Abstract
Targeting L858R/T790M and L858R/T790M/C797S mutant EGFR is a critical challenge in developing EGFR tyrosine kinase inhibitors to overcome drug resistance in non-small cell lung cancer (NSCLC). The discovery of next-generation EGFR tyrosine kinase inhibitors (TKIs) is therefore necessary. To this end, a series of furopyridine derivatives were evaluated for their EGFR-based inhibition and antiproliferative activities using computational and biological approaches. We found that several compounds derived from virtual screening based on a molecular docking and solvated interaction energy (SIE) method showed the potential to suppress wild-type and mutant EGFR. The most promising PD13 displayed strong inhibitory activity against wild-type (IC50 of 11.64 ± 1.30 nM), L858R/T790M (IC50 of 10.51 ± 0.71 nM), which are more significant than known drugs. In addition, PD13 revealed a potent cytotoxic effect on A549 and H1975 cell lines with IC50 values of 18.09 ± 1.57 and 33.87 ± 0.86 µM, respectively. The 500-ns MD simulations indicated that PD13 formed a hydrogen bond with Met793 at the hinge region, thus creating excellent EGFR inhibitory activity. Moreover, the binding of PD13 in the hinge region of EGFR was the major determining factor in stabilizing the interactions via hydrogen bonds and van der Waals (vdW). Altogether, PD13 is a promising novel EGFR inhibitor that could be further clinically developed as fourth-generation EGFR-TKIs.
1. Introduction
EGFR is a transmembrane protein tyrosine kinase that plays an essential role in cellular signaling for cell proliferation, invasion, metastasis, apoptosis, and angiogenesis [1,2,3,4,5,6]. Structurally, EGFR is composed of three domains—a transmembrane domain, an extracellular domain, and an intracellular tyrosine kinase (TK) domain—which are constructed of four important conserved regions: (i) the activation loop (A-loop), (ii) the glycine-rich loop (G-loop), (iii) the catalytic loop (C-loop), and (iv) the hinge region [7,8]. Deregulation of EGFR is caused by increased EGFR activity such as overexpression and mutations of EGFR [9,10,11]. Increased EGFR activity is associated with numerous malignant tumors, including esophageal cancers, glioblastoma, anal cancers, epithelial cancers of the head and neck, breast cancer, and lung cancers [12,13,14,15,16], especially non-small cell lung cancer (NSCLC). Lung cancer is a major cause of cancer death globally, and NSCLC is the most frequently diagnosed cancer [16,17,18], whereas activating mutations in the tyrosine kinase (TK) domain are recognized as the tumorigenic driver in NSCLC. As a matter of fact, extensive research has been executed to study EGFR function as a therapeutic target for NSCLC.
Several EGFR inhibitors have been developed and reported (Figure 1). Erlotinib [19] and gefitinib [20], first-generation reversible EGFR inhibitors, proved to have a strong response and to lead to prolonged survival in NSCLC patients. However, after 9–14 months of treatment, the secondary “gatekeeper” T790M mutation increased ATP-binding affinity and caused recurrence in most NSCLC patients [21]. Second-generation irreversible EGFR inhibitors targeting EGFR with the T790M activating mutation, such as afatinib [22] and dacomitinib [23], were developed to overcome that problem. Although the irreversible covalent bond with C797 confers enhanced sensitivity and selectivity, serious side effects of these TKIs, such as skin rash and diarrhea due to their activity against wild-type EGFR [22,23,24,25], were observed. To combat drug resistance and limit dose-related toxic side effects, Osimertinib (AZD9291), which is a pyrimidine scaffold, was developed as an irreversible third-generation inhibitor [26]. Osimertinib has a high affinity for the drug-resistant L855R/T790M double mutant [27] but no activity against the wild-type EGFR [27,28,29,30]. Additionally, clinical studies revealed that approximately 40% of patients who received osimertinib therapy developed a tertiary mutation (C797S) in the EGFR, resulting in a loss of covalent interactions between the irreversible inhibitors and the side chain of C797 [31]. Therefore, the discovery of potent inhibitors of both wild-type and mutant forms of EGFR is required. Since NSCLC is one of oncology’s “big killers” in the patient population and the EGFR signaling is a main driver for this type of tumor, it is a worthwhile endeavor to develop improved therapeutics in the clinical setting.
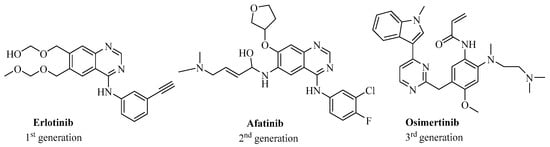
Figure 1.
Two-dimensional (2D) structure of three reference drugs targeting EGFR, including erlotinib, afatinib, and osimertinib.
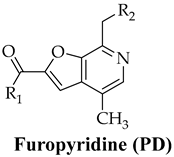
Furopyridine (PD, Table 1), a furo[2,3-c]pyridine compound, is a product of a structural modification of vitamin B6, which plays an extremely important biological role in living organisms, since it is involved in a wide range of biochemical reactions necessary for cellular metabolism and function. The latter explains why vitamin B6 and its derivatives are considered biologically privileged molecules with which to develop new therapeutics.

Table 1.
Chemical structure of furopyridine; PD and its derivatives.
Furthermore, PDs display a wide range of biological activities, including anticancer [32,33], antiviral [34], anti-inflammatory [35], and HIV protease inhibitory activities [36], and also inhibit α-glucosidase [37]. The products of the furan cyclization of pyridoxal under the action of hetarylamines 2-alkylamino-3-hetarylamino-4-hydroxymethylfuro[2,3-c]pyridines have an immunostimulating effect due to their selective activation of Toll-like TLR8 receptors [38]. Even though anticancer activity of PD has been reported, an understanding of the binding modes of action of PD and its derivatives against NSCLC remains elusive. Hung et al. [33] reported the antiproliferative activity of synthesized thienopyridine derivatives using the panel of NCI-60 cell lines with GI50 values in the low nanomolarity range, especially the melanoma cell line MDA-MD-435 (GI50—23 nM). Rahman et al. also evaluated the CDK2 inhibitory and antiproliferative activity of pyrazolopyridine, furopyridine, and pyridine derivatives. Furopyridine derivative (14) showed IC50 0.93 µM as a CDK2 inhibitor as well as significant inhibition on different human cancer cell lines (HCT-116, MCF-7, HepG2, and A549).
The main aim of this work was the synthesis and evaluation of novel classes of anti-EGFR agents in order to understand the mechanism of anticancer activity as well as their binding modes to the receptor. Herein, we have reported the synthesis of a number of novel furopyridine derivatives with pharmacologically relevant substituents and also used molecular docking and molecular dynamics (MD) simulations with free energy calculations to virtually screen 14 in-house PD compounds towards wild-type, L858R/T790M, and L858R/T790M/C797S EGFR. Moreover, in vitro EGFR inhibitory and anti-cancer activities against NSCLC cell lines in comparison with normal cell lines were studied. Subsequently, 500-ns MD simulations were carried out to investigate the binding behavior of the most promising PD against wild-type and mutant forms of EGFR. The most promising PD from this work could be further developed as a novel anticancer drug by targeting both wild-type and mutant forms of EGFR.
2. Results
2.1. Virtual Screening
To assess the potency of EGFR inhibitor(s) against wild-type, L858R/T790M, and L858R/T790M/C797S EGFR, molecular docking was carried out on 14 PD compounds. Three drugs—erlotinib, afatinib, and Osimertinib—were selected as a positive control for wild-type, L858R/T790M, and L858R/T790M/C797S EGFR, respectively. The GOLD docking results were plotted in Figure 2. We found that nine PD compounds, including PD3, PD4, PD6, PD8, and PD10-PD13, showed a higher fitness score than erlotinib. Moreover, those PDs were found to inhibit L858R/T790M double mutant EGFR. Interestingly, a series of four PDs (PD4, PD8, PD13, and PD14) exhibited a strong affinity with L858R/T790M/C797S, which is better than osimertinib. The first-round screening suggested that several PD derivatives could be effective in inhibiting both wild-type and mutant forms of EGFR.
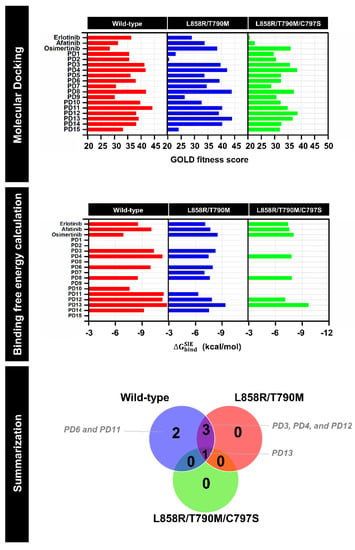
Figure 2.
Docking results (top), estimated binding free energy calculation based on SIE method (middle), and summarization of virtual screening (bottom) of furopyridine (PD) derivatives towards wild-type, L858R/T790M, L858R/T790M/C797S EGFR relative to three reference drugs (erlotinib, afatinib, and osimertinib).
