Catalytic, Theoretical, and Biological Investigations of Ternary Metal (II) Complexes Derived from L-Valine-Based Schiff Bases and Heterocyclic Bases
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Section
2.1.1. Reagents and Characterization
2.1.2. Preparation of Potassium (E)-2-((5-bromo-2-hydroxy benzylidene)amino)-3-methylbutanoate (HL)
2.2. Preparation of Complexes
2.2.1. Preparation of [Co(L)(phen)] (1a)
2.2.2. Preparation of [Ni(L)(phen)] (1b)
2.2.3. Preparation of [Cu(L)(phen)] (1c)
2.2.4. Preparation of [Zn(L)(phen)] (1d)
2.2.5. Preparation of [Co(L)(bpy)] (1e)
2.2.6. Preparation of [Ni(L)(bpy)] (1f)
2.2.7. Preparation of [Cu(L)(bpy)] (1g)
2.2.8. Preparation of [Zn(L)(bpy)] (1h)
2.3. In Vitro MTT Assay
2.4. Molecular Modeling
2.5. Computational Analysis
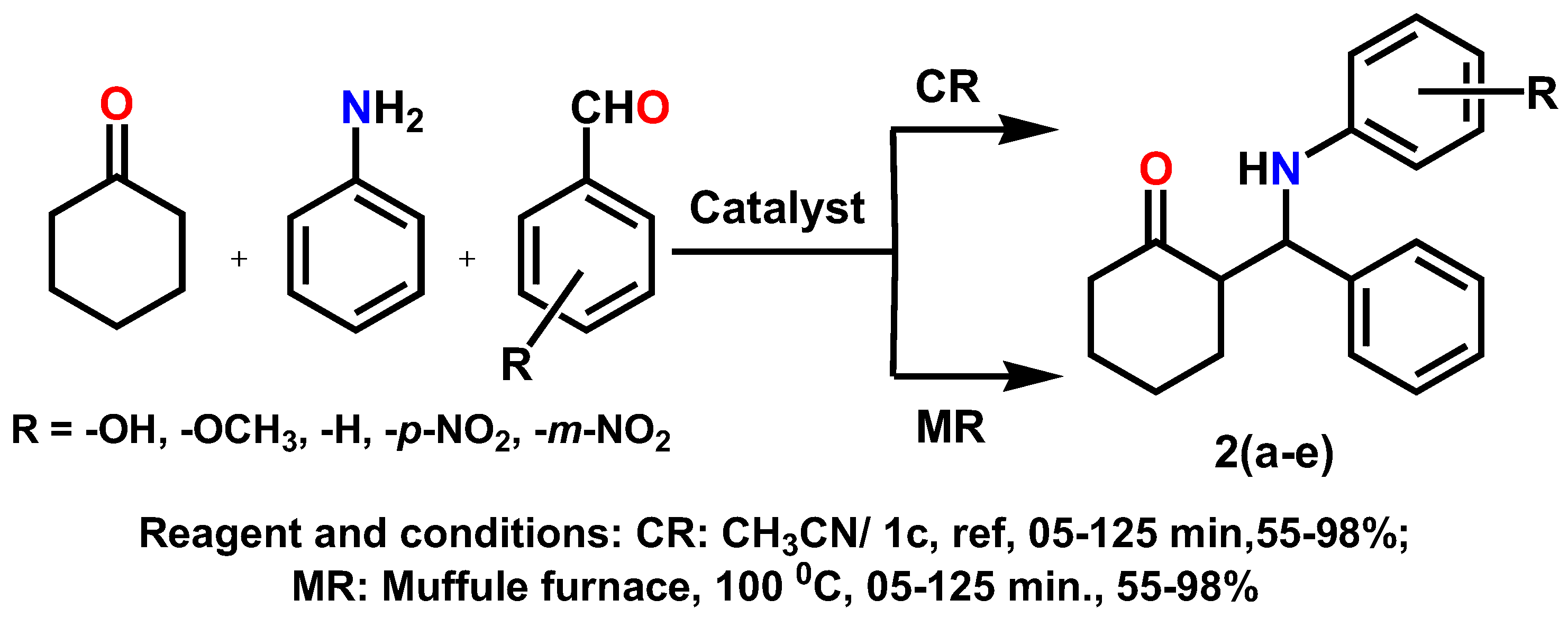
2.6. The General Process for the Synthesis of β-Amino Carbonyl Derivative
3. Results and Discussion
3.1. Results and Discussion
3.2. Spectral Characterization
3.2.1. FT-IR Spectral Analysis
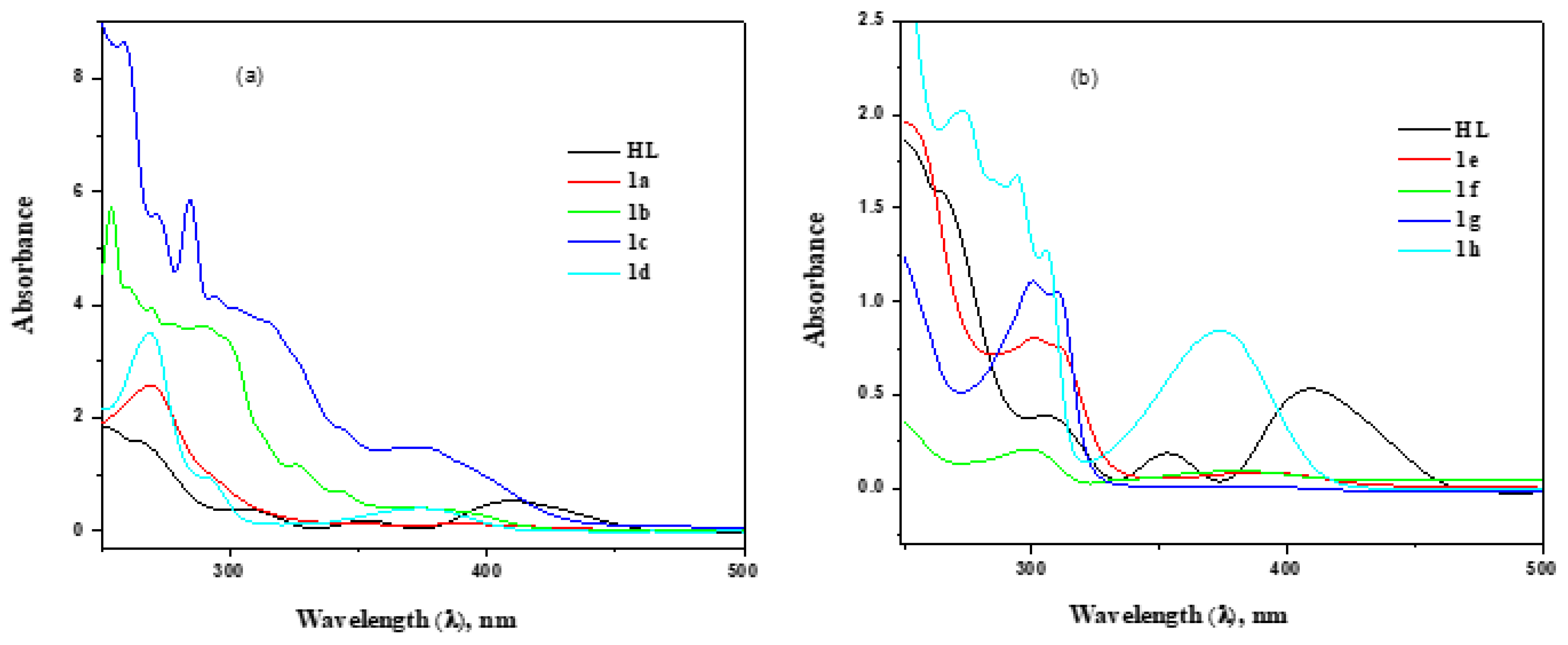
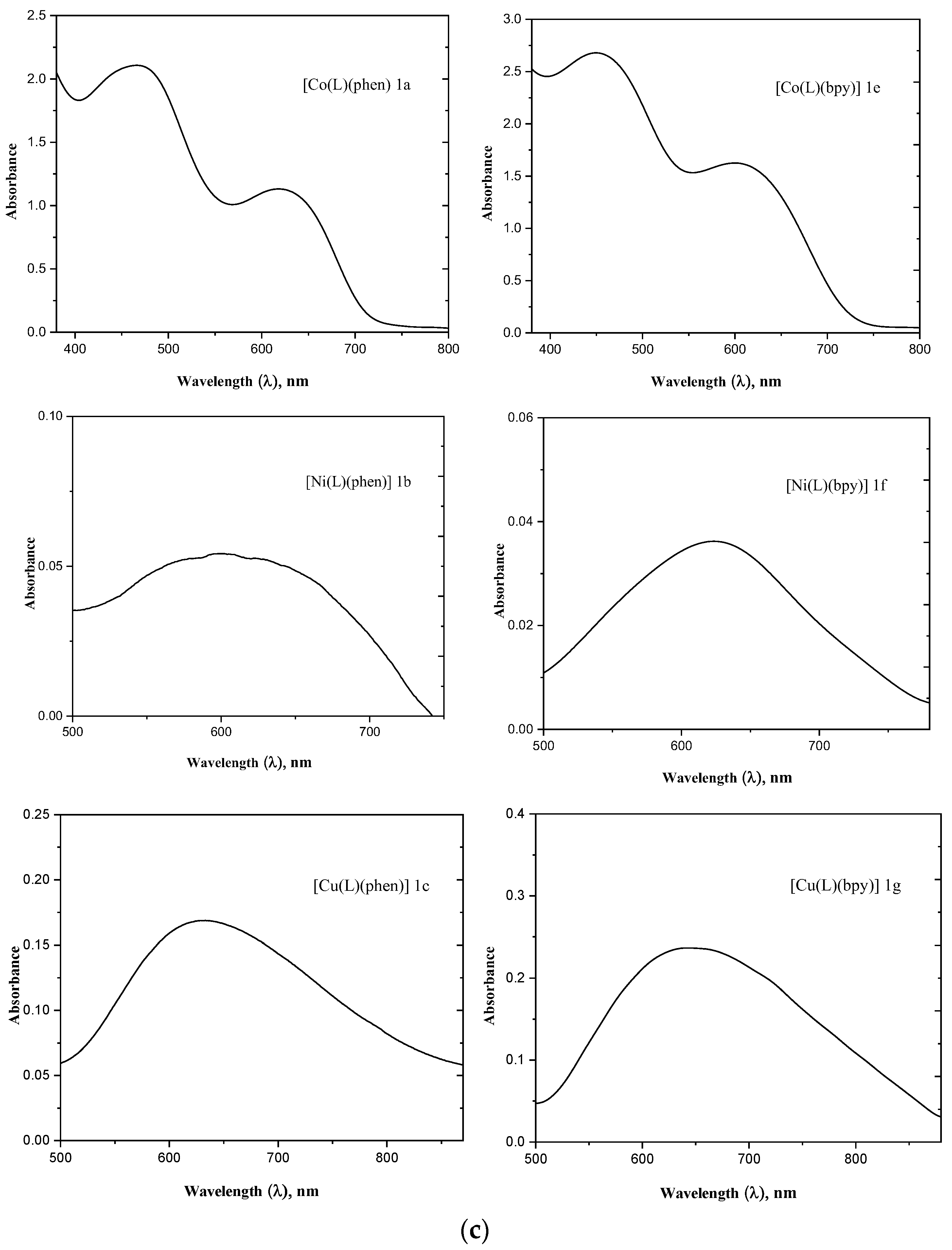
3.2.2. UV–Visible Spectral Analysis
3.2.3. Mass Spectral Analysis
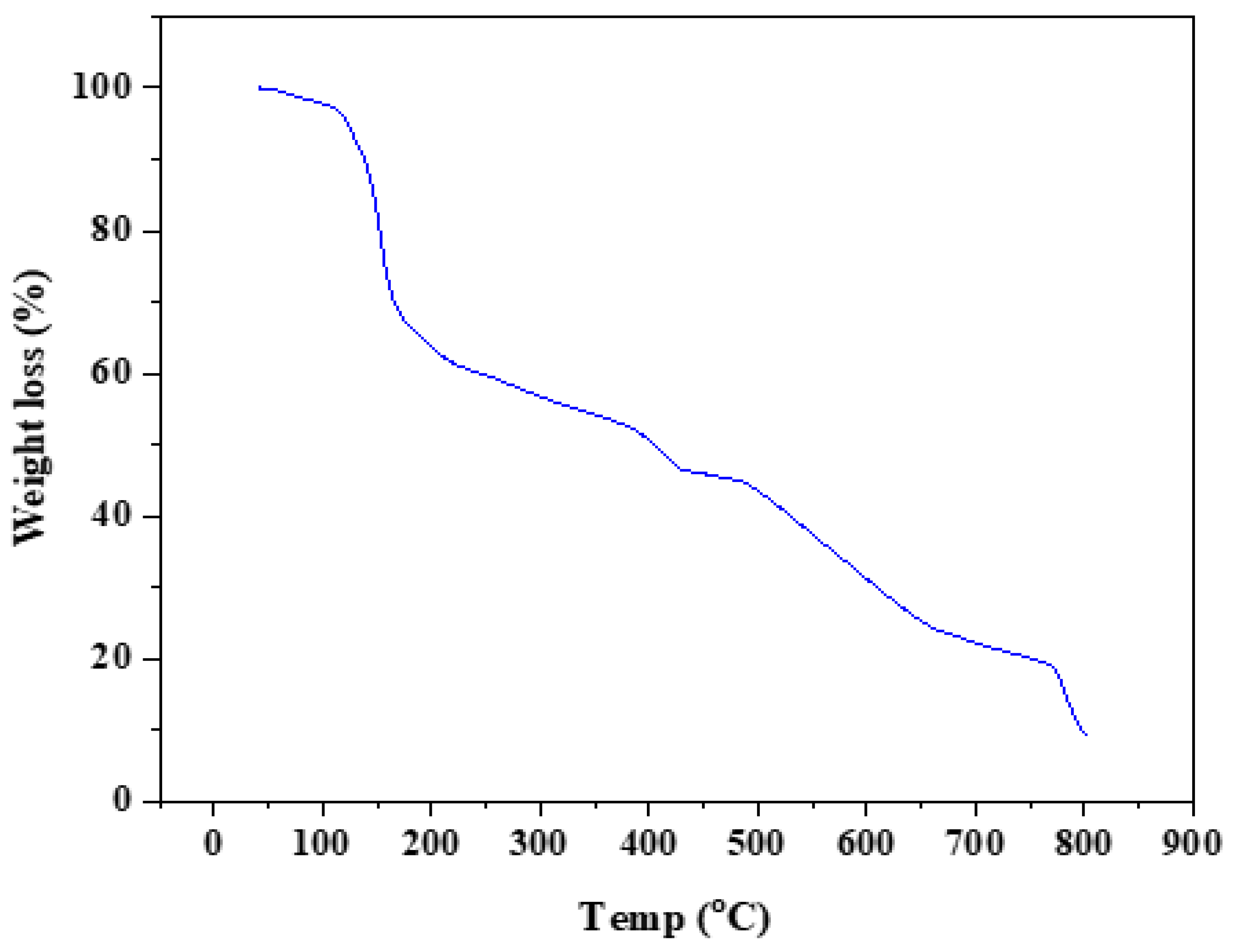
3.2.4. Thermal Analysis
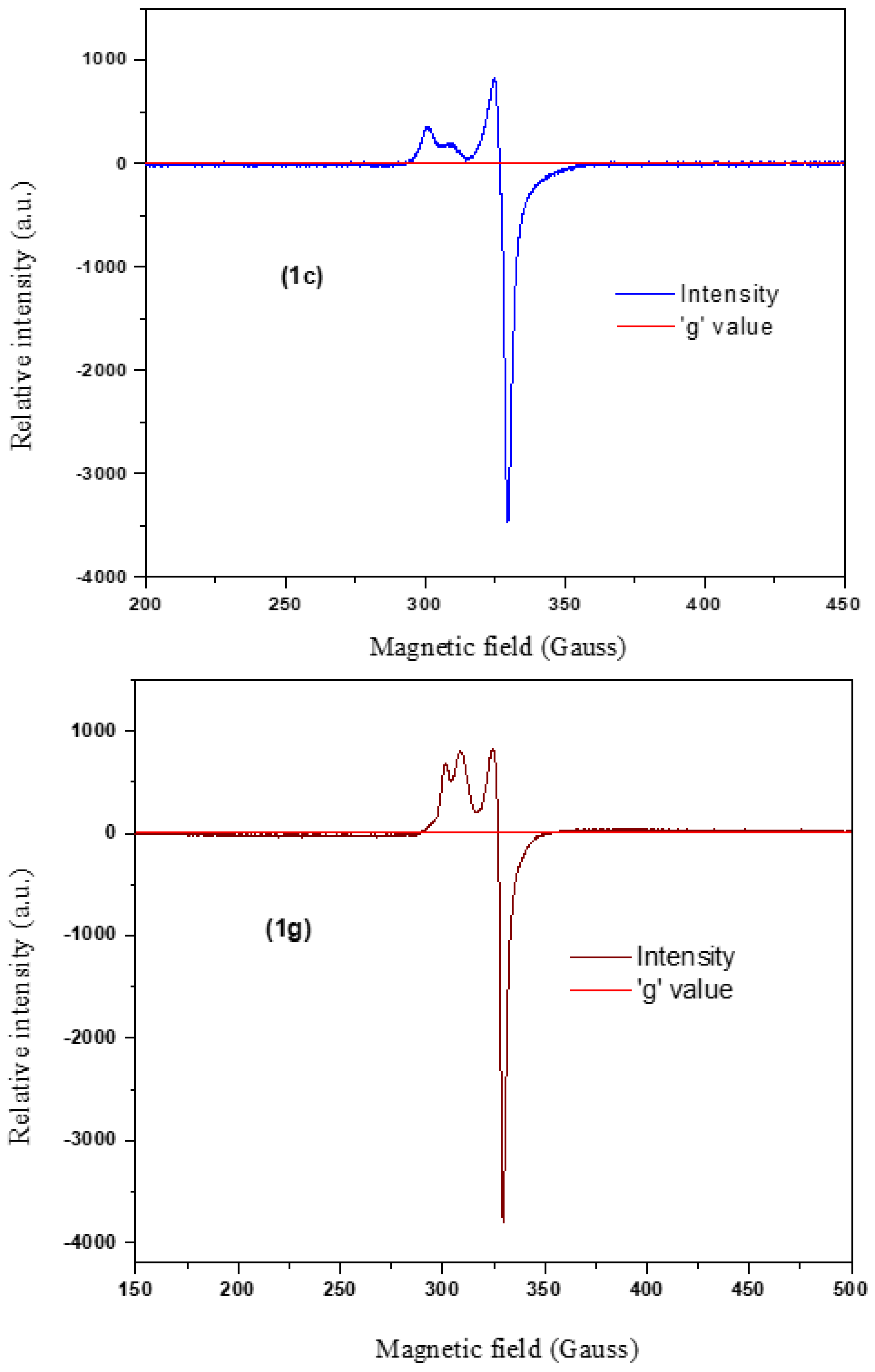
3.2.5. EPR Spectral Analysis
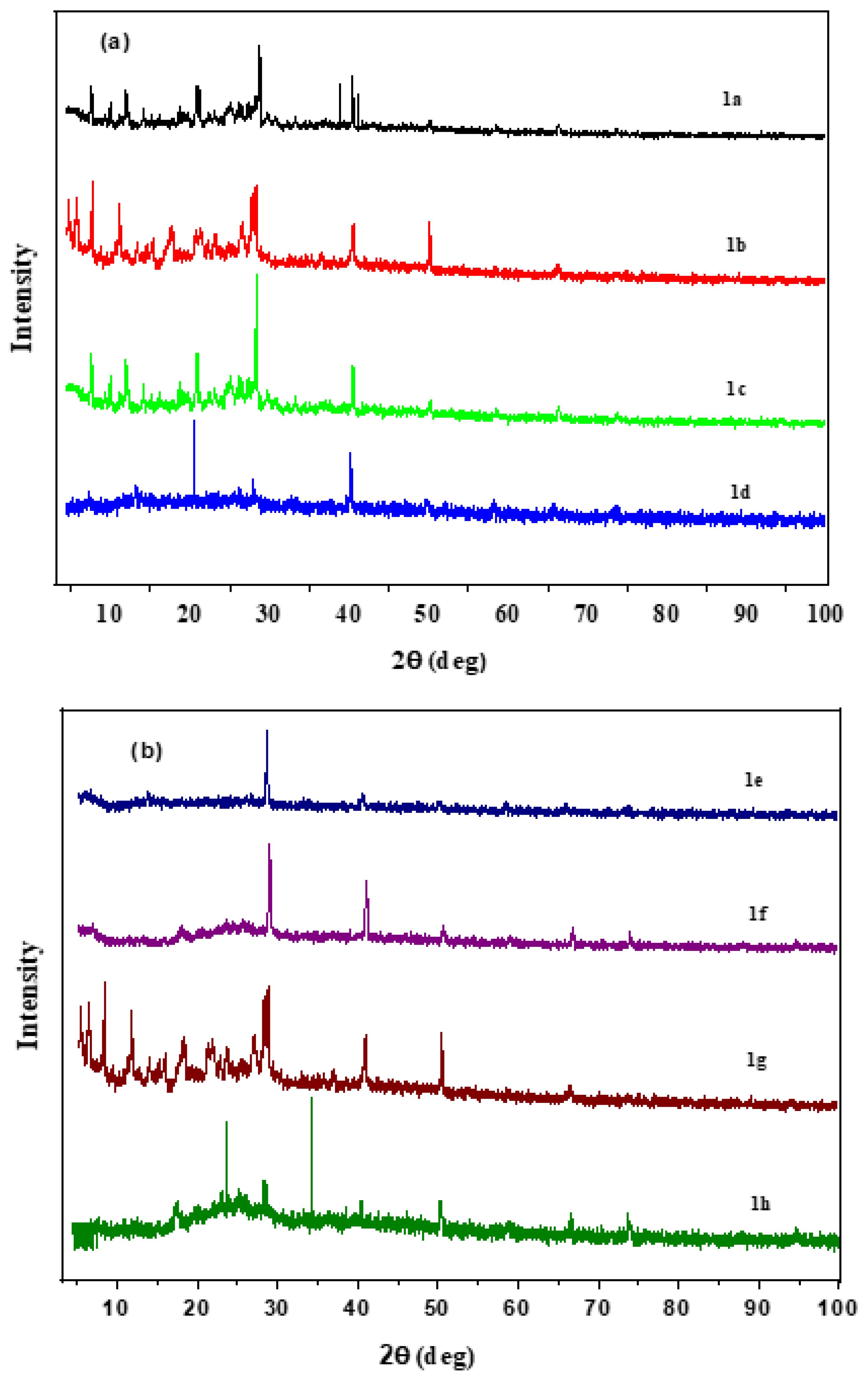
3.2.6. XRD Analysis
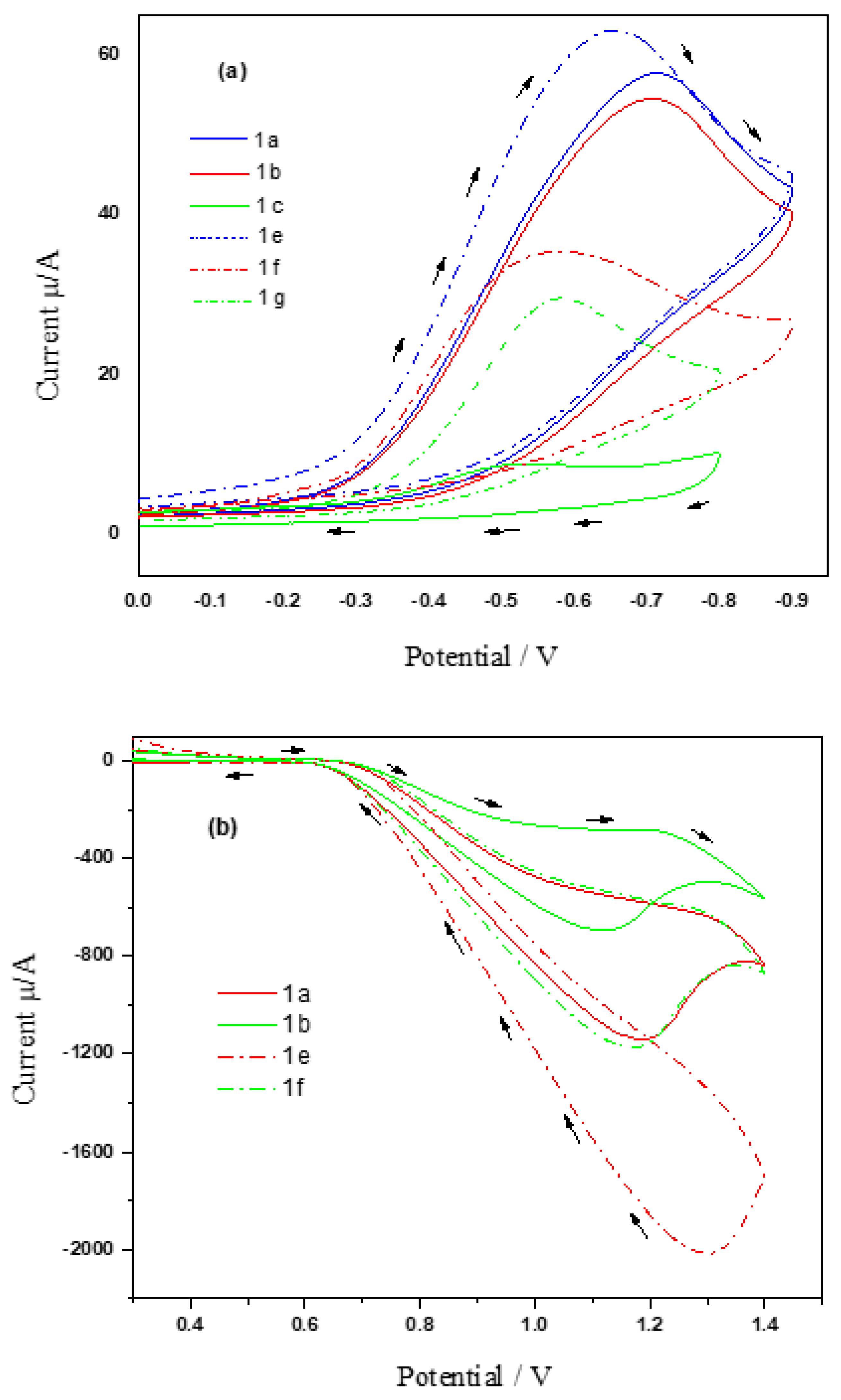
3.3. Electrochemical Studies
3.3.1. Reduction Process
3.3.2. Oxidation Process
3.4. Catalytic Activity
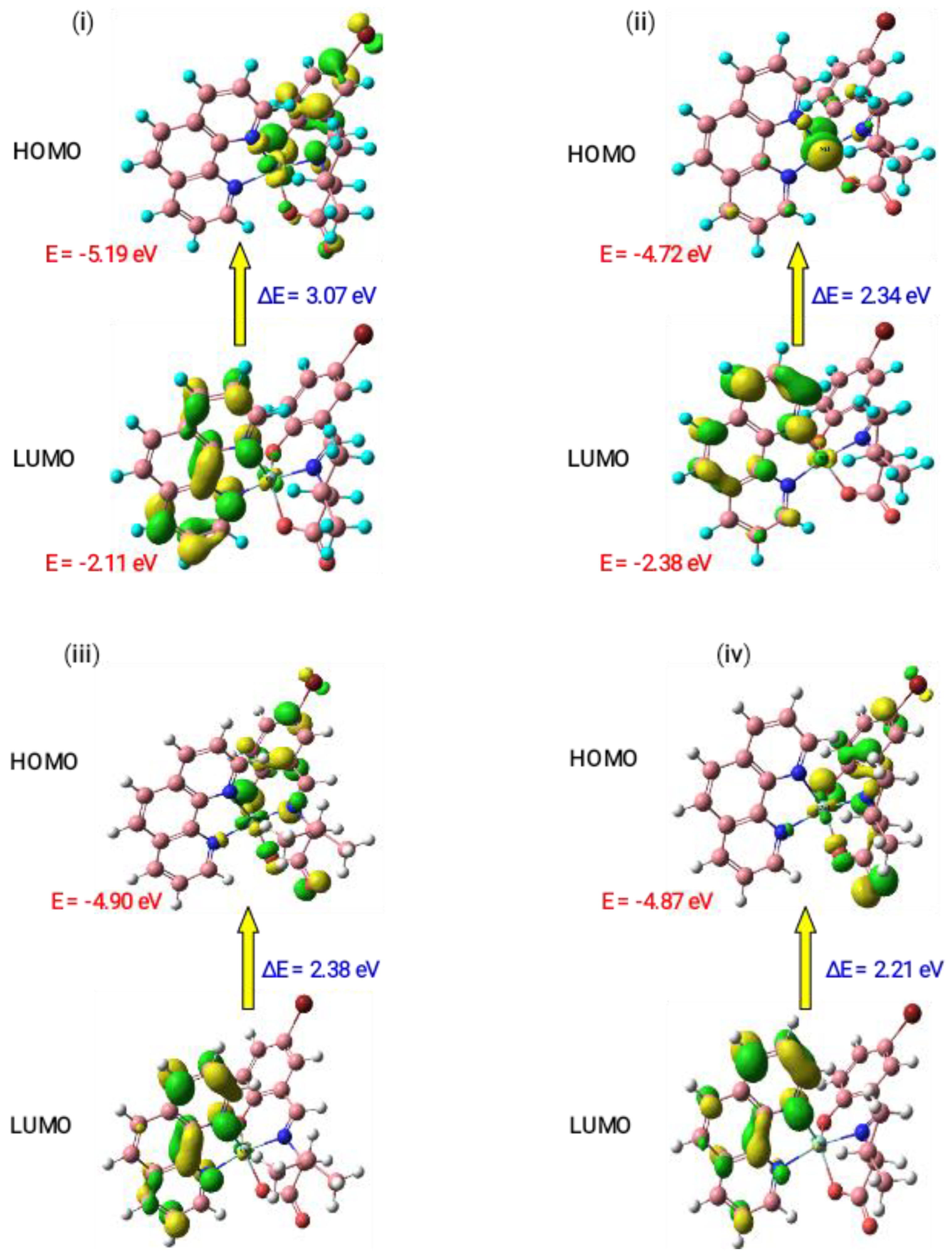
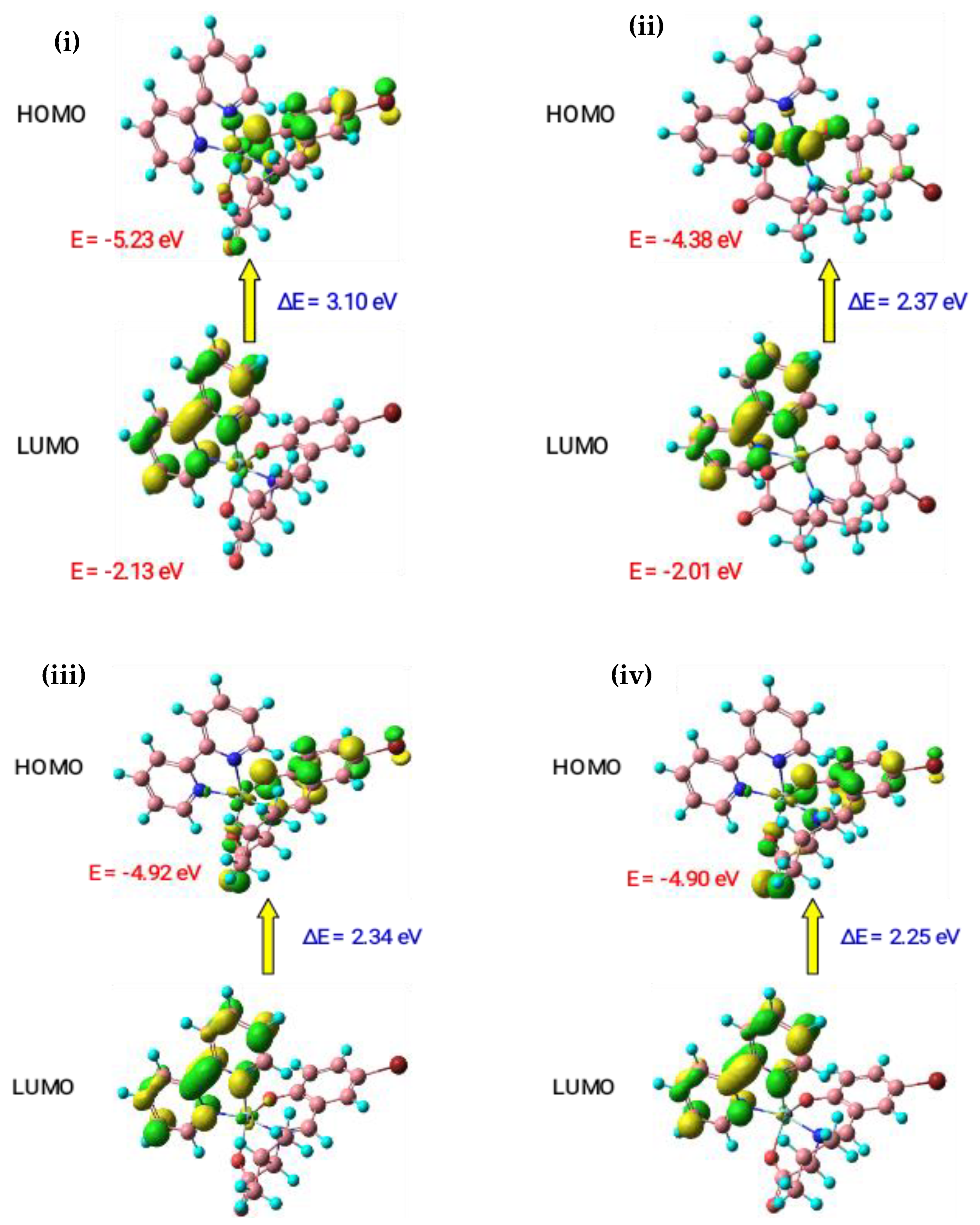
3.5. Theoretical Studies
3.5.1. Geometry Optimization
3.5.2. Molecular Orbital Analysis
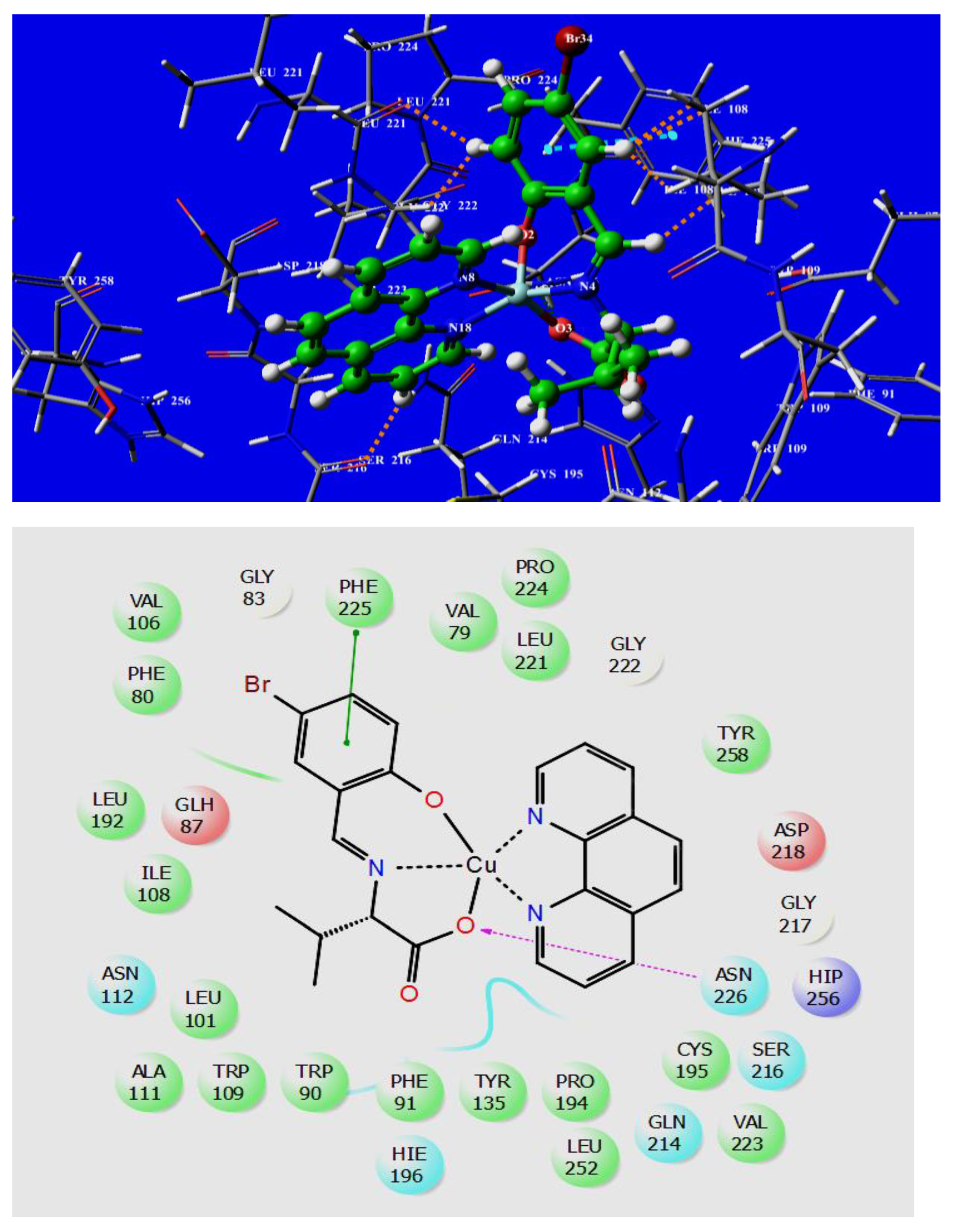
3.6. Molecular Docking Studies
Validation of the Active Site of Thymidylate Synthase
3.7. Biological Evaluation
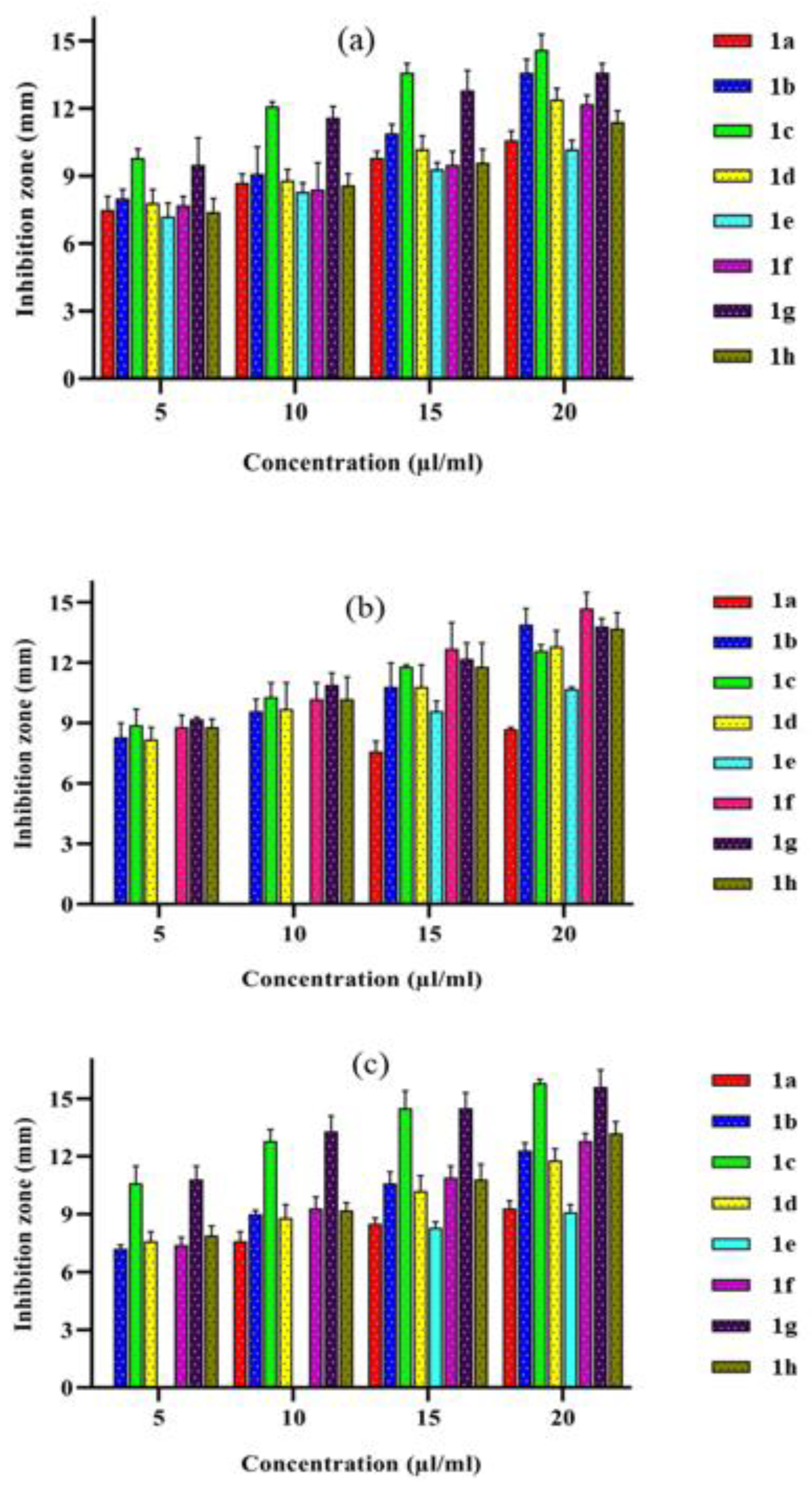
3.7.1. In Vitro Antibacterial Assay
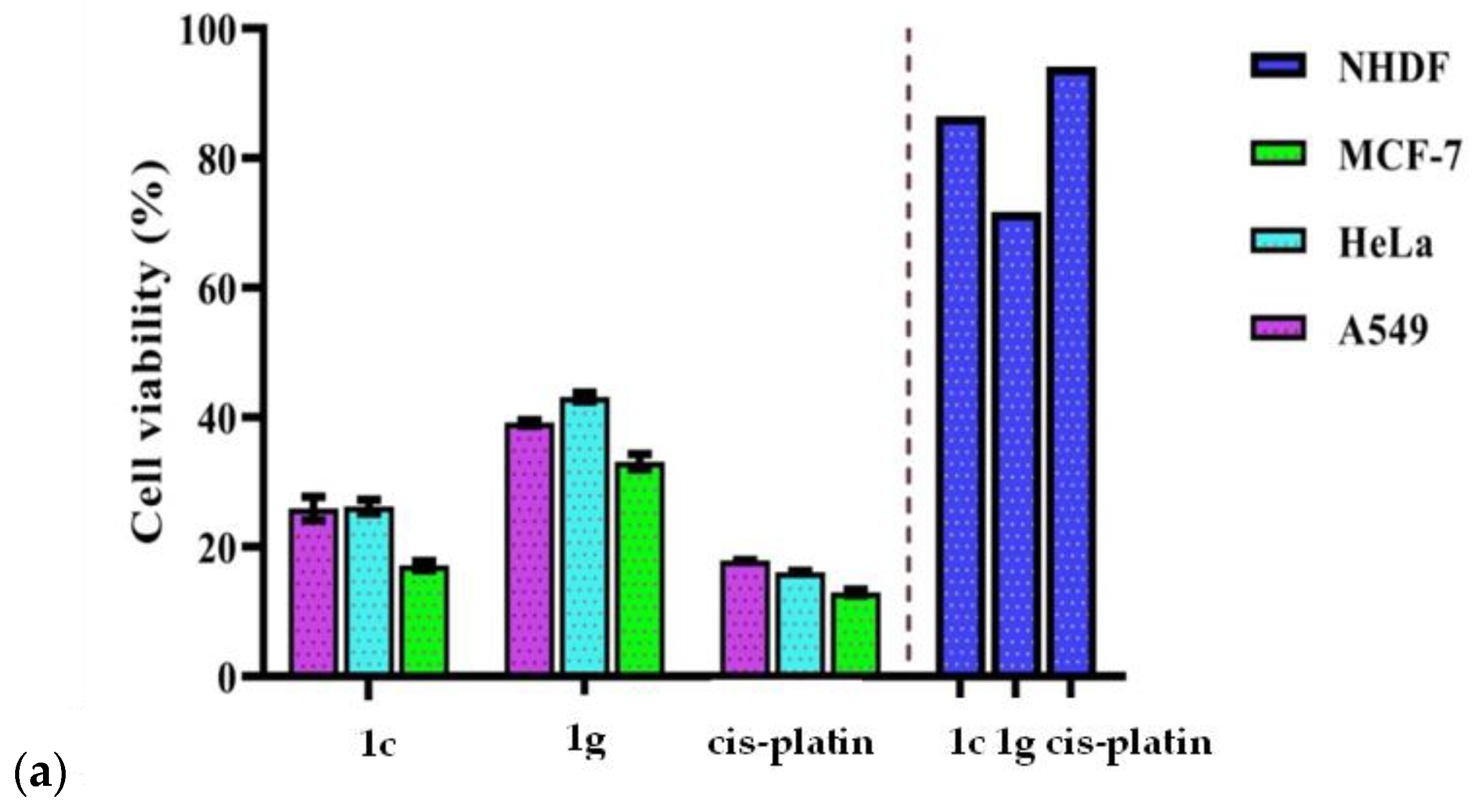
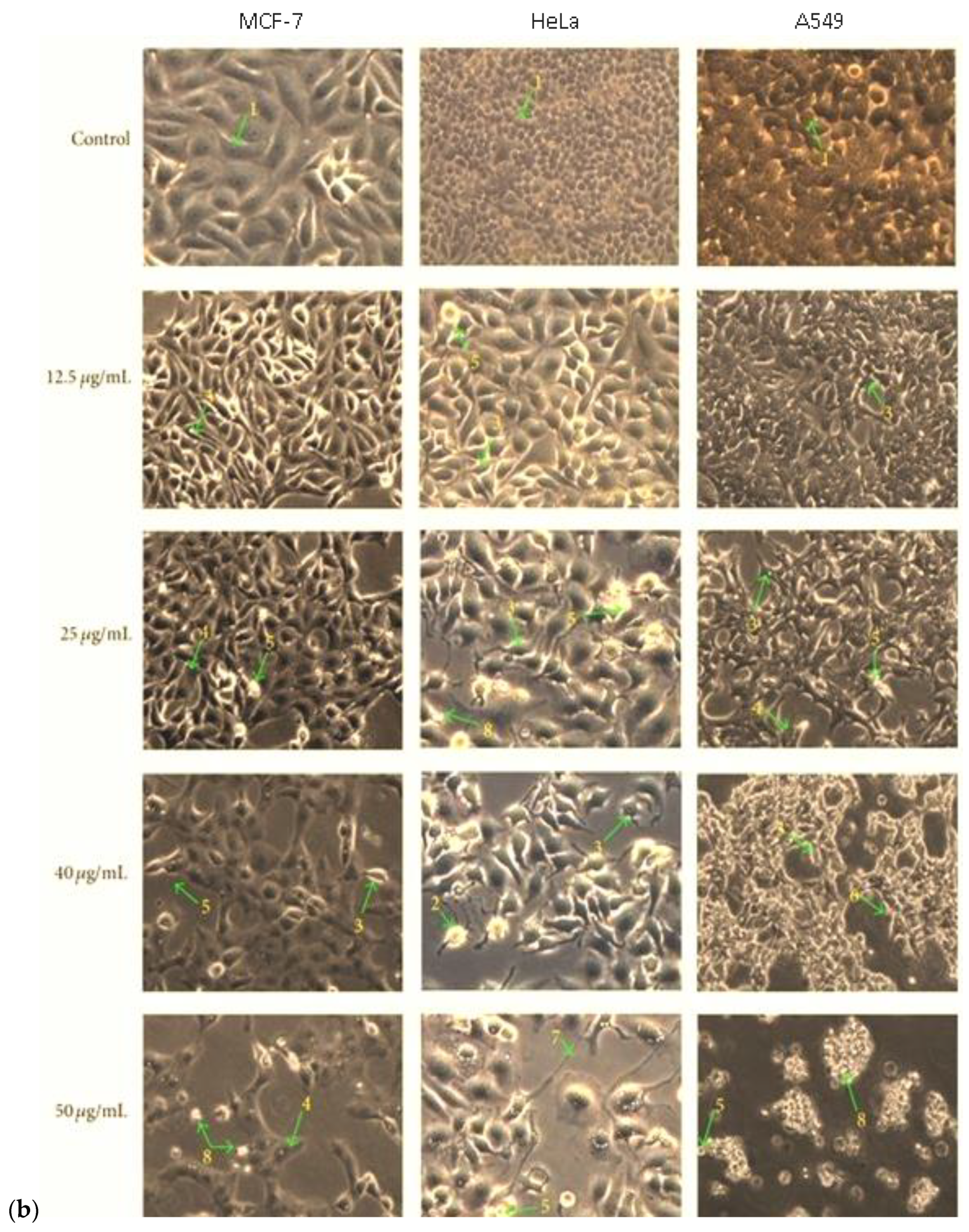
3.7.2. In Vitro Anticancer Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| bpy | 2,2′-Bipyridyl |
| DMF | N,N-Dimethylformamide |
| DPPH | 2,2′-Diphenyl-1-picrylhydrazyl |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| DFT | Density functional theory |
| EPR | Electron paramagnetic resonance spectroscopy |
| ESI-MS | Electrospray ionization mass spectroscopy |
| FBS | Fetal bovine serum |
| FMO | Frontier molecular orbital |
| HOMO | Highest occupied molecular orbital |
| IC50 | 50% of inhibitory concentration |
| LUMO | Lowest unoccupied molecular orbital |
| MTT | 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide |
| NHDF | Nontumorigenic human dermal fibroblasts |
| OPLS | Optimized potentials for liquid simulations |
| PDB | Protein data bank |
| phen | 1,10-Phenanthroline |
| RMSD | Root mean square deviation |
| TBAP | Tetra(n-butyl)ammonium perchlorate |
| UV–Vis | Ultraviolet–visible |
| XRD | X-ray diffraction |
| MCF-7 | Michigan Cancer Foundation |
| HeLa | Henrietta Lacks |
References
- Rosenberg, B.; VanCamp, L. The successful regression of large solid sarcoma 180 tumors by platinum compounds. Cancer Res. 1970, 30, 1799–1802. [Google Scholar]
- Rosenberg, B.; Vancamp, L.; Trosko, J.E.; Mansour, V.H. Platinum compounds: A new class of potent antitumour agents. Nature 1969, 222, 385–386. [Google Scholar] [CrossRef] [PubMed]
- Creaven, B.S.; Duff, B.; Egan, D.A.; Kavanagh, K.; Rosair, G.; Thangella, V.R.; Walsh, M. Anticancer and antifungal activity of copper (II) complexes of quinolin-2 (1H)-one-derived Schiff bases. Inorg. Chim. Acta 2010, 363, 4048–4058. [Google Scholar] [CrossRef]
- Sondhi, S.M.; Singh, N.; Kumar, A.; Lozach, O.; Meijer, L. Synthesis, anti-inflammatory, analgesic and kinase (CDK-1, CDK-5 and GSK-3) inhibition activity evaluation of benzimidazole/benzoxazole derivatives and some Schiff’s bases. Bioorganic Med. Chem. 2006, 14, 3758–3765. [Google Scholar] [CrossRef] [PubMed]
- Aboul-Fadl, T.; Bin-Jubair, F.A.; Aboul-Wafa, O. Schiff bases of indoline-2,3-dione (isatin) derivatives and nalidixic acid carbohydrazide, synthesis, antitubercular activity and pharmacophoric model building. Eur. J. Med. Chem. 2010, 45, 4578–4586. [Google Scholar] [CrossRef]
