKHKHKHKHKHKHKHMFAFQYLLVM | Cationic CPP combined with CTP | Seedlings and leaves of A. thaliana and Nicotiana tabacum | Endocytosis or direct membrane translocation (for cellular entry) Unknown (for plastid targeting) | – | [78,79] |
| BP100 | KKLFKKILKYL | Amphipathic | Leaves of A. thaliana, BY-2 cells | Endocytosis | – | [35,80,81] |
| MTP-KH9 | MLSLRQSIRFFKKHKHKHKHKHKHKHKHKH | Cationic CPP combined with MTS | Leaves of A. thaliana | Endocytosis (for cellular entry) Unknown (for mitochondrial targeting) | – | [81,82] |
| (LURL)3 | LURLLURLLURL | Amphipathic MTS | Onion bulbs | Gold nanoparticle biolistics (for cellular entry) Binding to mitochondrial import receptors Tom20 and Tom22 (for mitochondrial targeting) | Negligible toxicity | [83] |
| Delivery Methods | Genes | Targets | References |
|---|---|---|---|
| Non-CPP-based gene delivery | Proteinase inhibitor genes | Tobacco | [3] |
| Recombinant Bt toxic proteins | Vigna ungiguiculata | [4,5] | |
| α-Amylase inhibitors, plant lectins | Adzuki bean | [5] | |
| Antibiotic-resistant Ti plasmid (A. tumefaciens mediated transfection) | Tobacco | [7] | |
| Hygromycin resistance gene | Protoplasts of rice | [14] | |
| CPP-based gene delivery | p35S-RLuc-tNOS and p35S-GFP-tNOS plasmids | Leaves of A. thaliana | [34] |
| p35S-Nluc-tNOS or p35S-GFP-tNOS plasmid | Seedlings of A. thaliana | [38] | |
| pHBT-sGFP(S65T)-NOS plasmid | Roots of mung bean and soybean | [67] | |
| psbAp:GFP:SPECr:psbAt At plastid genome integration vector, cox2p: GFP:SPECr:cox2t At mitochondrial genome integration vector, and cox2t:SPECr:GFP:cox2p Nt mitochondrial genome integration vector | Seedlings and leaves of A. thaliana or N. tabacum | [78] | |
| PsbA-SPECr-sGFP-psbA, Prrn-aadA-sfGFP-Trps, PsbA-SPECr-sGFP-psbA, and Prrn-aadA-sfGFP-Trps | Leaves of A. thaliana | [79] | |
| pDONR-cox2:rluc and pDONR-cox2:gfp plasmids | Leaves of A. thaliana | [81] | |
| pAct-1GUS plasmid | Wheat immature embryos | [97] | |
| pPrrn::GFP(S65T)::TpsbA, pPrrn::DsRed::TpsbA, and pPpsbA::Rluc plasmids | Leaves of A. thaliana | [99] | |
| psfGN155-MxMT and psfGC155-MxMT plasmids | Leaves of N. benthamiana | [100] | |
| pBI221, pBI121, and pPpsbA::Rluc plasmids | Leaves of Arabidopsis, soybean, and tomato | [101] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.R.; Chen, C.-W.; Huang, Y.-W.; Lee, H.-J. Cell-Penetrating Peptides for Use in Development of Transgenic Plants. Molecules 2023, 28, 3367. https://doi.org/10.3390/molecules28083367
Liu BR, Chen C-W, Huang Y-W, Lee H-J. Cell-Penetrating Peptides for Use in Development of Transgenic Plants. Molecules. 2023; 28(8):3367. https://doi.org/10.3390/molecules28083367
Chicago/Turabian StyleLiu, Betty Revon, Chi-Wei Chen, Yue-Wern Huang, and Han-Jung Lee. 2023. "Cell-Penetrating Peptides for Use in Development of Transgenic Plants" Molecules 28, no. 8: 3367. https://doi.org/10.3390/molecules28083367
APA StyleLiu, B. R., Chen, C.-W., Huang, Y.-W., & Lee, H.-J. (2023). Cell-Penetrating Peptides for Use in Development of Transgenic Plants. Molecules, 28(8), 3367. https://doi.org/10.3390/molecules28083367