To verify the first-round screening, the screened PDs/EGFRs complexes derived from molecular docking were subsequently studied on 100 snapshots extracted from the last 10 ns of 100-ns molecular dynamics (MD) simulations. The predicted binding affinity based on the solvated interaction energy (SIE) method of screened PD compounds and three positive controls (erlotinib, afatinib, and osimertinib) is shown in Figure 2. PD6 (ΔGbind of −9.96 kcal/mol) and PD11 (−11.30 kcal/mol) displayed greater binding affinity than erlotinib (−8.50 kcal/mol) in the wild-type EGFR system, suggesting that both PD6 and PD11 were specific for the wild-type EGFR. Moreover, the three PDs (PD3, PD4, and PD12) exhibited the most efficient binding to both wild-type and L858R/T790M EGFR, whereas that against L858R/T790M/C797S EGFR showed poor binding affinities. Surprisingly, the most promising, PD13, could be able to suppress both wild-type and mutant EGFR. The ΔGbind calculations of PD13 based on the SIE method gave ΔGbind of wild-type, L858R/T790M, and L858R/T790M/C797S EGFR as follows: −11.81, −9.39 and −9.7 kcal/mol. Hence, the screened compounds with estimated binding free energies based on the SIE method that were higher than those of the positive controls from each strain of EGFR were synthesized and chosen for in vitro evaluation.
2.2. Chemistry
All the target compounds were synthesized as outlined in Scheme 1.
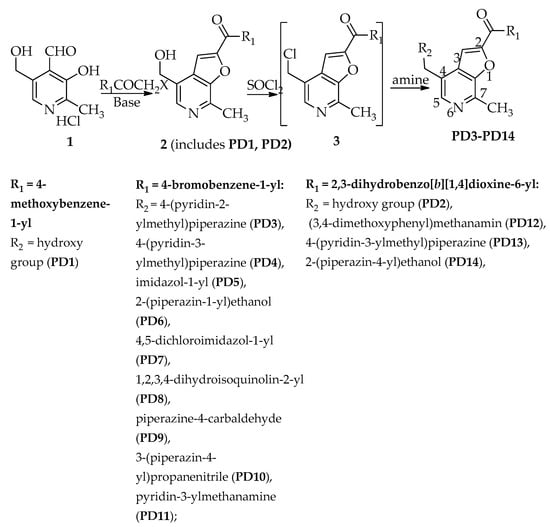
Scheme 1.
Synthesis of target compounds.
The cyclization of pyridoxal hydrochloride 1 in aqueous acetonitrile at 50–60 °C used potassium carbonate as a catalyst soft base, and proceeded fairly quickly in all cases—within 2–3 h—and led to the synthesis of 4-hydroxymethyl-7-methylfuro[2,3-c]pyridines (PD1, PD2). The yields of compounds PD1, PD2 were 55% and 79%, respectively.
The replacement of a hydroxyl group by a chlorine atom in compounds PD1, PD2, by the action of thionyl chloride in DMF furnished the target compounds 2 [39]. Further, the reaction of the resulting chloromethyl derivatives 3 with alkyl-and dialkylamines resulted in the formation of the corresponding 4-aminomethylfuro[2,3-c]pyridines PD3-PD14. It was more convenient to carry out the reactions in EtOH (for PD3-PD6, PD9-PD11, 25 °C—reflux, 3–6 h) or DMF (for PD7, PD8, PD12, PD13, 30–40 °C, 3–5 h).
The R1 and R2 substituents were chosen considering that the biological activity of the compounds, as well as their pharmacodynamics and pharmacokinetics, are significantly affected by both electronic factors (electron donation or electron acceptor of substituents) and their physical properties (lipophilicity, hydrophilicity, polarizability, etc.)
The structure of the obtained furo[2,3-c]pyridines was confirmed by 1H NMR spectroscopy. 1H NMR data for compounds PD1, PD2 can be illustrated by the example of 4-hydroxymethylfuro[2,3-c]pyridines PD1. The 1H NMR of this ketone showed the proton signals of H-3 and H-5 of the bicyclic system (singlets at 7.79 and 8.24 ppm, respectively), the benzoyl group (two-proton, somewhat distorted doublet at 8.10 ppm (H-2′, H-6′, J 8.7 Hz) as well as multiplate at 7.08–7.14 (2H, H-3′, H-5′)), the hydroxymethyl group (two-proton and single-proton singlets at 4.79 and 5.24 ppm, respectively), and the methyl group (singlet at 2.76 ppm) (see Supplementary Files, 1H-NMR spectra) (SI).
In the spectra of amino derivatives PD3-PD14, there was no signal of the CH2OH group, while in the hydrochlorides of these compounds (PD3-PD5, PD9, PD11-PD14), a broadened singlet of the NH+ group appeared in the region of 5.01–5.71 ppm. The aromatic proton signals of these compounds (PD3-PD14) correspond to the proposed structures and differ from the signals of 4-hydroxymethylfuro[2,3-c]pyridine (PD1) by 01–03 ppm (see Supplementary Files, 1H-NMR spectra). The 13C-NMR of all these compounds was in agreement with their structure (see Supplementary Materials, 13C-NMR).
2.3. Kinase Inhibitory Activities of EGFR
Potent compounds obtained from in silico screening were selected to elucidate their inhibitory EGFR activity against different types of EGFR (wild-type, L858R/T790M and L858R/T790M/C797S). Note that erlotinib, afatinib, and osimertinib, which are FDA-approved drugs targeting EGFR, were used as the positive control. The selected compounds and drugs were first screened to observe the sign of EGFR using a single 1 µM dose concentration. The PD compounds that present a percentage value of EGFRs inhibition <50 were selected to calculate the IC50 values. The relative inhibition of EGFR and IC50 curves were analyzed and are plotted in Figure 3, and the IC50 values of focused PD and drugs are summarized in Table 2. It was found that PD13 was active in wild-type and L858R/T790M forms of EGFR, whereas PD4 was only against wild-type EGFR.
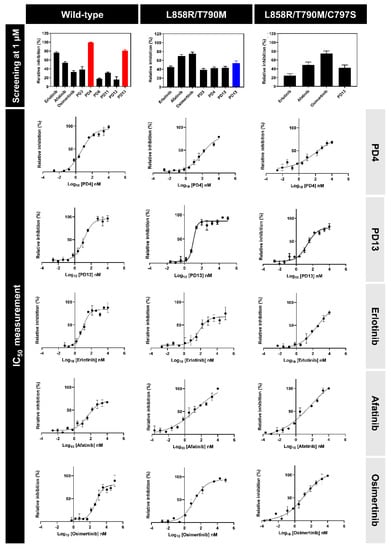
Figure 3.
Three forms of EGFR (wild-type, L858R/T790M, and L858R/T790M/C797S) inhibitory screening of PD derivatives at 1 µM concentration (top) and IC50 curves of compounds that present more than 50% PD4 and PD13 in comparison with reference drugs (bottom).

Table 2.
EGFR inhibitory activities of focused PD and drugs in different forms of EGFR.
The IC50 values of PD4 and PD13 were determined in comparison with reference drugs (Table 2). The IC50 values of two PD compounds and drugs inhibiting three strains of EGFR were in the nanomolar range. PD4/wild-type (IC50 of 4.78 ± 0.73 nM) displayed EGFR inhibitory activity >2-fold compared to erlotinib (of 14.11 ± 0.19 nM). On the other hand, PD4 complexed with L858R/T790M (91.02 ± 2.01 nM) and L858R/T790M/C797S EGFR (103.70 ± 3.62 nM) gave the IC50 value of >100 nM, suggesting that PD4 is specific against wild-type EGFR. PD13 showed the lowest IC50 values for both wild-type (11.64 ± 1.30 nM) and L858R/T790M (10.51 ± 0.71 nM). While PD13 inhibited L858R/T790M/C797S EGFR with IC50 values in the double-digit nanomolar range, it had lower potencies than third-generation EGFR inhibitor osimertinib. Furthermore, the PD13/triple mutant EGFR appeared to be a more promising candidate compound than previously reported L858R/T790M/C797S EGFR inhibitors such as 2,9-disubstituted-8-phenylthio/phenyl-sulfinyl-9H-purines (IC50 of 114 nM) [40], quinoline derivative (IC50 of 113 nM) [41], or 5-methylpyrimidopyridone derivative (IC50 of 27.5 nM) [42].
2.4. Antiproliferative Activities on NSCLC and Normal Cell Lines
The antiproliferative activity of two focused PD compounds and drugs were evaluated against NSCLC cell lines, namely the wild-type NSCLC cell line A549 as well as L858R/T790M mutant NSCLC cell line H1975. Erlotinib, afatinib, and osimertinib were used as positive controls. The results are summarized in Table 3. We found that both PD4 and PD13 exhibited strong anti-cancer activity by inhibiting the proliferation of A549 rather than H1975 cell lines. In addition, PD13 exhibited stronger antiproliferative activities than afatinib and osimertinib against the A549 cell line. Moreover, PD13 showed cell growth inhibition in H1975 cell cultures at a similar level to afatinib.

Table 3.
In vitro anticancer activity of focused PD and drugs against NSCLC and normal cell lines.
Furthermore, PD4, PD13, and positive controls were then investigated for their cytotoxicity to the normal cell line Vero (monkey kidney epithelial cells). We found that the IC50 value of PD13 was >100 µM in Vero cells, whereas for PD4 IC50 was <100 µM. These results suggested that PD13 showed low cytotoxicity against the normal cell. Therefore, PD13 was chosen in order to study its binding affinity by MD simulations.