- Mohammed, D.F.; Madlool, H.A.; Faris, M.; Shalan, B.H.; Hasan, H.H.; Azeez, N.F.; Abbas, F.H. Harnessing inorganic nanomaterials for chemodynamic cancer therapy. Nanomedicine 2022, 17, 1891–1906. [Google Scholar] [CrossRef]
- Obaid, R.F.; Nada, K.K.H.; Samah, A.K.; Lubab, A.J.A.; Abduladheem, T.J. Antibacterial activity, anti-adherence and anti-biofilm activities of plants extracts against Aggregatibacter actinomycetemcomitans: An in vitro study in Hilla City, Iraq. Caspian J. Environ. Sci. 2022, 20, 367–372. [Google Scholar]
- Aboul-Fadl, T.; Mohammed, F.A.-H.; Hassan, E.A.-S. Synthesis, antitubercular activity and pharmacokinetic studies of some Schiff bases derived from 1-alkylisatin and isonicotinic acid hydrazide (INH). Arch. Pharmacal Res. 2003, 26, 778–784. [Google Scholar] [CrossRef]
- Ollevier, T.; Nadeau, E.; Guay-Bégin, A.-A. Direct-type catalytic three-component Mannich reaction in aqueous media. Tetrahedron Lett. 2006, 47, 8351–8354. [Google Scholar] [CrossRef]
- Manabe, K.; Mori, Y.; Wakabayashi, T.; Nagayama, S.; Kobayashi, S. Organic synthesis inside particles in water: Lewis acid− surfactant-combined catalysts for organic reactions in water using colloidal dispersions as reaction media. J. Am. Chem. Soc. 2000, 122, 7202–7207. [Google Scholar] [CrossRef]
- Akiyama, T.; Takaya, J.; Kagoshima, H. One-pot Mannich-type reaction in water: HBF4 catalyzed condensation of aldehydes, amines, and silyl enolates for the synthesis of β-amino carbonyl compounds. Synlett 1999, 1999, 1426–1428. [Google Scholar] [CrossRef]
- Manabe, K.; Mori, Y.; Kobayashi, S. Three-component carbon–carbon bond-forming reactions catalyzed by a Brønsted acid–surfactant-combined catalyst in water. Tetrahedron 2001, 57, 2537–2544. [Google Scholar] [CrossRef]
- Zhao, G.; Jiang, T.; Gao, H.; Han, B.; Huang, J.; Sun, D. Mannich reaction using acidic ionic liquids as catalysts and solvents. Green Chem. 2004, 6, 75–77. [Google Scholar] [CrossRef]
- Chang, T.; He, L.; Bian, L.; Han, H.; Yuan, M.; Gao, X. Brønsted acid-surfactant-combined catalyst for the Mannich reaction in water. RSC Adv. 2014, 4, 727–731. [Google Scholar] [CrossRef]
- Jin, Y.; Chen, D.; Zhang, X.R. Direct Asymmetric anti-Mannich-Type Reactions Catalyzed by Cinchona Alkaloid Derivatives. Chirality 2014, 26, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Wiemer, B. DD Perrin and WLF Armarego: Purification of Laboratory Chemicals. 3. Aufl., Oxford.; Pergamon Press, 1988, 391 S., zahlr. Tab., $37,50, ISBN 0-08-034714-2; Wiley Online Library: New York, NY, USA, 1989. [Google Scholar]
- Safat, S.; Buazar, F.; Albukhaty, S.; Matroodi, S. Enhanced sunlight photocatalytic activity and biosafety of marine-driven synthesized cerium oxide nanoparticles. Sci. Rep. 2021, 11, 14734. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, D.S.; Mohammed, M.K. Studying the bactericidal ability and biocompatibility of gold and gold oxide nanoparticles decorating on multi-wall carbon nanotubes. Chem. Pap. 2020, 74, 4033–4046. [Google Scholar] [CrossRef]
- Mahmood, R.I.; Kadhim, A.A.; Ibraheem, S.; Albukhaty, S.; Mohammed-Salih, H.S.; Abbas, R.H.; Jabir, M.S.; Mohammed, M.K.; Nayef, U.M.; AlMalki, F.A. Biosynthesis of copper oxide nanoparticles mediated Annona muricata as cytotoxic and apoptosis inducer factor in breast cancer cell lines. Sci. Rep. 2022, 12, 16165. [Google Scholar] [CrossRef]
- Alhomaidi, E.; Jasim, S.A.; Amin, H.I.M.; Lima Nobre, M.A.; Khatami, M.; Jalil, A.T.; Hussain Dilfy, S. Biosynthesis of silver nanoparticles using Lawsonia inermis and their biomedical application. IET Nanobiotechnol. 2022, 16, 284–294. [Google Scholar] [CrossRef]
- Alijani, H.Q.; Fathi, A.; Amin, H.I.M.; Lima Nobre, M.A.; Akbarizadeh, M.R.; Khatami, M.; Jalil, A.T.; Naderifar, M.; Dehkordi, F.S.; Shafiee, A. Biosynthesis of core–shell α-Fe2O3@ Au nanotruffles and their biomedical applications. Biomass Convers. Biorefinery 2022, 1–15. [Google Scholar] [CrossRef]
- Choudhary, M.I.; Thomsen, W.J. Bioassay Techniques for Drug Development; CRC Press: Boca Raton, FL, USA, 2001. [Google Scholar]