2.5. System Stability
The stability of PD13 and the reference drugs erlotinib, afatinib, and osimertinib was determined using all-atom root mean square displacement (RMSD) to observe the system stability. The RMSD values of each system were obtained from three independent simulations. As shown in Figure 4, the RMSD values of the erlotinib system dramatically increased in the first 100 ns and were maintained at the fluctuation of ~2.0–3.0 Å until the end of the simulation for all independent runs. The wild-type/PD13 complexes reached three runs after 300 ns. The RMSD values of afatinib complexed with L858R/T790M EGFR fluctuated ~1.5–3.0 Å and were maintained at the fluctuation of ~2.0–3.0 Å until the end of the simulation. In the case of PD13/double mutant, EGFR showed similar patterns to the three independent simulations. The RMSD value of osimertinib continuously increased in the first 100 ns and was stably maintained at a fluctuation of 3.0–4.0 Å. L858R/T790M/C797S complexed with PD13 appeared to be quite stable after 400 ns for all simulated systems, while the RMSD value of PD13 and known drugs complexed with EGFRs of all systems tended to be stable after 200 ns. Altogether, the MD trajectories from 400 to 500 ns of each simulation were extracted for further analysis in terms of: (i) key binding residues; (ii) protein-ligand hydrogen bonding; and (iii) binding affinity.
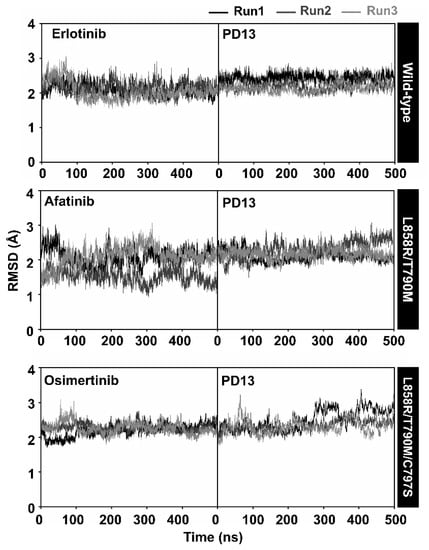
Figure 4.
All-atom RMSD of EGFRs with PD13 and drugs during 500 ns of simulation from three independent simulations.
2.6. Key Binding Residues
To explore the binding affinity of PD13 and reference drugs, the per-residue free energy decomposition () based on the MM/PBSA method was calculated on 100 snapshots extracted from the last 100 ns of all simulations. Note that, among the 695–1018 residues of EGFR, only the energy stabilization of ≤−1.0 kcal/mol for the 700–900 residues of all systems was plotted in Figure 5. The compounds were shaded based on their highest and lowest energies, with white representing the highest energies and blue representing the lowest energies.
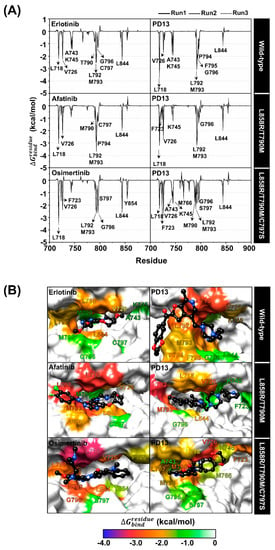
Figure 5.
(A) Per-residue decomposition free energy of EGFRs for the binding of PD13 and drugs from three independent simulations (top). (B) The binding orientation of PD13 and drugs within the binding pocket of EGFR was drawn from the last snapshot. The lowest and highest energies are shaded from blue to white (bottom).
The wild-type/erlotinib showed important residue for binding ten residues including L718, V726, A743, K745, T790, L792, M793, G796, C797, and L844. Interestingly, the PD13/wild-type complex displayed a similar binding pattern to that of reported EGFR inhibitors and was stabilized by L718 (−3.74 kcal/mol, dark red) at the hinge region, which is in good agreement with a previous study that identified L718, A743, L792, M793, G796, and L844 as key residues involved in the main binding of EGFR inhibitors [43,44,45]. In addition, the residue M793 of L858R/T790M EGFR exhibited the highest stabilization for PD13 (−3.43 kcal/mol, dark red) compared to the afatinib (−3.21 kcal/mol, red) system. This may be one of the reasons that explains why PD13 could inhibit L858R/T790M EGFR better than afatinib. The binding of PD13 with L858R/T790M/C797S EGFR displayed a higher number of amino acid residues than Osimertinib; L718, V726, A743, K745, M766, L788, M790, L792, M793, G796, S797, and L844 are all involved in ligand stabilization. Among all the residues, the moderate L858R/T790M/C797S EGFR inhibitory potency of PD13 may be due to steric clashing of M790 with the phenyl ring of the inhibitor.
2.7. Protein-Ligand Hydrogen Bonding
The hydrogen bond is one of the main factors for determining the binding strength of the protein–ligand complex. We further calculated the hydrogen bond occupations between EGFRs with PD13 and reference drugs using the two following criteria: (i) distance between hydrogen donor (HD) and acceptor (HA) ≤ 3.5 Å; and (ii) the angle between HD and HA being over 120° (HD−H⋯HA ≥ 120°). The percentages of hydrogen bond occupations for the studied ligands were observed and are compared in Figure 6.
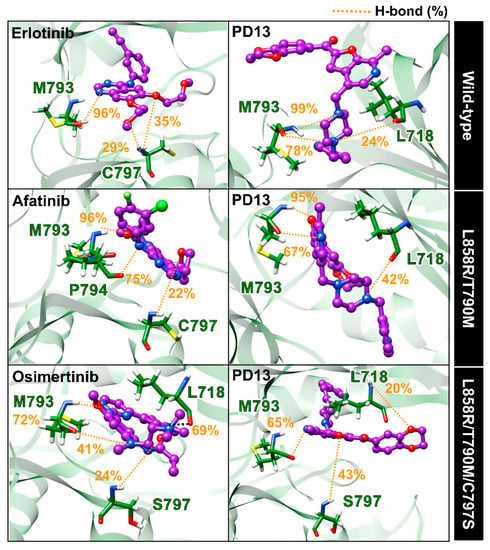
Figure 6.
The three-dimensional representative structures showing a percentage of the H-bond occupations of PD13 and drugs with EGFRs, drawn from the last snapshot of run1. The yellow dashed lines indicate H-bond formation.
The hydrogen bond formations of wild-type/PD13 indicated strong stabilization with the M793 (78–98%) and L718 (24%), and are similar to that found in wild-type/erlotinib complex. In the case of L858R/T790M double EGFR, this compound formed a hydrogen bond with M793 (67–95%) and L178 (42%), in good agreement with the per-residue decomposition analysis mentioned above. PD13 complexed with triple mutant EGFR showed similar H-bond formations to those of other forms of EGFR (except S797). We speculate that S797 residue that is mutated EGFR could lead to moderate inhibitory activity of L858R/T790M/C797S EGFR due to more favorable interactions with S797 via a hydrogen bond. Therefore, the inhibitory activity of PD13/EGFRs was induced by the two key residues, which are M793 and S797, at the hinge region. The main EGFR residues (L718, M793, and S797) evidenced crucial binding via hydrogen bonds, which is similar to other reported EGFR tyrosine kinases such as tupichinol E [46], quinazoline [47], and 9-heterocyclyl substituted 9H-purine derivatives [48].
2.8. Binding Affinity
The binding efficiency of PD13 and reference drugs was calculated using the MM/PBSA approach on 100 snapshots extracted from the last 100 ns of three independent simulations. The values of the averaged ∆Gbind over the three independent simulations are listed in Table 4 in comparison with the half-maximal inhibitory concentration (IC50).

Table 4.
The average ΔGbind and its energy component (kcal/mol) of PD13 and drugs in complexes with EGFRs calculated with the MM/PBSA method, compared to the half-maximal inhibitory concentration (IC50) values. Data are shown as mean ± standard deviation (SD) of three independent simulations.
In the gas term, we found that vdW interactions were three- to fourfold higher than electrostatic interactions in all studied ligands. Therefore, vdW interactions play an important role in the recognition of PD13 and reference drugs. These interaction findings are strongly consistent with previous studies on several small-molecule therapeutic kinase inhibitors such as olmutinib, lapatinib, and icotinib [49,50,51]. From the ∆Gbind calculation, the predicted binding values produced similar phenomena to that of experimental ∆Gbind values converted from the IC50 values, indicating that the MM/PBSA approach produced reliable ∆Gbind results. It was found that PD13 complexed with the wild-type and L858R/T790M system had a better binding affinity than erlotinib or afatinib, respectively. While PD13 complexed with L858R/T790M/C797S displayed lower binding affinity than osimertinib, the results agree well with in vitro L858R/T790M/C797S EGFR inhibitory activities, showing that this compound could inhibit wild-type and L858R/T790M better than L858R/T790M/C797S EGFR.
3. Materials and Methods
3.1. General Information
NMR 1H (300 MHz) and 13C (125 MHz) spectra of newly synthesized compounds were recorded on a spectrometer Bruker AC-300 (300 MHz) and BRUKER AVANCE NEO 500 MHz FT spectrometers in DMSO-d6. Chemical shifts of nuclei 1H were measured relative to the residual signals of deuterosolvent (δ = 2.50 ppm). Coupling constants (J) are reported in Hz. Melting points were determined by using Fisher-Johns Melting Point Apparatus (Fisher Scientific) and are uncorrected. Elemental analysis was performed using the classical method of microanalysis. The reaction and purity of the obtained compounds were monitored by TLC (plates with Al2O3 III activity grade, eluent CHCl3, development of TLC plates by exposition to iodine vapors in “iodine chamber”).
3.2. Chemistry
The reaction conditions and yields of compounds PD3–PD14 are presented in Table 5.

Table 5.
Reaction conditions and yields of compounds PD3–PD14.
Compound PD2 was synthesized as described previously [39].
[4-Hydroxymethyl-7-methylfuro[2,3-c]pyridin-2-yl](4-methoxyphenyl)methanone (PD1).
The mixture of pyridoxal hydrochloride 1 (1.02 g, 0.05 mol) and 2-bromo-1-(4-methoxyphenyl)ethan-1-one (1.0 g, 0.05 mol) with a saturated aqueous solution of K2CO3 (10 mL), CH3CN (10 mL) and 0.05 g TEBA was stirred vigorously at 20–25 °C for 1 h and at 50–60 °C for 3 h. The mixture was cooled, water (50 mL) was added, and the formed precipitate was filtered off and washed with water (15 mL). Yield: 0.76 g (57%). Light beige crystals, m.p. 147–148 °C (CH3CN). 1H NMR (DMSO-d6: δ ppm 2.76 (s, 3H, Me), 3.93 (s, 3H, OCH3), 4.79 (s, 2H, CH2), 5.24 (br. s, 1H, OH), 7.08–7.14 (m, 2H, H-3′, H-5′), 7.79 (s, 1H, H-3), 8.10 (d, 2H, J 8.7, H-2′, H-6′), 8.24 (s, 1H, H-5)). 13C NMR (DMSO-d6) δ ppm: 18.322, 55.607, 58.928, 113.455, 114.154, 128.664, 129.946, 130.748, 131.870, 139.910, 142.353, 150.118, 152.76, 163.576, 181.882 (DMSO-d6: 38.795–40.045). Anal.Calc. for C17H15NO4; (%). C, 68.68; H, 5.09; N, 4.71%. Found: C, 68.53; H, 5.00; N, 4.67%.
General Procedure for the Synthesis of 7-Methylfuro[2,3-c]pyridine Derivatives PD3–PD14
A mixture of corresponding 4-chloromethyl derivative 3 (3 mmol) [41], saturated K2CO3 solution (3 mL) and corresponding amine (3 mmol) in solvent was stirred. Conditions for the reaction are given in Table 1. Then, 50 mL of water was added, and the formed precipitate was filtered off and washed with water (15 mL). To convert the amines PD3–PD5, PD9, and PD11–PD14 into their hydrochlorides, they were treated with a saturated solution of HCl in isopropanol until a weakly acidic reaction occurred (pH~3).
(4-Bromophenyl){7-methyl-4-[(4-(pyridin-2-ylmethyl)piperazin-1-yl]methyl}furo[2,3-c]pyridin-2-yl)methanone dihydrochloride (PD3).
Yield: 1.08 g (62%). Light beige crystals, m.p. 224–225 °C (EtOH). 1H NMR (DMSO-d6): δ ppm 2.98 (s, 3H, CH3), 3.48 (s, 4H, H-3′, H-5′), 3.60 (s, 4H, H-2′, H-6′), 4.45 (s, 2H, CH2), 4.84 (s, 2H, CH2), 6.54 (br. s, 2H, 2NH+), 7.59–7.65 (m, 1H, H-3), 7.79–7.82 (m, 2H, H-3‴, H-5‴), 7.91 (t, J 7.8, 1H, H-4‴), 8.11–8.15 (m, 3H, H-2″, H-6″, H-6‴), 8.60 (s, 1H, H-3″), 8.70–7.72 (m, H-5″), 8.85 (s, 1H, H-5). 13C NMR (DMSO-d6) δ ppm: 15.936, 47.989, 52.125, 57.351, 115.179, 125.002, 126.285, 128.292, 131.703, 132.094, 134.511, 137.749, 140.949, 143.629, 146.405, 149.727, 150.445, 155.023, 182.261 (DMSO-d6: 38.872–40.128) Anal.Calc. for C26H27BrCl2N4O2;(%): C, 54.00; H, 4.71; Br, 13.82; Cl, 12.26; N, 9.69%. Found: C, 54.11; H, 4.62; Br+Cl, 26.21; N, 9.57%.
(4-Bromophenyl){7-methyl-4-[(4-(pyridin-3-ylmethyl)piperazin-1-yl]methyl}furo[2,3-c]pyridin-2-yl)methanone dihydrochloride (PD4).
Yeild: 1.15 g (66%). Light beige crystals, m.p. 243–247 °C (EtOH). 1H NMR (DMSO-d6): δ ppm 2.96 (s, 3H, CH3), 3.47–4.54 (m, 8H, H-2′, H-3′, H-5′, H-6′), 4.50 (s, 2H, CH2), 4.77 (s, 2H, CH2), 5.01 (br. s, 2H, 2NH+), 7.09 (s, 1H, H-3), 7.81 (d, J 8.3, 2H, H-2″, H-6″), 7.94 (s, 1H, H-5‴), 8.13 (d, J 8.6, 2H, H-3″, H-5″), 8.53 (s, 1H, H-6‴), 8.76–8.86 (m, 2H, H-5, H-2‴), 9.13 (s, 1H, H-4‴).). 13C NMR (75 MHz, DMSO-d6) δ 183.53 (C=O), 154.28 (C-O), 149.52, 149.14, 148.43, 144.52, 143.19, 135.46, 135.30, 132.46, 131.94 (2C), 131.70 (2C), 131.41, 126.45, 124.62, 123.68, 108.85, 61.71, 58.29, 52.21 (2C), 49.11 (2C), 18.49 (CH3). Anal.Calc for C26H27BrCl2N4O2;(%).: C, 54.00; H, 4.71; Br, 13.82; Cl, 12.26; N, 9.69%. Found: C, 54.06; H, 4.64; Br+Cl, 26.11; N, 9.57%.
{4-[(1H-imidazol-1-yl)methyl]-7-methylfuro[2,3-c]pyridin-2-yl}(4-bromophenyl)methanone hydrochloride (PD5).
Yield: 0.61 g (47%). Light beige crystals, m.p. 278–280 °C (EtOH). 1H NMR (DMSO-d6): δ ppm 2.87 (s, 3H, CH3), 5.89 (s, 2H, CH2), 7.10 (s, 1H, H-3), 7.60 (s, 1H, H-4′), 7.82 (d, J 8.4, 2H, H-3″, H-5″), 7.89 (s, 1H, NH+), 8.04 (d, J 8.5, 2H, H-2″, H-6″), 8.39 (s, 1H, H-5′), 8.72 (s, 1H, H-2′), 9.60 (s, 1H, H-5). 13C NMR (DMSO-d6) δ ppm: 16.267, 46.364, 113.426, 120.075, 120.736, 121.986, 124.275, 128.174, 131.565, 131.976, 134.457, 135.624, 138.606, 143.633, 149.820, 155.206, 182.194 (DMSO-d6: 38.831–40.082 Anal.Calc for C19H15BrClN3O2; (%).: C, 52.74; H, 3.49; Br, 18.47; Cl, 8.19; N, 9.71%. Found: C, 52.81; H, 3.29; Br+Cl, 26.80; N, 9.64%.
(4-Bromophenyl)(4-{[4-(2-hydroxyethyl)piperazin-1-yl]methyl}-7-methylfuro[2,3-c]pyridin-2-yl)methanone (PD6).
Yield: 1.00 g (73%). Light beige crystals, m.p. 105–107 °C (EtOH). 1H NMR (DMSO-d6): δ ppm 2.54 (s, 8H, H-2′, H-3′, H-5′, H-6′), 2.75 (s, 6H, CH3, CH2, OH), 3.54 (s, 2H, CH2), 3.79 (s, 2H, CH2), 7.77–7.82 (m, 2H, H-3″, H-5″), 7.87 (s, 1H, H-3), 7.98–8.03 (m, 2H, H-2″, H-6″), 8.23 (s, 1H, H-5). 13C NMR (DMSO-d6) δ ppm: 16.667, 47.502, 52.632, 115.417, 128.247, 131.716, 132.107, 134.697, 136.832, 141.641, 144.212, 149.804, 154.369, 161.108, 182.485 (DMSO-d6: 38.872–40.122). Anal.Calc for C22H24BrN3O3;(%).: C, 57.65; H, 5.28; Br, 17.43; N, 9.17%. Found: C, 57.58; H, 5.11; Br, 17.80; N, 9.04%.
(4-Bromophenyl){4-[(4,5-dichloro-1H-imidazol-1-yl)methyl]-7-methylfuro[2,3-c]pyridin-2-yl}methanone (PD7).