- Alyamani, A.A.; Al-Musawi, M.H.; Albukhaty, S.; Sulaiman, G.M.; Ibrahim, K.M.; Ahmed, E.M.; Jabir, M.S.; Al-Karagoly, H.; Aljahmany, A.A.; Mohammed, M.K. Electrospun Polycaprolactone/Chitosan Nanofibers Containing Cordia myxa Fruit Extract as Potential Biocompatible Antibacterial Wound Dressings. Molecules 2023, 28, 2501. [Google Scholar] [CrossRef]
- Faris, A.H.; Hamid, K.J.; Naji, A.M.; Mohammed, M.K.; Nief, O.A.; Jabir, M.S. Novel Mo-doped WO3/ZnO nanocomposites loaded with polyvinyl alcohol towards efficient visible-light-driven photodegradation of methyl orange. Mater. Lett. 2022, 334, 133746. [Google Scholar] [CrossRef]
- Olegovich Bokov, D.; Jalil, A.T.; Alsultany, F.H.; Mahmoud, M.Z.; Suksatan, W.; Chupradit, S.; Qasim, M.T.; Delir Kheirollahi Nezhad, P. Ir-decorated gallium nitride nanotubes as a chemical sensor for recognition of mesalamine drug: A DFT study. Mol. Simul. 2022, 48, 438–447. [Google Scholar] [CrossRef]
- Jihad, M.A.; Noori, F.T.M.; Jabir, M.S.; Albukhaty, S.; AlMalki, F.A.; Alyamani, A.A. Polyethylene Glycol Functionalized Graphene Oxide Nanoparticles Loaded with Nigella sativa Extract: A Smart Antibacterial Therapeutic Drug Delivery System. Molecules 2021, 26, 3067. [Google Scholar] [CrossRef]
- Zhong, G.-Q.; Zhong, Q. Solid–solid synthesis, characterization, thermal decomposition and antibacterial activities of zinc (II) and nickel (II) complexes of glycine–vanillin Schiff base ligand. Green Chem. Lett. Rev. 2014, 7, 236–242. [Google Scholar] [CrossRef]
- Shivankar, V.; Vaidya, R.; Dharwadkar, S.; Thakkar, N. Synthesis, characterization, and biological activity of mixed ligand Co (II) complexes of 8-hydroxyquinoline and some amino acids. Synth. React. Inorg. Met.-Org. Chem. 2003, 33, 1597–1622. [Google Scholar] [CrossRef]
- Trzesowska-Kruszynska, A. Copper complex of glycine Schiff base: In situ ligand synthesis, structure, spectral, and thermal properties. J. Mol. Struct. 2012, 1017, 72–78. [Google Scholar] [CrossRef]
- Al Rugaie, O.; Jabir, M.S.; Mohammed, M.K.; Abbas, R.H.; Ahmed, D.S.; Sulaiman, G.M.; Mohammed, S.A.; Khan, R.A.; Al-Regaiey, K.A.; Alsharidah, M. Modification of SWCNTs with hybrid materials ZnO–Ag and ZnO–Au for enhancing bactericidal activity of phagocytic cells against Escherichia coli through NOX2 pathway. Sci. Rep. 2022, 12, 17203. [Google Scholar] [CrossRef]
- Ghosh, T.; Mondal, B.; Ghosh, T.; Sutradhar, M.; Mukherjee, G.; Drew, M.G. Synthesis, structure, solution chemistry and the electronic effect of para substituents on the vanadium center in a family of mixed-ligand [VVO (ONO)(ON)] complexes. Inorg. Chim. Acta 2007, 360, 1753–1761. [Google Scholar] [CrossRef]
- Somov, N.; Chausov, F.; Zakirova, R.; Fedotova, I. Synthesis and structure of cobalt (II) complexes with nitrilotris (methylenephosphonic) acid [Co(H2O)3{NH (CH2PO3H)3}] and Na4[Co{N(CH2PO3)3}]·13H2O. Russ. J. Coord. Chem. 2015, 41, 798–804. [Google Scholar] [CrossRef]
- Kunishita, A.; Doi, Y.; Kubo, M.; Ogura, T.; Sugimoto, H.; Itoh, S. Ni (II)/H2O2 reactivity in bis [(pyridin-2-yl)methyl] amine tridentate ligand system. Aromatic hydroxylation reaction by bis (μ-oxo) dinickel (III) complex. Inorg. Chem. 2009, 48, 4997–5004. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, J.; Gurumoorthy, P.; Karthick, C.; Rahiman, A.K. Mononuclear zinc (II) complexes of 2-((2-(piperazin-1-yl)ethylimino)methyl)-4-substituted phenols: Synthesis, structural characterization, DNA binding and cheminuclease activities. J. Mol. Struct. 2014, 1062, 147–157. [Google Scholar] [CrossRef]
- Billing, D.; Dudley, R.; Hathaway, B.; Tomlinson, A. Single-crystal electronic and electron spin resonance spectra of dichloroaquo-(2,9-dimethyl-1,10-phenanthroline) copper (II). J. Chem. Soc. A Inorg. Phys. Theor. 1971, 691–696. [Google Scholar] [CrossRef]
- Hathaway, B.; Tomlinson, A. Copper (II) ammonia complexes. Coord. Chem. Rev. 1970, 5, 1–43. [Google Scholar] [CrossRef]
- Maki, A.; McGarvey, B. Electron spin resonance in transition metal chelates. I. Copper (II) bis-acetylacetonate. J. Chem. Phys. 1958, 29, 31–34. [Google Scholar] [CrossRef]
- Wasson, J.R.; Trapp, C. Electron spin resonance study of copper (II) O-alkyl-1-amidinourea complexes. Properties of the CuN42-chromophore. J. Phys. Chem. 1969, 73, 3763–3772. [Google Scholar] [CrossRef]
- Mohammed, M.M.; Ali, N.S.M.; Alalwan, H.A.; Alminshid, A.H.; Aljaafari, H.A. Synthesis of ZnO-CoO/Al2O3 nanoparticles and its application as a catalyst in ethanol conversion to acetone. Results Chem. 2021, 3, 100249. [Google Scholar] [CrossRef]
- Naji, A.M.; Kareem, S.H.; Faris, A.H.; Mohammed, M.K. Polyaniline polymer-modified ZnO electron transport material for high-performance planar perovskite solar cells. Ceram. Int. 2021, 47, 33390–33397. [Google Scholar] [CrossRef]
- Jasim, S.A.; Hachem, K.; Abed Hussein, S.; Turki Jalil, A.; Hameed, N.M.; Dehno Khalaji, A. New chitosan modified with epichlohydrin and bidentate Schiff base applied to removal of Pb2+ and Cd2+ ions. J. Chin. Chem. Soc. 2022, 69, 1051–1059. [Google Scholar] [CrossRef]
- Hammadi, A.H.; Jasim, A.M.; Abdulrazzak, F.H.; Al-Sammarraie, A.M.; Cherifi, Y.; Boukherroub, R.; Hussein, F.H. Purification for carbon nanotubes synthesized by flame fragments deposition via hydrogen peroxide and acetone. Materials 2020, 13, 2342. [Google Scholar] [CrossRef]
- El-Sonbati, A.; Diab, M.; El-Bindary, A.; Eldesoky, A.; Morgan, S.M. Correlation between ionic radii of metals and thermal decomposition of supramolecular structure of azodye complexes. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 135, 774–791. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.K.; Al-Mousoi, A.K.; Khalaf, H.A. Deposition of multi-layer graphene (MLG) film on glass slide by flame synthesis technique. Optik 2016, 127, 9848–9852. [Google Scholar] [CrossRef]
- Al-Mousoi, A.K.; Mohammed, M.K.; Khalaf, H.A. Preparing and characterization of indium arsenide (InAs) thin films by chemical spray pyrolysis (CSP) technique. Optik 2016, 127, 5834–5840. [Google Scholar] [CrossRef]
- Benzekri, A.; Dubourdeaux, P.; Latour, J.-M.; Rey, P.; Laugier, J. Binuclear copper (II) complexes of a new sulphur-containing binucleating ligand: Structural and physicochemical properties. J. Chem. Soc. Dalton Trans. 1991, 12, 3359–3365. [Google Scholar] [CrossRef]
- Santra, B.K.; Reddy, P.A.; Nethaji, M.; Chakravarty, A.R. Structural model for the CuB site of dopamine β-hydroxylase: Crystal structure of a copper (II) complex showing N3OS coordination with an axial sulfur ligation. Inorg. Chem. 2002, 41, 1328–1332. [Google Scholar] [CrossRef]
- Azevedo, F.; de CT Carrondo, M.A.; de Castro, B.; Convery, M.; Domingues, D.; Freire, C.; Duarte, M.T.; Nielsen, K.; Santos, I.C. Electrochemical and structural studies of nickel (II) complexes with N2O2 Schiff base ligands 2. Crystal and molecular structure of N,N′-l,2-ethane-1,2-diyl-bis(2-hydroxyacetophenonylideneiminate) nickel (II), N, N′-1,2-cis cyclohexane-1,2-diyl-bis(2-hydroxyacetophenonylideneiminate)-nickel (II) and N,N′-1,2-benzene-1,2-diyl-bis(3,5-dichlorosalicylideneiminate) nickel (II). Inorg. Chim. Acta 1994, 219, 43–54. [Google Scholar]
- Addison, A.W.; Rao, T.N.; Reedijk, J.; van Rijn, J.; Verschoor, G.C. Synthesis, structure, and spectroscopic properties of copper (II) compounds containing nitrogen–sulphur donor ligands; the crystal and molecular structure of aqua [1, 7-bis (N-methylbenzimidazol-2′-yl)-2, 6-dithiaheptane] copper (II) perchlorate. J. Chem. Soc. Dalton Trans. 1984, 7, 1349–1356. [Google Scholar] [CrossRef]
- Kivelson, D.; Neiman, R. ESR studies on the bonding in copper complexes. J. Chem. Phys. 1961, 35, 149–155. [Google Scholar] [CrossRef]
- El-Ghamaz, N.; Ghoneim, M.; El-Sonbati, A.; Diab, M.; El-Bindary, A.; Abd El-Kader, M. Synthesis and optical properties studies of antipyrine derivatives thin films. J. Saudi Chem. Soc. 2017, 21, S339–S348. [Google Scholar] [CrossRef]
- Sasikumar, G.; Arulmozhi, S.; Ashma, A.; Sudha, A. Mixed ligand ternary complexes of Co (II), Ni (II), Cu (II) and Zn (II) and their structural characterization, electrochemical, theoretical and biological studies. J. Mol. Struct. 2019, 1187, 108–120. [Google Scholar] [CrossRef]
- Al-Awsi, G.R.L.; Alsudani, A.A.; Omran, F.K. The Antibacterial Activity of Althaea Officinalis L. Methanolic Extract against Some Nosocomial Pathogens In Vitro and In Vivo; IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2021; p. 012013. [Google Scholar]
- Alsudani, A.A.; Mohammed, G.J.; Al-Awsi, G.R.L. In Vitro, the Antimicrobial Activity of Some Medicinal Plant Extracts on the Growth of Some Bacterial and Fungal Pathogens. J. Phys. Conf. Ser. 2019, 1294, 062099. [Google Scholar] [CrossRef]