Yield: 1.30 g (93%). Light beige crystals, m.p. 183–185 °C (EtOAc: CH3CN 1:1). 1H NMR (DMSO-d6): δ ppm 2.77 (s, 3H, CH3), 5.56 (s, 2H, CH2), 7.78–7.82 (m, 2H, H-2″, H-6″), 7.91 (s, 1H, H-3), 7.98–8.01 (m, 2H, H-3″, H-5″), 7.60 (s, 1H, H-2′), 8.29 (s, 1H, H-5).). 13C NMR (DMSO-d6) δ ppm: 18.499, 44.750, 112.463, 113.385, 123.238, 124.937, 127.818, 131.117, 131.460, 131.917, 135.064, 136.543, 141.801, 144.369, 150.175, 152.896, 182.751 (DMSO-d6: 38.875–40.125). Anal.Calc. for C19H12BrCl2N3O2;(%).: C, 49.06; H, 2.60; Br, 17.18; Cl, 15.24; N, 9.03%. Found: C, 49.00; H, 2.41; Br+Cl, 32.51; N, 8.83%.
(4-Bromophenyl){4-[(3,4-dihydroisoquinolin-2(1H)-yl)methyl]-7-methylfuro[2,3-c]pyridin-2-y})methanone (PD8).
Yield: 1.04 g (75%). Light beige crystals, m.p. 134–135 °C (EtOAc). 1H NMR (DMSO-d6): δ ppm 2.77–2.85 (m, 7H, CH3, H-3′, H-4′), 3.65 (s, 2H, CH2), 3.97 (s, 1H, H-1′), 6.97 (s, 1H, H-3), 7.08–7.10 (m, 3H, H-5′–H-7′), 7.73 (d, J 8.1, 2H, H-2″, H-6″), 7.84–8.01 (m, 3H, H-8′, H-3″, H-5″), 8.31 (s, 1H, H-5).). 13C NMR (DMSO-d6) δ ppm: 18.424, 28.709, 50.176, 55.260, 56.652, 114.820, 125.471, 125.977, 126.387, 126.503, 127.663, 128.439, 131.395, 131.870, 132.248, 133.998, 134.614, 135.185, 142.052, 412.872, 150.381, 152.291, 182.851 (DMSO-d6: 38.878–40.128). Anal.Calc for C25H21BrN2O2; (%): C, 65.08; H, 4.59; Br, 17.32; N, 6.07%. Found: C, 65.01; H, 4.40; Br, 17.48; N, 6.03%.
4-{[2-(4-Bromobenzoyl)-7-methylfuro[2,3-c]pyridin-4-yl]methyl}piperazine-1-carbaldehyde hydrochloride (PD9).
Yield: 0.98 g (68%). Light beige crystals, m.p. 233–239 °C (EtOH). 1H NMR (DMSO-d6): δ ppm 2.91 (s, 3H, CH3), 3.51 (s, 8H, H-2′, H-3′, H-5′, H-6′), 4.74 (s, 2H, CH2), 7.81 (d, J 8.5, 2H, H-2″, H-6″), 8.10–8.14 (m, 2H, H-3″, H-5″), 8.45 (s, 1H, H-3), 8.75 (s, 1H, CH), 10.09 (s, 1H, H-5). 13C NMR (DMSO-d6) δ ppm: 18.309, 52.458, 53.228, 56.754, 58.447, 60.172, 114.820, 126.343, 127.612, 131.331, 131.838, 132.120, 135.191, 141.962, 142.661, 150.278, 152.189, 182.774 (DMSO-d6: 38.878–40.122). Anal.Calc. for C21H21BrClN3O3;(%).: C, 52.68; H, 4.42; Br, 16.69; Cl, 7.41; N, 8.78%. Found: C, 52.45; H, 4.29; Br+Cl, 24.21; N, 8.71%.
3-{4-[(2-(4-bromobenzoyl)-7-methylfuro[2,3-c]pyridin-4-yl)methyl]piperazin-1-yl}propanenitrile (PD10).
Yield: 1.18 g (84%). Light beige crystals, m.p. 129–132 °C (EtOH). 1H NMR (DMSO-d6): δ ppm 2.44–2.61 (m, 14H, CH3, H-2′, H-3′, H-5′, H-6′, DMF), 2.75 (s, 4H, 2CH2), 3.78 (s, 2H, CH2), 7.80 (d, J 8.3, 2H, H-2″, H-6″), 7.86 (s, 1H, H-3), 8.01 (d, J 8.2, 2H, H-3″, H-5″), 8.23 (s, 1H, H-5).13C NMR (DMSO-d6) δ ppm: 14.877, 18.288, 52.117–52.220(d), 52.585, 56.554, 114.768, 119.833, 126.200, 127.585, 131.310, 131.817, 132.150, 135.164, 142.005, 142.672, 150.245, 152.175, 182.766 (DMSO-d6: 38.832–40.082). Anal.Calc. for C23H23BrN4O2;(%).: C, 59.11; H, 4.96; Br, 17.10; N, 11.99%. Found: C, 59.03; H, 4.89; Br, 17.29; N, 11.72%.
(4-Bromophenyl)(7-methyl-4-{[(pyridin-3-ylmethyl)amino]methyl}furo[2,3-c]pyridin-2-yl)methanone dihydrochloride (PD11).
Yield: 0.67 g (44%). Light beige crystals, m.p. 245–248 °C (EtOH). 1H NMR (DMSO-d6): δ ppm 2.93 (s, 3H, CH3), 4.48 (s, 2H, CH2), 4.66 (s, 2H, CH2), 5.69 (br. s, 2H, 2NH+), 7.77–7.83 (m, 2H, H-2″, H-6″), 7.88 (s, 1H, H-3), 8.12–8.17 (m, 2H, H-3″, H-5″), 8.48 (s, 1H, H-5′), 8.74–8.82 (m, 3H, H-2′, H-4′, H-6′), 9.11 (s, 1H, H-5). 13C NMR (75 MHz, DMSO-d6) δ 164.38 (C=O), 163.41 (2C), 143.04 (2C), 141.77 (2C), 141.14, 140.42, 139.64, 129.99 (3C), 126.22 (4C), 115.70, 111.85, 57.36 (CH2-NH), 56.48 (NH-CH2), 18.98 (CH3). Anal.Calc. for C22H20BrCl2N3O2; (%).: C, 51.89; H, 3.96; Br, 15.69; Cl, 13.92; N, 8.25%. Found: C, 51.73; H, 3.71; Br+Cl, 29.81; N, 8.04%.
(2,3-Dihydrobenzo[d][1,4]dioxin-6-yl)(4-{[(3,4-dimethoxybenzyl)amino]methyl}-7-methylfuro[2,3-c]pyridin-2-yl)methanone hydrochloride (PD12).
Yield: 0.94 g (61%). Light beige crystals, m.p. 208–211 °C (EtOH). 1H NMR (DMSO-d6): δ ppm 2.97 (s, 3H, CH3), 3.77 (s, 3H, OCH3), 3.80 (s, 3H, OCH3), 4.16 (s, 2H, CH2), 4.33–4.42 (m, 4H, H-2″, H-3″), 4.57 (s, 2H, CH2), 5.69 (br. s, 1H, NH+), 6.86 (d, J 8.2, 1H, H-8″), 7.03–7.06 (m, 2H, H-5′, H-6′), 7.63 (d, J 2.1, 1H, H-2′), 7.78 (dd, 8.5, J 2.2, 1H, H-7″), 8.32 (s, 1H, H-3), 8.82 (s, 1H, H-5″), 10.41 (s, 1H, H-5). 13C NMR (75 MHz, DMSO-d6) δ 186.65 (C=O), 169.44 (C-C=O), 164.38, 162.00 (C-CH3), 157.81, 142.07 (2C), 140.19 (2C), 127.65 (4C), 122.99 (4C), 105.07, 96.84, 95.57, 57.90 (CH2-O-O-CH2), 56.89 (CH2-O-O-CH2), 56.47 (4C), 18.99 (CH3). Anal.Calc. for C27H27ClN2O6;(%).: C, 63.47; H, 5.33; Cl, 6.94; N, 5.48%. Found: C, 63.30; H, 5.16; Cl, 6.99; N, 5.33%.
(2,3-Dihydrobenzo[d][1,4]dioxin-6-yl){7-methyl-4-[(4-(pyridin-3-ylmethyl)piperazin-1-yl)methyl]furo[2,3-c]pyridin-2-yl}methanone dihydrochloride (PD13).
Yield: 0.97 g (58%). Light beige crystals, m.p. 224–227 °C (EtOH). 1H NMR (DMSO-d6): δ ppm 2.96 (s, 3H, CH3), 3.39–3.46 (m, 8H, H-2′, H-3′, H-5′, H-6′), 4.33–4.43 (m, 6H, H-2″, H-3″, CH2), 4.70 (m, 2H, CH2), 5.48 (br. s, 2H, 2NH+), 7.00–7.10 (m, 2H, H-3, H-8″), 7.62 (d, J 2.1, 1H, H-5″), 7.76 (dd, 8.5, J 2.1, 1H, H-7″), 8.39 (s, 1H, H-5‴), 8.65 (s, 1H, H-4‴), 8.77 (s, 1H, H-2‴), 8.82 (d, J 5.2, H-6‴), 9.06 (s, 1H, H-5). 13C NMR (DMSO-d6) δ ppm: 15.728, 47.653, 63.875, 64.645, 113.920, 117.511, 118.498, 124.179, 126.519, 128.545, 137.823, 143.107, 143.376, 144.678, 146.819, 148.987, 149.487, 155.700, 180.995 (DMSO-d6: 38.741–39.997). Anal.Calc. for C28H30Cl2N4O4; (%).: C, 60.33; H, 5.42; Cl, 12.72; N, 10.05%. Found: C, 60.16; H, 5.27; Cl, 12.87; N, 9.83%.
(2,3-Dihydrobenzo[d][1,4]dioxin-6-yl)(4-{[4-(2-hydroxyethyl)piperazin-1-yl]methyl}-7-methylfuro[2,3-c]pyridin-2-yl)methanone dihydrochloride (PD14).
Yield: 0.97 g (75%). Light beige crystals, m.p. 252–255 °C (EtOH). 1H NMR (DMSO-d6): δ ppm 3.00 (s, 3H, CH3), 3.27 (t, J 4.9, 2H, CH2), 3.62–3.71 (m, 8H, H-2′, H-3′, H-5′, H-6′), 3.85 (t. J 5.0, 2H, CH2), 4.34–4.42 (m, 4H, H-3″, H-8″), 4.82 (s, 2H, CH2), 5.71 (br. s, 3H, OH, 2NH+), 7.04 (d, J 8.5, 1H, H-8″), 7.62 (d, J 2.1, 1H, H-5″), 7.78 (dd, J 8.5, 2.1, 1H, H-7″), 8.46 (s, 1H, H-3), 8.85 (1H, H-5. 13C NMR (400 MHz, DMSO-d6) δ 15.89, 55.09, 63.87, 64.64, 113.98, 117.53, 118.49, 124.16, 128.60, 137.53, 143.26, 143.38, 148.97, 149.50, 155.52, 181.05. Anal.Calc. for C24H29Cl2N3O5;(%).: C, 56.48; H, 5.73; Cl, 13.89; N, 8.23%. Found: C, 56.31; H, 5.61; Cl, 13.97; N, 8.10%.
3.3. Computational Studies
3.3.1. System Preparation
The three-dimensional structure of wild-type (PDB ID: 1M17) [52], L858R/T790M (PDB ID: 4I22) [53] and L858R/T790M/C797S EGFR (PDB ID: 6LUD) [52] were obtained from the Protein Data Bank. These crystal structures complexed with known drugs and subsequently known drugs were extracted. A series of 14 furo[2,3-c]pyridine derivatives was built using the Gaussview 09 program. All the ligands were optimized using the Gaussian 09 program (HF/6-31G*) and, subsequently, the protonation states of all ionizable amino acids were characterized using ChemAxon at pH 7.0. Molecular docking studies were performed using the GOLD program. The setting of the docking was studied using the following parameter as 10 Å with a sphere and 100 docking poses. The docking runs in each complex were sorted and selected based on GOLD fitness score. Then, the docking results were visualized for interaction using the UCSF Chimera package. The docked PDs/EGFRs with the highest GOLD fitness scores were chosen as the initial structures for performing MD simulations.
3.3.2. Molecular Dynamics Simulation and Free Energy Calculation Based on Solvated Interaction Energy (SIE) Method
The initial structures of PDs/EGFRs derived from molecular docking were carried out using pmemd CUDA in the AMBER 16 program [54] under periodic boundary conditions with the isobaric isothermal (NPT) ensemble in triplicate. The general AMBER force fields GAFF [55] and FF14SB [56] were applied for the ligand and protein, respectively. The partial charges of ligand were generated as follows: (i) the 3D structure of PD derivatives were optimized using the HF/6-31G* method [57] in the Gaussian09 program; (ii) the electrostatic potential (ESP) charge and restrained ESP (RESP) charge of the ligand were obtained by the AMBER16 program. All studied complexes were solvated by the TIP3P water model [58]. All missing hydrogen atoms of the protein and ligand were added by the LEaP module and subsequently minimized. The sodium ions were randomly neutralized in the simulated systems by Cl− counter ions. The added hydrogen atoms and water molecules were then minimized using 2500 steps of the steepest descents (SD), followed by 2500 steps of the conjugated gradient (CG), before starting the MD simulations. The SHAKE algorithm was used to fix all covalent bonds involving hydrogen atoms [59]. The systems were heated up to 310 K for 100 ps. Finally, the whole system was performed under the NPT ensemble (310 K, 1 atm) until reaching 100 ns. The root mean square deviation (RMSD) of each system was calculated using all-atom, and then the MD trajectories from the last 10-ns simulation were extracted for analysis in terms of total binding free energy (ΔGbind) based on the solvated interaction energy (SIE) method. The potent PD derivatives with the lowest binding free energy were selected to determine EGFR inhibitory as well as anticancer activity. After finding the more potent PD derivative obtained from in silico screening and in vitro evaluation, the 500-ns MD simulation was performed. In addition, the per-residue decomposition free energy (ΔGbind,residue) calculations were carried out using the MMPBSA.py module in AMBER16 [60]. The CPPTRJ module was used to calculate intermolecular hydrogen bonding.
3.4. Biological Studies
3.4.1. EGFR Inhibitory ACTIVITY
The IC50 values of the potent PD derivatives obtained from the SIE method were measured using the ADP-GloTM kinase assay kit as previously reported [19]. Firstly, 8 µL of buffer (40 mM Tris-HCl pH 7.5, 20 mM MgCl2, and 0.1 mg/mL bovine serum albumin) were added to a 384-well plate (Promega, solid white). Secondly, 5 µL of 1.25 ng/µL EGFRs enzyme (wild-type, L858R/T790M, and L858R/T790M/C797S EGFR) and 2 µL of 1 μM of compounds/known drugs were added separately to individual cell cultures, followed by 10 µL of a mixture of 5 µM ATP and 2.5 µM poly(glu-tyr), and were incubated for 1 h at room temperature. Thirdly, 5 µL of the ADP-Glo reagent was added and incubated for 40 min. Finally, 10 µL of kinase detection reagent was added and incubated at room temperature for 30 min and was detected by measuring the luminescence using a microplate reader (Infinite M200 microplate reader, Tecan, Männedorf, Switzerland). A triplicate assay was performed on all potent PD derivatives and known drugs. The percentage relative inhibition (%) of potent compounds was measured compared to the control with no inhibitor as shown in Equation (1):
3.4.2. Cell Viability Assay
The cytotoxicity activities of potent PD derivatives obtained from the SIE method against A549 and H1975, as well as Vero (which are adherent cells), were assessed using MTT (3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide) assays. The A549 and Vero cells were grown in complete DMEM medium whereas H1975 cells were grown in complete RPMI medium supplemented with 10% (v/v) FBS, 100 U/mL penicillin and 100 µg/mL streptomycin. All cells were routinely cultured at 37 °C in a 5% (v/v) CO2, 95% (v/v) air humidified incubator. A549 (5 × 103 cells per well), H1975 (5 × 103 cells per well), and Vero (2 × 103 cells per well) cells were seeded into 96-well plates and incubated overnight. After cell attachment, cells were replenished using fresh medium with 0.1% FBS containing various concentrations of potent PD derivatives and known drugs and then incubated at 37 °C for 72 h. After that, fresh medium containing MTT solution (5 mg/mL) was added and incubated at 37 °C. After 3 h, the formed formazan crystal was dissolved with 100 μL of dimethyl sulfoxide (DMSO). Finally, the absorbance of formazan solution was measured at a wavelength 570 nm using a microplate reader (Infinite M200 microplate reader, Tecan, Männedorf, Switzerland). The independent treatments were repeated in triplicate. The IC50 determination of compounds was achieved using GraphPad Prism version 9.0.
4. Conclusions
In this work, a combination of in silico and in vitro studies were performed to screen for novel EGFR inhibitors that could inhibit both wild-type and mutant forms of EGFR. We found that potent compounds from virtual screening exhibited promising EGFR inhibitory activity. In particular, PD13 showed strong inhibition of wild-type and L858R/T790M double mutant EGFR, while L858R/T790M/C797S showed moderate inhibition. In addition, the most potent PD13 revealed high cytotoxicity against A549 and H1975. In 500-ns MD simulations, the model of PD13 and known drugs/EGFRs was stable. The binding affinity of PD13 complexed with wild-type and mutant forms of EGFR was significantly higher than that of known drugs (except for the L858R/T790M/C797S system), which was consistent with the hot-spot residues and EGFR inhibitory activity. The calculations revealed that M793 and S797 are the main residues in the hinge region for the binding of PD13 with three EGFR strains via H-bond formations. In addition, the binding of all studied PD compounds is driven mainly by vdW interaction. Our results showed that PD13 could potentially act as a promising non-mutant and mutant EGFR drug for cancer therapy.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28073014/s1, 1H NMR Spectra of compounds PD1–PD14.