- Tweedy, B. Plant extracts with metal ions as potential antimicrobial agents. Phytopathology 1964, 55, 910–914. [Google Scholar]
- Erkkila, K.E.; Odom, D.T.; Barton, J.K. Recognition and reaction of metallointercalators with DNA. Chem. Rev. 1999, 99, 2777–2796. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.-N.; Zou, X.-H.; Liu, J.-G. Shape-and enantioselective interaction of Ru (II)/Co (III) polypyridyl complexes with DNA. Coord. Chem. Rev. 2001, 216, 513–536. [Google Scholar] [CrossRef]

| Complexes | Formula | Color | Mol.Wt. gm/mol | M.p. (°C) | Yield (%) | Found (calc.) (%) | ||
|---|---|---|---|---|---|---|---|---|
| C | H | N | ||||||
| [Co(L)(phen)] 1a | C24H20BrCoN3O3 | Brown | 537.08 | >300 | 80 | 53.63 (53.65) | 3.72 (3.75) | 7.81 (7.82) |
| [Ni(L)(phen)] 1b | C24H20BrN3NiO3 | Pale green | 536.92 | >300 | 80 | 53.66 (53.68) | 3.73 (3.75) | 7.80 (7.82) |
| [Cu(L)(phen)] 1c | C24H20BrCuN3O3 | green | 541.78 | >300 | 88 | 53.19 (53.20) | 3.70 (3.72) | 7.72 (7.75) |
| [Zn(L)(phen)] 1d | C24H20BrN3O3Zn | Pale yellow | 543.28 | >300 | 67 | 52.98 (53.02) | 3.69 (3.71) | 7.72 (7.73) |
| [Co(L)(bpy)] 1e | C22H20BrCoN3O3 | Brown | 513.03 | >300 | 84 | 51.46 (51.48) | 3.92 (3.93) | 8.18 (8.19) |
| [Ni(L)(bpy)] 1f | C22H20BrN3NiO3 | Pale green | 512.89 | >300 | 79 | 51.49 (51.51) | 3.91 (3.93) | 8.17 (8.19) |
| [Cu(L)(bpy)] 1g | C22H20BrCuN3O3 | green | 517.69 | >300 | 83 | 51.00 (51.02) | 3.88 (3.89) | 8.09 (8.11) |
| [Zn(L)(bpy) ] 1h | C22H20BrN3O3Zn | Pale yellow | 519.38 | >300 | 67 | 50.81 (50.84) | 3.86 (3.88) | 8.06 (8.09) |
| Complexes | FT-IR Spectral Data (cm−1) | UV–Visible Spectral Data (λmax/nm (ε/M−1cm−1dm3) | |||||
|---|---|---|---|---|---|---|---|
| ν(-C=N-) | νas(COO) | νs(COO) | ν(M-O) | ν(M-N) | d-d | Charge Transfer | |
| HL | 1643 | 1594 | 1405 | -- | -- | -- | 409 (500), 353 (180), 264 (1590) |
| [Co(L)(phen)] 1a | 1636 | 1587 | 1371 | 549 | 457 | 494 (152) 634 (58) | 228 (33,895) (π–π*) 269 (25,834) (n–π*) 337 (1209) (LMCT) |
| [Ni(L)(phen)] 1b | 1612 | 1586 | 1380 | 542 | 460 | 623 (6) | 254 (57,302) (π–π*) 288 (36,331) (n–π*) 389 (3657) (LMCT) |
| [Cu(L)(phen)] 1c | 1611 | 1562 | 1356 | 554 | 486 | 637 (17) | 259 (864,222) (π–π*) 284 (58,665) (n–π*) 372 (1502) (LMCT) |
| [Zn(L)(phen)] 1d | 1622 | 1557 | 1376 | 549 | 502 | -- | 270 (35,064) (π–π*) 292 (9154) (n–π*) 373 (4019) (LMCT |
| [Co(L)(bpy)] 1e | 1623 | 1575 | 1369 | 562 | 488 | 463 (127) 619 (65) | 249 (19,579)(π–π*) 301 (8048) (n–π*) 389 (854) (LMCT) |
| [Ni(L)(bpy)] 1f | 1634 | 1570 | 1379 | 542 | 485 | 625 (4) | 238 (4056) (π–π*) 298 (2094) (n–π*) 379 (920) (LMCT) |
| [Cu(L)(bpy)] 1g | 1594 | 1568 | 1358 | 553 | 461 | 642 (24) | 240 (13,846) (π–π*) 300 (11,085) (n–π*) 375 (144) (LMCT |
| [Zn(L3)(bpy)] 3h | 1635 | 1577 | 1370 | 548 | 497 | -- | 274 (20,198) (π–π*) 294 (16,748) (n–π*) 374 (8425) (LMCT) |
| Complexes | Epc (V) | Epa (V) | ∆Ep (V) | Ipc (μA) | Ipa (μA) | Ipc/Ipa |
|---|---|---|---|---|---|---|
| [Co(L)(phen)] 1a | −0.71 | +1.19 | 1.90 | 57.63 | −1138 | 0.050 |
| [Ni(L)(phen)] 1b | −0.71 | +1.12 | 1.83 | 54.50 | −694 | 0.078 |
| [Cu(L)(phen)] 1c | −0.54 | -- | -- | 8.90 | -- | -- |
| [Zn(L)(phen)] 1d | NA * | NA | -- | -- | -- | -- |
| [Co(L)(bpy)] 1e | −0.66 | +1.30 | 1.96 | 62.97 | −2015 | 0.031 |
| [Ni(L)(bpy)] 1f | −0.58 | +1.17 | 1.75 | 35.40 | −1174 | 0.030 |
| [Cu(L)(bpy)] 1g | −0.58 | -- | -- | 29.55 | -- | -- |
| [Zn(L)(bpy)] 1h | NA | NA | -- | -- | -- | -- |
| S.No | Catalyst | Deri- vative | Concentration of Substituted Phenanthroline Metal Salts | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.0367 mmol | 0.0735 mmol | 0.1103 mmol | |||||||||||||
| CR | MR | CR | MR | CR | MR | ||||||||||
| T | Y | T | Y | T | Y | T | Y | T | Y | T | Y | ||||
| 1 | Substituted phenanthroline metal (II) salts | 1a | 2a | 85 | 65 | 40 | 75 | 75 | 76 | 30 | 82 | 65 | 88 | 20 | 97 |
| 2b | 95 | 64 | 45 | 71 | 85 | 74 | 35 | 79 | 75 | 86 | 25 | 94 | |||
| 2c | 75 | 66 | 35 | 75 | 70 | 77 | 30 | 82 | 60 | 92 | 20 | 90 | |||
| 2d | 65 | 73 | 30 | 80 | 60 | 83 | 25 | 84 | 50 | 91 | 15 | 93 | |||
| 2e | 55 | 76 | 25 | 80 | 45 | 88 | 20 | 87 | 40 | 90 | 10 | 95 | |||