Author Contributions
A.G., K.C., V.K. and T.R.; methodology, K.C., T.R. and A.Z.; software, K.C., T.R. and A.P.; validation, D.T., A.Z., V.K., T.A., P.M., A.P., A.G., I.Y., K.C. and T.R.; formal analysis, D.T., A.Z., V.K., T.A., P.M., A.P., A.G., I.Y., K.C. and T.R.; investigation, D.T. and V.C., L.D.; data curation, D.T., A.G. and T.R.; writing—original draft preparation, D.T., A.Z., V.K., T.A., P.M., A.P., A.G., I.Y., K.C. and T.R.; writing—review and editing, D.T., A.Z., V.K., T.A., P.M., A.P., A.G., I.Y., K.C. and T.R.; visualization, D.T., A.Z., V.K., T.A., P.M., A.P., A.G., I.Y., K.C. and T.R.; supervision, P.M., A.G., K.C. and T.R.; funding acquisition, A.Z., K.C. and T.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work has been supported by the Ministry of Science and Higher Education of the Russian Federation (Southern Federal University, 2022, project FENW-2023-0011), Fundamental Scientific Research of the State Academies of Sciences for 2022–2025 (grant no. 0710-2019-0044.), and the Thailand Science Research and Innovation Fund, Chulalongkorn University (HEA662300073).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
D.T. thanks the 90th Anniversary of Chulalongkorn University (CU) Fund (Ratchadaphiseksomphot Endowment Fund, GCUGR1125642021M), and the Potential Development in Research for Graduate student Project, Faculty of Science, Chulalongkorn university.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are available from the authors.
References
- Engel, J.; Richters, A.; Getlik, M.; Tomassi, S.; Keul, M.; Termathe, M.; Lategahn, J.; Becker, C.; Mayer-Wrangowski, S.; Grütter, C. Targeting drug resistance in EGFR with covalent inhibitors: A structure-based design approach. J. Med. Chem. 2015, 58, 6844–6863. [Google Scholar] [CrossRef]
- Peters, S.; Zimmermann, S.; Adjei, A.A. Oral epidermal growth factor receptor tyrosine kinase inhibitors for the treatment of non-small cell lung cancer: Comparative pharmacokinetics and drug–drug interactions. Cancer Treat. Rev. 2014, 40, 917–926. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Matsuoka, Y.; Funahashi, A.; Kitano, H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Bbiol. 2005, 1, 2005.0010. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.R.; Dudek, H.; Tao, X.; Masters, S.; Fu, H.; Gotoh, Y.; Greenberg, M.E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 1997, 91, 231–241. [Google Scholar] [CrossRef]
- Chen, P.; Xie, H.; Sekar, M.C.; Gupta, K.; Wells, A. Epidermal growth factor receptor-mediated cell motility: Phospholipase C activity is required, but mitogen-activated protein kinase activity is not sufficient for induced cell movement. J. Cell Biol. 1994, 127, 847–857. [Google Scholar] [CrossRef]
- Kharitonenkov, A.; Chen, Z.; Sures, I.; Wang, H.; Schilling, J.; Ullrich, A. A family of proteins that inhibit signalling through tyrosine kinase receptors. Nature 1997, 386, 181–186. [Google Scholar] [CrossRef]
- Sternberg, M.J.; Gullick, W.J. A sequence motif in the transmembrane region of growth factor receptors with tyrosine kinase activity mediates dimerization. PEDS 1990, 3, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H. Osimertinib making a breakthrough in lung cancer targeted therapy. Onco Targets Ther. 2016, 9, 5489. [Google Scholar] [CrossRef] [PubMed]
- Bethune, G.; Bethune, D.; Ridgway, N.; Xu, Z. Epidermal growth factor receptor (EGFR) in lung cancer: An overview and update. J. Thorac. Dis. 2010, 2, 48–51. [Google Scholar] [PubMed]
- Li, X.; Fan, X.-X.; Jiang, Z.-B.; Loo, W.T.; Yao, X.-J.; Leung, E.L.-H.; Chow, L.W.; Liu, L. Shikonin inhibits gefitinib-resistant non-small cell lung cancer by inhibiting TrxR and activating the EGFR proteasomal degradation pathway. Pharmacol. Res. 2017, 115, 45–55. [Google Scholar] [CrossRef]
- Normanno, N.; De Luca, A.; Bianco, C.; Strizzi, L.; Mancino, M.; Maiello, M.R.; Carotenuto, A.; De Feo, G.; Caponigro, F.; Salomon, D.S. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006, 366, 2–16. [Google Scholar] [CrossRef]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Perez, E.A. The role of adjuvant monoclonal antibody therapy for breast cancer: Rationale and new studies. Curr. Oncol. Rep. 2001, 3, 516–522. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, S.; MacLennan, G.T.; Eble, J.N.; Lopez-Beltran, A.; Yang, X.J.; Pan, C.-X.; Zhou, H.; Montironi, R.; Cheng, L. Epidermal growth factor receptor protein expression and gene amplification in small cell carcinoma of the urinary bladder. Clin. Cancer Res. 2007, 13, 953–957. [Google Scholar] [CrossRef]
- Shia, J.; Klimstra, D.S.; Li, A.R.; Qin, J.; Saltz, L.; Teruya-Feldstein, J.; Akram, M.; Chung, K.Y.; Yao, D.; Paty, P.B. Epidermal growth factor receptor expression and gene amplification in colorectal carcinoma: An immunohistochemical and chromogenic in situ hybridization study. Mod. Pathol. 2005, 18, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, F.A.; Rodrigues Pereira, J.; Ciuleanu, T.; Tan, E.H.; Hirsh, V.; Thongprasert, S.; Campos, D.; Maoleekoonpiroj, S.; Smylie, M.; Martins, R. Erlotinib in previously treated non–small-cell lung cancer. NEJM 2005, 353, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Soler, R.; Chachoua, A.; Hammond, L.A.; Rowinsky, E.K.; Huberman, M.; Karp, D.; Rigas, J.; Clark, G.M.; Santabárbara, P.; Bonomi, P. Determinants of tumor response and survival with erlotinib in patients with non-small-cell lung cancer. J. Clin. Oncol. 2004, 22, 3238–3247. [Google Scholar] [CrossRef]
- Kris, M.G.; Natale, R.B.; Herbst, R.S.; Lynch Jr, T.J.; Prager, D.; Belani, C.P.; Schiller, J.H.; Kelly, K.; Spiridonidis, H.; Sandler, A. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non–small cell lung cancer: A randomized trial. JAMA 2003, 290, 2149–2158. [Google Scholar] [CrossRef]
- Michalczyk, A.; Klüter, S.; Rode, H.B.; Simard, J.R.; Grütter, C.; Rabiller, M.; Rauh, D.; Fritsche, A.; Elfringhoff, A.S.; Fabian, J. Structural insights into how irreversible inhibitors can overcome drug resistance in EGFR. Bioorg. Med. Chem. 2008, 16, 3482–3488. [Google Scholar] [CrossRef] [PubMed]
- Heuckmann, J.M.; Rauh, D.; Thomas, R.K. Epidermal growth factor receptor (EGFR) signaling and covalent EGFR inhibition in lung cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2012, 30, 3417–3420. [Google Scholar] [CrossRef] [PubMed]
- Kalgutkar, A.S.; Dalvie, D.K. Drug discovery for a new generation of covalent drugs. Expert Opin. Drug Discov. 2012, 7, 561–581. [Google Scholar] [CrossRef] [PubMed]
- Copeland, R.A.; Pompliano, D.L.; Meek, T.D. Drug–target residence time and its implications for lead optimization. Nat. Rev. Drug Discov. 2006, 5, 730–739. [Google Scholar] [CrossRef] [PubMed]
- Kwak, E.L.; Sordella, R.; Bell, D.W.; Godin-Heymann, N.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Driscoll, D.R.; Fidias, P.; Lynch, T.J. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc. Natl. Acad. Sci. USA 2005, 102, 7665–7670. [Google Scholar] [CrossRef] [PubMed]
- Cross, D.A.; Ashton, S.E.; Ghiorghiu, S.; Eberlein, C.; Nebhan, C.A.; Spitzler, P.J.; Orme, J.P.; Finlay, M.R.; Ward, R.A.; Mellor, M.J.; et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014, 4, 1046–1061. [Google Scholar] [CrossRef]
- Lelais, G.; Epple, R.; Marsilje, T.H.; Long, Y.O.; McNeill, M.; Chen, B.; Lu, W.; Anumolu, J.; Badiger, S.; Bursulaya, B. Discovery of (R, E)-N-(7-Chloro-1-(1-[4-(dimethylamino) but-2-enoyl] azepan-3-yl)-1 H-benzo [d] imidazol-2-yl)-2-methylisonicotinamide (EGF816), a Novel, Potent, and WT Sparing Covalent Inhibitor of Oncogenic (L858R, ex19del) and Resistant (T790M) EGFR Mutants for the Treatment of EGFR Mutant Non-Small-Cell Lung Cancers. J.Med.Chem. 2016, 59, 6671–6689. [Google Scholar]