| 2 | 1b | 2a | 95 | 62 | 50 | 71 | 85 | 74 | 40 | 80 | 75 | 86 | 30 | 94 | |
| 2b | 105 | 60 | 55 | 73 | 95 | 71 | 45 | 74 | 85 | 83 | 35 | 91 | |||
| 2c | 85 | 61 | 45 | 72 | 80 | 75 | 40 | 80 | 70 | 90 | 30 | 88 | |||
| 2d | 75 | 71 | 40 | 78 | 70 | 78 | 35 | 77 | 60 | 88 | 25 | 89 | |||
| 2e | 65 | 72 | 35 | 76 | 55 | 82 | 30 | 85 | 50 | 86 | 20 | 91 | |||
| 3 | 1c | 2a | 105 | 60 | 60 | 69 | 95 | 70 | 50 | 78 | 85 | 82 | 40 | 92 | |
| 2b | 115 | 57 | 65 | 70 | 105 | 67 | 55 | 71 | 95 | 80 | 45 | 88 | |||
| 2c | 95 | 59 | 55 | 68 | 90 | 72 | 50 | 77 | 80 | 82 | 40 | 90 | |||
| 2d | 85 | 67 | 50 | 79 | 80 | 76 | 45 | 78 | 70 | 82 | 35 | 89 | |||
| 2e | 75 | 72 | 45 | 76 | 65 | 82 | 40 | 85 | 60 | 86 | 30 | 91 | |||
| 4 | 1d | 2a | 115 | 59 | 70 | 64 | 105 | 69 | 60 | 72 | 95 | 80 | 50 | 90 | |
| 2b | 125 | 57 | 75 | 70 | 115 | 67 | 65 | 71 | 105 | 80 | 55 | 88 | |||
| 2c | 105 | 59 | 55 | 65 | 90 | 70 | 50 | 75 | 80 | 80 | 40 | 88 | |||
| 2d | 95 | 65 | 50 | 77 | 80 | 73 | 45 | 75 | 70 | 81 | 35 | 88 | |||
| 2e | 75 | 70 | 45 | 72 | 65 | 79 | 40 | 83 | 60 | 82 | 30 | 90 | |||
| S.No | Catalyst | Deri- vative | Concentration of Substituted Pyridine Metal Salts | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.0384 mmol | 0.0769 mmol | 0.1154 mmol | |||||||||||||
| CR | MR | CR | MR | CR | MR | ||||||||||
| T | Y | T | Y | T | Y | T | Y | T | Y | T | Y | ||||
| 1 | Substituted 2,2-bipyidyl metal (II) salts | 1e | 2a | 75 | 68 | 35 | 78 | 70 | 79 | 20 | 85 | 55 | 91 | 15 | 96 |
| 2b | 85 | 67 | 40 | 74 | 80 | 77 | 25 | 82 | 65 | 89 | 20 | 92 | |||
| 2c | 65 | 69 | 30 | 79 | 65 | 80 | 20 | 85 | 50 | 95 | 15 | 95 | |||
| 2d | 55 | 76 | 25 | 83 | 55 | 86 | 15 | 87 | 40 | 94 | 10 | 97 | |||
| 4e | 45 | 79 | 20 | 84 | 40 | 91 | 10 | 90 | 30 | 93 | 05 | 98 | |||
| 2 | 1f | 2a | 85 | 65 | 45 | 74 | 75 | 77 | 30 | 83 | 65 | 89 | 25 | 92 | |
| 2b | 95 | 63 | 50 | 77 | 85 | 73 | 35 | 77 | 75 | 86 | 30 | 90 | |||
| 2c | 75 | 64 | 40 | 75 | 70 | 78 | 30 | 83 | 60 | 93 | 25 | 91 | |||
| 2d | 65 | 74 | 35 | 81 | 60 | 81 | 25 | 81 | 50 | 91 | 20 | 89 | |||
| 2e | 55 | 75 | 30 | 79 | 45 | 85 | 20 | 88 | 40 | 89 | 15 | 91 | |||
| 3 | 1g | 2a | 95 | 63 | 55 | 71 | 85 | 73 | 40 | 81 | 75 | 85 | 35 | 90 | |
| 2b | 105 | 60 | 60 | 73 | 95 | 70 | 45 | 74 | 85 | 83 | 40 | 89 | |||
| 2c | 85 | 62 | 50 | 71 | 80 | 75 | 40 | 80 | 70 | 85 | 35 | 89 | |||
| 2d | 75 | 70 | 45 | 82 | 70 | 79 | 25 | 81 | 60 | 86 | 30 | 90 | |||
| 2e | 65 | 75 | 40 | 79 | 55 | 85 | 30 | 88 | 50 | 89 | 25 | 92 | |||
| 4 | 1h | 2a | 105 | 63 | 65 | 68 | 95 | 72 | 50 | 75 | 85 | 85 | 45 | 91 | |
| 2b | 115 | 61 | 70 | 73 | 105 | 71 | 55 | 74 | 95 | 83 | 50 | 90 | |||
| 2c | 95 | 62 | 50 | 65 | 80 | 73 | 40 | 78 | 70 | 84 | 35 | 89 | |||
| 2d | 85 | 69 | 45 | 77 | 70 | 76 | 35 | 81 | 60 | 86 | 30 | 92 | |||
| 2e | 65 | 73 | 40 | 72 | 55 | 82 | 30 | 87 | 60 | 85 | 25 | 90 | |||
| Catalyst | Percentage of Recyclability of Catalyst | |||
|---|---|---|---|---|
| First Cycle | Second Cycle | Third Cycle | Fourth Cycle | |
| 1a | 91 | 91 | 90 | 89 |
| 1b | 89 | 89 | 87 | 87 |
| 1c | 87 | 87 | 87 | 85 |
| 1d | 92 | 91 | 91 | 90 |
| 1e | 90 | 89 | 89 | 88 |
| 1f | 88 | 88 | 88 | 86 |
| 1g | 90 | 89 | 89 | 88 |
| 1h | 89 | 88 | 88 | 86 |
| Bond Angle (deg) B3LYP/LACVP++ | |||||||
|---|---|---|---|---|---|---|---|
| [Co(L)(phen)] 1a | [Ni(L)(phen)] 1b | [Cu(L)(phen)] 1c | [Zn(L1)(phen)] 1d | ||||
| N(18)-Co(1)-N(8) | 84.536 | N(18)-Ni(1)-N(8) | 85.326 | N(18)-Cu(1)-N(8) | 81.547 | N(18)-Zn(1)-N(8) | 79.945 |
| N(18)-Co(1)-N(4) | 171.246 | N(18)-Ni(1)-N(4) | 153.435 | N(18)-Cu(1)-N(4) | 154.944 | N(18)-Zn(1)-N(4) | 151.161 |
| N(18)-Co(1)-O(3) | 85.923 | N(18)-Ni(1)-O(3) | 86.310 | N(18)-Cu(1)-O(3) | 85.065 | N(18)-Zn(1)-O(3) | 81.925 |
| N(18)-Co(1)-O(2) | 98.447 | N(18)-Ni(1)-O(2) | 113.717 | N(18)-Cu(1)-O(2) | 108.729 | N(18)-Zn(1)-O(2) | 113.265 |
| N(8)-Co(1)-N(4) | 98.118 | N(8)-Ni(1)-N(4) | 94.750 | N(8)-Cu(1)-N(4) | 95.945 | N(8)-Zn(1)-N(4) | 98.121 |
| N(8)-Co(1)-O(3) | 126.249 | N(8)-Ni(1)-O(3) | 156.955 | N(8)-Cu(1)-O(3) | 149.119 | N(8)-Zn(1)-O(3) | 135.705 |
| N(8)-Co(1)-O(2) | 101.038 | N(8)-Ni(1)-O(2) | 94.179 | N(8)-Cu(1)-O(2) | 95.100 | N(8)-Zn(1)-O(2) | 103.250 |