- Zhou, W.; Ercan, D.; Chen, L.; Yun, C.H.; Li, D.; Capelletti, M.; Cortot, A.B.; Chirieac, L.; Iacob, R.E.; Padera, R.; et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature 2009, 462, 1070–1074. [Google Scholar] [CrossRef]
- Zhou, W.; Ercan, D.; Janne, P.A.; Gray, N.S. Discovery of selective irreversible inhibitors for EGFR-T790M. Bioorg. Med. Chem. Lett. 2011, 21, 638–643. [Google Scholar] [CrossRef]
- Planken, S.; Behenna, D.C.; Nair, S.K.; Johnson, T.O.; Nagata, A.; Almaden, C.; Bailey, S.; Ballard, T.E.; Bernier, L.; Cheng, H.; et al. Discovery of n-((3 r, 4 r)-4-fluoro-1-(6-((3-methoxy-1-methyl-1 h-pyrazol-4-yl) amino)-9-methyl-9 h-purin-2-yl) pyrrolidine-3-yl) acrylamide (pf-06747775) through structure-based drug design: A high affinity irreversible inhibitor targeting oncogenic egfr mutants with selectivity over wild-type egfr. J. Med. Chem. 2017, 60, 3002–3019. [Google Scholar]
- Eberlein, C.A.; Stetson, D.; Markovets, A.A.; Al-Kadhimi, K.J.; Lai, Z.; Fisher, P.R.; Meador, C.B.; Spitzler, P.; Ichihara, E.; Ross, S.J.; et al. Acquired Resistance to the Mutant-Selective EGFR Inhibitor AZD9291 Is Associated with Increased Dependence on RAS Signaling in Preclinical Models. Cancer Res. 2015, 75, 2489–2500. [Google Scholar] [CrossRef] [PubMed]
- Hung, J.M.; Arabshahi, H.J.; Leung, E.; Reynisson, J.; Barker, D. Synthesis and cytotoxicity of thieno [2, 3-b] pyridine and furo [2, 3-b] pyridine derivatives. Eur. J. Med. Chem. 2014, 86, 420–437. [Google Scholar] [CrossRef] [PubMed]
- Garamvölgyi, R.; Dobos, J.; Sipos, A.; Boros, S.; Illyés, E.; Baska, F.; Kékesi, L.; Szabadkai, I.; Szántai-Kis, C.; Kéri, G. Design and synthesis of new imidazo [1, 2-a] pyridine and imidazo [1, 2-a] pyrazine derivatives with antiproliferative activity against melanoma cells. Eur. J. Med. Chem. 2016, 108, 623–643. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.M.; Al-Refai, M.; Azmi, M.N.; Osman, H.; Bakar, M.H.A.; Geyer, A. Synthesis, characterization and cytotoxicity of new nicotinonitriles and their furo [2, 3-b] pyridine derivatives. JICS 2019, 16, 715–722. [Google Scholar] [CrossRef]
- Rai, S.K.; Singh, P.; Khanam, S.; Tewari, A.K. Polymorphic study and anti-inflammatory activity of a 3-cyano-2-pyridone based flexible model. NJC 2016, 40, 5577–5587. [Google Scholar] [CrossRef]
- Dorsey, B.D.; McDonough, C.; McDaniel, S.L.; Levin, R.B.; Newton, C.L.; Hoffman, J.M.; Darke, P.L.; Zugay-Murphy, J.A.; Emini, E.A.; Schleif, W.A. Identification of MK-944a: A second clinical candidate from the hydroxylaminepentanamide isostere series of HIV protease inhibitors. J. Med. Chem. 2000, 43, 3386–3399. [Google Scholar] [CrossRef]
- Agarwal, R.; Jha, K.K.; Munshi, P.; Adepally, U.; Singh, A.; Charyd, M.T.; Sen, S. Substituted furopyridinediones as novel inhibitors of a-glucosidase. RSC Adv. 2015, 5, 90374. [Google Scholar]
- Salunke, D.B.; Yoo, E.; Shukla, N.M.; Balakrishna, R.; Malladi, S.S.; Serafin, K.J.; Day, V.W.; Wang, X.; David, S.A. Structure–activity relationships in human Toll-like receptor 8-active 2, 3-diamino-furo [2, 3-c] pyridines. J. Med. Chem. 2012, 55, 8137–8151. [Google Scholar] [CrossRef]
- Zubenko, A.; Divaeva, L.; Morkovnik, A.; Fetisov, L.; Sochnev, V.; Kononenko, K.; Bodryakov, A.; Klimenko, A. Structural Modification of Pyridoxal. Synthesis and Evaluation of Anti-Infective Activity of New 4-Chloro-and 4-Alkyl (dialkyl) aminomethyl-2-hetaryl (hetaroyl) furo[2, 3-c] pyridines. Russ. J. Gen. Chem. 2020, 90, 2242–2247. [Google Scholar] [CrossRef]
- Hei, Y.-Y.; Shen, Y.; Wang, J.; Zhang, H.; Zhao, H.-Y.; Xin, M.; Cao, Y.-X.; Li, Y.; Zhang, S.-Q. Synthesis and evaluation of 2, 9-disubstituted 8-phenylthio/phenylsulfinyl-9H-purine as new EGFR inhibitors. Bioorg. Med. Chem. 2018, 26, 2173–2185. [Google Scholar] [CrossRef]
- Karnik, K.S.; Sarkate, A.P.; Tiwari, S.V.; Azad, R.; Burra, P.V.; Wakte, P.S. Computational and Synthetic approach with Biological Evaluation of Substituted Quinoline derivatives as small molecule L858R/T790M/C797S triple mutant EGFR inhibitors targeting resistance in Non-Small Cell Lung Cancer (NSCLC). Bioorg. Chem. 2021, 107, 104612. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhang, T.; Zhu, S.-J.; Sun, M.; Tong, L.; Lai, M.; Zhang, R.; Xu, W.; Wu, R.; Ding, J. Structure-based design of 5-methylpyrimidopyridone derivatives as new wild-type sparing inhibitors of the epidermal growth factor receptor triple mutant (EGFRL858R/T790M/C797S). J. Med. Chem. 2019, 62, 7302–7308. [Google Scholar] [CrossRef] [PubMed]
- Rajith, B.; Chakraborty, C.; NagaSundaram, N.; Ali, S.K.; Zhu, H. Structural signature of the G719S-T790M double mutation in the EGFR kinase domain and its response to inhibitors. Sci. Rep. 2014, 4, 5868. [Google Scholar]
- Martínez-Jiménez, F.; Overington, J.P.; Al-Lazikani, B.; Marti-Renom, M.A. Rational design of non-resistant targeted cancer therapies. Sci. Rep. 2017, 7, 46632. [Google Scholar] [CrossRef]
- Stamos, J.; Sliwkowski, M.X.; Eigenbrot, C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 2002, 277, 46265–46272. [Google Scholar] [CrossRef]
- Sepay, N.; Mondal, R.; Al-Muhanna, M.K.; Saha, D. Identification of natural flavonoids as novel EGFR inhibitors using DFT, molecular docking, and molecular dynamics. NJC 2022, 46, 9735–9744. [Google Scholar] [CrossRef]
- Zhang, R.; Chen, S.; Zhang, X.; Yu, R.; Wan, S.; Geng, M.; Jiang, T. Synthesis and evaluation of novel non-covalent binding quinazoline glycoside derivatives targeting the L858R and T790M variants of EGFR. RSC Adv. 2016, 6, 36857–36862. [Google Scholar] [CrossRef]
- Lei, H.; Fan, S.; Zhang, H.; Liu, Y.-J.; Hei, Y.-Y.; Zhang, J.-J.; Zheng, A.-Q.; Xin, M.; Zhang, S.-Q. Discovery of novel 9-heterocyclyl substituted 9H-purines as L858R/T790M/C797S mutant EGFR tyrosine kinase inhibitors. Eur. J. Med. Chem. 2020, 186, 111888. [Google Scholar] [CrossRef]
- Bello, M.; Saldaña-Rivero, L.; Correa-Basurto, J.; García, B.; Sánchez-Espinosa, V.A. Structural and energetic basis for the molecular recognition of dual synthetic vs. natural inhibitors of EGFR/HER2. Int. J. Biol. Macromol. 2018, 111, 569–586. [Google Scholar] [CrossRef]
- Kou, S.-B.; Lin, Z.-Y.; Wang, B.-L.; Shi, J.-H.; Liu, Y.-X. Evaluation of the binding behavior of olmutinib (HM61713) with model transport protein: Insights from spectroscopic and molecular docking studies. J. Mol. Struct. 2021, 1224, 129024. [Google Scholar] [CrossRef]
- Zhang, H.-X.; Xiong, H.-X.; Li, L.-W. Investigation on the protein-binding properties of icotinib by spectroscopic and molecular modeling method. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2016, 161, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Kashima, K.; Kawauchi, H.; Tanimura, H.; Tachibana, Y.; Chiba, T.; Torizawa, T.; Sakamoto, H. CH7233163 overcomes osimertinib resistant EGFR-Del19/T790M/C797S mutation. Mol. Cancer Ther. 2020, 19, 2288–2297. [Google Scholar] [CrossRef]
- Gajiwala, K.S.; Feng, J.; Ferre, R.; Ryan, K.; Brodsky, O.; Weinrich, S.; Kath, J.C.; Stewart, A. Insights into the aberrant activity of mutant EGFR kinase domain and drug recognition. Structure 2013, 21, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Gotz, A.W.; Williamson, M.J.; Xu, D.; Poole, D.; Le Grand, S.; Walker, R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J. Chem. Theory Comput. 2012, 8, 1542–1555. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Frisch, A. Gaussian 09w Reference; Gaussian, Inc.: Wallingford, CT, USA, 2009; 25p. [Google Scholar]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Miller III, B.R.; McGee Jr, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA. py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).