| N(4)-Co(1)-O(3) | 84.163 | N(4)-Ni(1)-O(3) | 83.387 | N(4)-Cu(1)-O(3) | 82.697 | N(4)-Zn(1)-O(3) | 79.707 |
| N(4)-Co(1)-O(2) | 92.301 | N(4)-Ni(1)-O(2) | 92.797 | N(4)-Cu(1)-O(2) | 94.313 | N(4)-Zn(1)-O(2) | 95.274 |
| O(3)-Co(1)-O(2) | 123.606 | O(3)-Ni(1)-O(2) | 108.843 | O(3)-Cu(1)-O(2) | 112.778 | O(3)-Zn(1)-O(2) | 121.039 |
| Bond distance (Å) | |||||||
| Co(1)-N(18) | 1.870 | Ni(1)-N(18) | 1.836 | Cu(1)-N(18) | 1.934 | Zn(1)-N(18) | 2.029 |
| Co(1)-N(8) | 1.918 | Ni(1)-N(8) | 1.884 | Cu(1)-N(8) | 2.059 | Zn(1)-N(8) | 2.049 |
| Co(1)-N(4) | 1.871 | Ni(1)-N(4) | 1.845 | Cu(1)-N(4) | 1.912 | Zn(1)-N(4) | 2.004 |
| Co(1)-O(3) | 1.837 | Ni(1)-O(3) | 1.822 | Cu(1)-O(3) | 1.882 | Zn(1)-O(3) | 1.937 |
| Co(1)-O(2) | 1.918 | Ni(1)-O(2) | 2.014 | Cu(1)-O(2) | 1.932 | Zn(1)-O(2) | 1.873 |
| Bond Angle (deg) B3LYP/LACVP++ | |||||||
|---|---|---|---|---|---|---|---|
| [Co(L)(bpy)] 1e | [Ni(L)(bpy)] 1f | [Cu(L)(bpy)] 1g | [Zn(L)(bpy)] 1h | ||||
| N(16)-Co(1)-N(8) | 83.244 | N(16)-Ni(1)-N(8) | 78.400 | N(16)-Cu(1)-N(8) | 80.539 | N(16)-Zn(1)-N(8) | 78.477 |
| N(16)-Co(1)-N(4) | 172.888 | N(16)-Ni(1)-N(4) | 120.752 | N(16)-Cu(1)-N(4) | 152.716 | N(16)-Zn(1)-N(4) | 151.914 |
| N(16)-Co(1)-O(3) | 86.817 | N(16)-Ni(1)-O(3) | 79.803 | N(16)-Cu(1)-O(3) | 85.620 | N(16)-Zn(1)-O(3) | 82.922 |
| N(16)-Co(1)-O(2) | 97.946 | N(16)-Ni(1)-O(2) | 105.384 | N(16)-Cu(1)-O(2) | 113.072 | N(16)-Zn(1)-O(2) | 112.970 |
| N(8)-Co(1)-N(4) | 99.500 | N(8)-Ni(1)-N(4) | 158.557 | N(8)-Cu(1)-N(4) | 96.460 | N(8)-Zn(1)-N(4) | 98.792 |
| N(8)-Co(1)-O(3) | 125.104 | N(8)-Ni(1)-O(3) | 87.110 | N(8)-Cu(1)-O(3) | 148.646 | N(8)-Zn(1)-O(3) | 134.815 |
| N(8)-Co(1)-O(2) | 101.888 | N(8)-Ni(1)-O(2) | 88.367 | N(8)-Cu(1)-O(2) | 97.421 | N(8)-Zn(1)-O(2) | 104.122 |
| N(4)-Co(1)-O(3) | 83.670 | N(4)-Ni(1)-O(3) | 87.159 | N(4)-Cu(1)-O(3) | 81.407 | N(4)-Zn(1)-O(3) | 79.510 |
| N(4)-Co(1)-O(2) | 92.045 | N(4)-Ni(1)-O(2) | 94.828 | N(4)-Cu(1)-O(2) | 94.212 | N(4)-Zn(1)-O(2) | 94.892 |
| O(3)-Co(1)-O(2) | 122.850 | O(3)-Ni(1)-O(2) | 162.290 | O(3)-Cu(1)-O(2) | 117.930 | O(3)-Zn(1)-O(2) | 121.051 |
| Bond distance (Å) | |||||||
| Co(1)-N(16) | 1.855 | Ni(1)-N(16) | 2.289 | Cu(1)-N(16) | 1.929 | Zn(1)-N(16) | 2.026 |
| Co(1)-N(8) | 1.897 | Ni(1)-N(8) | 1.865 | Cu(1)-N(8) | 2.002 | Zn(1)-N(8) | 2.030 |
| Co(1)-N(4) | 1.880 | Ni(1)-N(4) | 1.790 | Cu(1)-N(4) | 1.928 | Zn(1)-N(4) | 2.013 |
| Co(1)-O(3) | 1.840 | Ni(1)-O(3) | 1.805 | Cu(1)-O(3) | 1.890 | Zn(1)-O(3) | 1.933 |
| Co(1)-O(2) | 1.917 | Ni(1)-O(2) | 1.804 | Cu(1)-O(2) | 1.938 | Zn(1)-O(2) | 1.873 |
| Complexes | HOMO (eV) | LUMO (eV) | ΔE (eV) | χ | η | σ | Pi | S | ω | ΔN Max |
|---|---|---|---|---|---|---|---|---|---|---|
| [Co(L)(phen)] 1a | −5.19 | −2.11 | 3.07 | 3.65 | 1.54 | 0.65 | −3.65 | 0.33 | 10.24 | −2.38 |
| [Ni(L)(phen)] 1b | −4.72 | −2.38 | 2.34 | 3.55 | 1.17 | 0.85 | −3.55 | 0.43 | 7.36 | −3.03 |
| [Cu(L)(phen)] 1c | −4.90 | −2.52 | 2.38 | 3.71 | 1.19 | 0.84 | −3.71 | 0.42 | 8.18 | −3.11 |
| [Zn(L)(phen)] 1d | −4.87 | −2.67 | 2.21 | 3.77 | 1.10 | 0.91 | −3.77 | 0.45 | 7.85 | −3.41 |
| [Co(L)(bpy)] 1e | −5.23 | −2.13 | 3.10 | 3.68 | 1.55 | 0.64 | −3.68 | 0.32 | 10.52 | −2.37 |
| [Ni(L)(bpy)] 1f | −4.38 | −2.01 | 2.37 | 3.19 | 1.19 | 0.84 | −3.19 | 0.42 | 6.05 | −2.69 |
| [Cu(L)(bpy)] 1g | −4.92 | −2.58 | 2.34 | 3.75 | 1.17 | 0.85 | −3.75 | 0.43 | 8.25 | −3.20 |
| [Zn(L)(bpy)] 1h | −4.90 | −2.66 | 2.25 | 3.78 | 1.12 | 0.89 | −3.78 | 0.45 | 8.02 | −3.37 |
| Complexes | Docking Score kcal.mol−1 | Active Sites with a Mode of Interaction | ||
|---|---|---|---|---|
| H-bond | Π–π Stacking | Hydrophobic Interactions (Cutoff at 5Å) | ||
| [Co(L)(phen)] 1a | −5.700 | -- | PHE 225 | PHE 80, PHE 91, ILE 108, TRP 109, TYR 135, LEU 192, CYS 195, LEU 221, VAL 223, PRO 224, PHE 225, TYR 258 |
| [Ni(L)(phen)] 1b | −5.615 | -- | PHE 225 | PHE 80, PHE 91, ILE 108, TRP 109, TYR 135, LEU 192, CYS 195, LEU 221, VAL 223, PRO 224, PHE 225, TYR 258 |
| [Cu(L)(phen)] 1c | −5.791 | ASN 226 | PHE 225 | PHE 80, PHE 91, ILE 108, TRP 109, TYR 135, LEU 192, PRO 193, PRO 194, CYS 195, VAL 223, PHE 225, VAL 238, TYR 258 |
| [Zn(L)(phen)] 1d | −5.367 | ASN 226 | -- | PHE 80, PHE 91, ILE 108, TRP 109, TYR 135, LEU 192, CYS 195, LEU 221, VAL 223, PHE 225, TYR 258 |
| [Co(L)(bpy)] 1e | −5.49 | ASN 226 | -- | PHE 80, TRP 90, LEU 101, ALA 111 ILE 108, TRP 109, ALA 111, TYR 135, LEU 192, CYS 195, LEU 221, VAL 223, PHE 225 |
| [Ni(L)(bpy)] 1f | −4.97 | -- | TRP 109 | PHE 80, TRP 90, LEU 101, ALA 111 ILE 108, TRP 109, ALA 111, TYR 135, LEU 192, PRO 193, PRO 194, CYS 195, LEU 221, VAL 223, PHE 225 |
| [Cu(L)(bpy)] 1g | −5.026 | -- | PHE 225 | PHE 80, PHE 91, ILE 108, TRP 109, ALA 111, TYR 135, LEU 192, CYS 195, LEU 221, VAL 223, PRO 224, PHE 225, TYR 258 |
| [Zn(L)(bpy)] 1h | −5.223 | ASN 226 | -- | PHE 80, PHE 91, ILE 108, TRP 109, TYR 135, LEU 192, CYS 195, LEU 221, VAL 223, PHE 225, TYR 258 |
| Complexes | Inhibition Zone Measured (mm) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli | Klebsiella pneumonia | Staphylococcus aureus | ||||||||||
| Concentration (µL/mL) | ||||||||||||
| 5 | 10 | 15 | 20 | 5 | 10 | 15 | 20 | 5 | 10 | 15 | 20 | |
| [Co(L)(phen)] 1a | 7.5 ± 0.6 | 8.7 ± 0.4 | 9.8 ± 0.3 | 10.6 ± 0.4 | -- | -- | 7.6 ± 0.5 | 8.7 ± 0.1 | -- | 7.6 ± 0.5 | 8.5 ± 0.3 | 9.3 ± 0.4 |
| [Ni(L)(phen)] 1b | 8.0 ± 0.4 | 9.1 ± 1.2 | 10.9 ± 0.4 | 13.6 ± 0.6 | 8.3 ± 0.7 | 9.6 ± 0.6 | 10.8 ± 1.2 | 13.9 ± 0.8 | 7.2 ± 0.2 | 9.0 ± 0.2 | 10.6 ± 0.6 | 12.3 ± 0.4 |
| [Cu(L)(phen)] 1c | 9.8 ± 0.4 | 12.1 ± 0.2 | 13.6 ± 0.4 | 14.6 ± 0.7 | 8.9 ± 0.8 | 10.3 ± 0.7 | 11.8 ± 0.1 | 12.6 ± 0.3 | 10.6 ± 0.9 | 12.8 ± 0.6 | 14.5 ± 0.9 | 15.8 ± 0.2 |
| [Zn(L)(phen)] 1d | 7.8 ± 0.6 | 8.8 ± 0.5 | 10.2 ± 0.6 | 12.4 ± 0.5 | 8.2 ± 0.6 | 9.7 ± 1.3 | 10.8 ± 1.1 | 12.8 ± 0.8 | 7.6 ± 0.5 | 8.8 ± 0.7 | 10.2 ± 0.8 | 11.8 ± 0.6 |
| [Co(L)(bpy)] 1e | 7.2 ± 0.6 | 8.3 ± 0.4 | 9.3 ± 0.3 | 10.2 ± 0.4 | -- | -- | 9.6 ± 0.5 | 10.7 ± 0.1 | -- | -- | 8.3 ± 0.3 | 9.1 ± 0.4 |
| [Ni(L)(bpy)] 1f | 7.7 ± 0.4 | 8.4 ± 1.2 | 9.5 ± 0.6 | 12.2 ± 0.4 | 8.8 ± 0.6 | 10.2 ± 0.8 | 12.7 ± 1.3 | 14.7 ± 0.8 | 7.4 ± 0.4 | 9.3 ± 0.6 | 10.9 ± 0.6 | 12.8 ± 0.4 |
| [Cu(L)(bpy)] 1g | 9.5 ± 1.2 | 11.6 ± 0.5 | 12.8 ± 0.9 | 13.6 ± 0.4 | 9.2 ± 0.1 | 10.9 ± 0.6 | 12.2 ± 0.8 | 13.8 ± 0.4 | 10.8 ± 0.7 | 13.3 ± 0.8 | 14.5 ± 0.8 | 15.6 ± 0.9 |
| [Zn(L)(bpy)] 1h | 7.4 ± 0.6 | 8.6 ± 0.5 | 9.6 ± 0.6 | 11.4 ± 0.5 | 8.8 ± 0.4 | 10.2 ± 1.1 | 11.8 ± 1.2 | 13.7 ± 0.8 | 7.9 ± 0.5 | 9.2 ± 0.4 | 10.8 ± 0.8 | 13.2 ± 0.6 |
| S. No | Complexes | Cell lines Tested | |||
|---|---|---|---|---|---|
| IC50 (μM) * | |||||
| A549 | HeLa | MCF-7 | NHDF | ||
| 1. | [Cu(L)(phen)] 1c | 25.95 ± 1.82 | 26.26 ± 1.06 | 17.13 ± 0.74 | 86.46 |
| 2. | [Cu(L)(bpy)] 1g | 39.18 ± 0.43 | 43.14 ± 0.65 | 33.18 ± 1.14 | 71.73 |
| 3. | cisplatin | 17.91 ± 0.12 | 16.13 ± 0.16 | 13.01 ± 0.44 | 94.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sasikumar, G.; Subramani, A.; Tamilarasan, R.; Rajesh, P.; Sasikumar, P.; Albukhaty, S.; Mohammed, M.K.A.; Karthikeyan, S.; Al-aqbi, Z.T.; Al-Doghachi, F.A.J.; et al. Catalytic, Theoretical, and Biological Investigations of Ternary Metal (II) Complexes Derived from L-Valine-Based Schiff Bases and Heterocyclic Bases. Molecules 2023, 28, 2931. https://doi.org/10.3390/molecules28072931
Sasikumar G, Subramani A, Tamilarasan R, Rajesh P, Sasikumar P, Albukhaty S, Mohammed MKA, Karthikeyan S, Al-aqbi ZT, Al-Doghachi FAJ, et al. Catalytic, Theoretical, and Biological Investigations of Ternary Metal (II) Complexes Derived from L-Valine-Based Schiff Bases and Heterocyclic Bases. Molecules. 2023; 28(7):2931. https://doi.org/10.3390/molecules28072931
Chicago/Turabian StyleSasikumar, Gopalakrishnan, Annadurai Subramani, Ramalingam Tamilarasan, Punniyamurthy Rajesh, Ponnusamy Sasikumar, Salim Albukhaty, Mustafa K. A. Mohammed, Subramani Karthikeyan, Zaidon T. Al-aqbi, Faris A. J. Al-Doghachi, and et al. 2023. "Catalytic, Theoretical, and Biological Investigations of Ternary Metal (II) Complexes Derived from L-Valine-Based Schiff Bases and Heterocyclic Bases" Molecules 28, no. 7: 2931. https://doi.org/10.3390/molecules28072931
APA StyleSasikumar, G., Subramani, A., Tamilarasan, R., Rajesh, P., Sasikumar, P., Albukhaty, S., Mohammed, M. K. A., Karthikeyan, S., Al-aqbi, Z. T., Al-Doghachi, F. A. J., & Taufiq-Yap, Y. H. (2023). Catalytic, Theoretical, and Biological Investigations of Ternary Metal (II) Complexes Derived from L-Valine-Based Schiff Bases and Heterocyclic Bases. Molecules, 28(7), 2931. https://doi.org/10.3390/molecules28072931












