Five New Cantharidin Derivatives from the Insect Mylabris cichorii L. and Their Potential against Kidney Fibrosis In Vitro
Abstract
1. Introduction
2. Results and Discussion
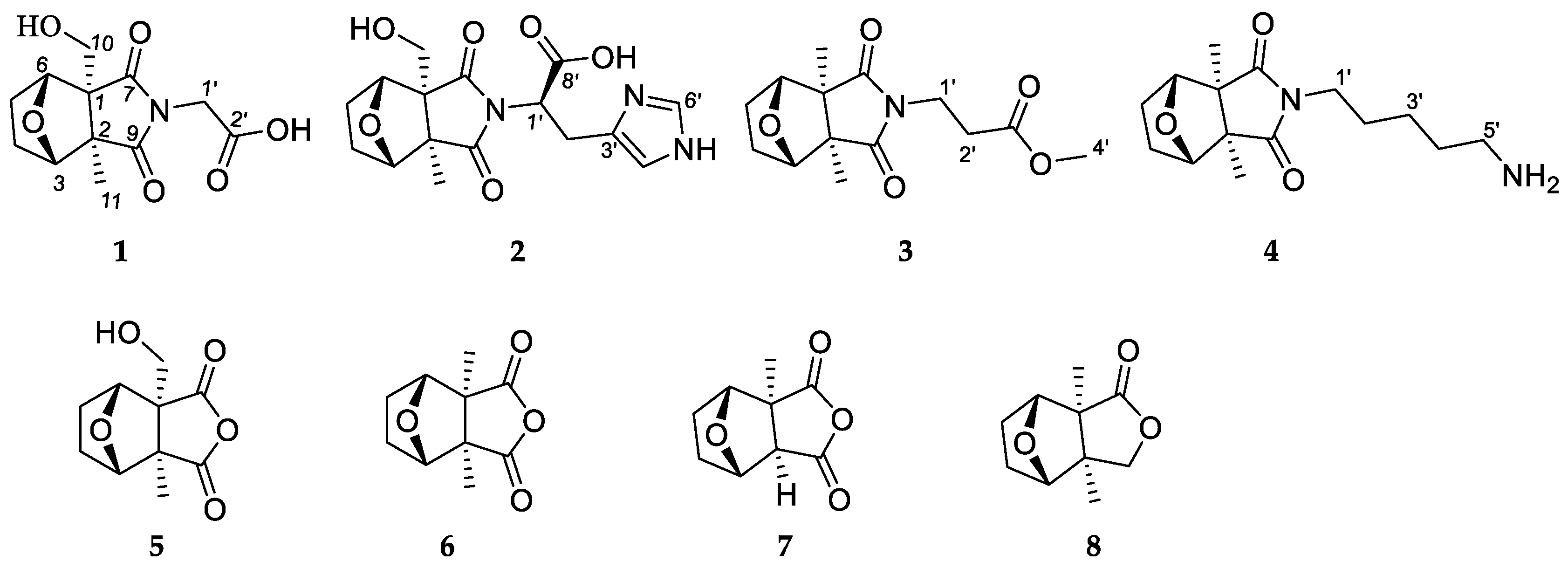
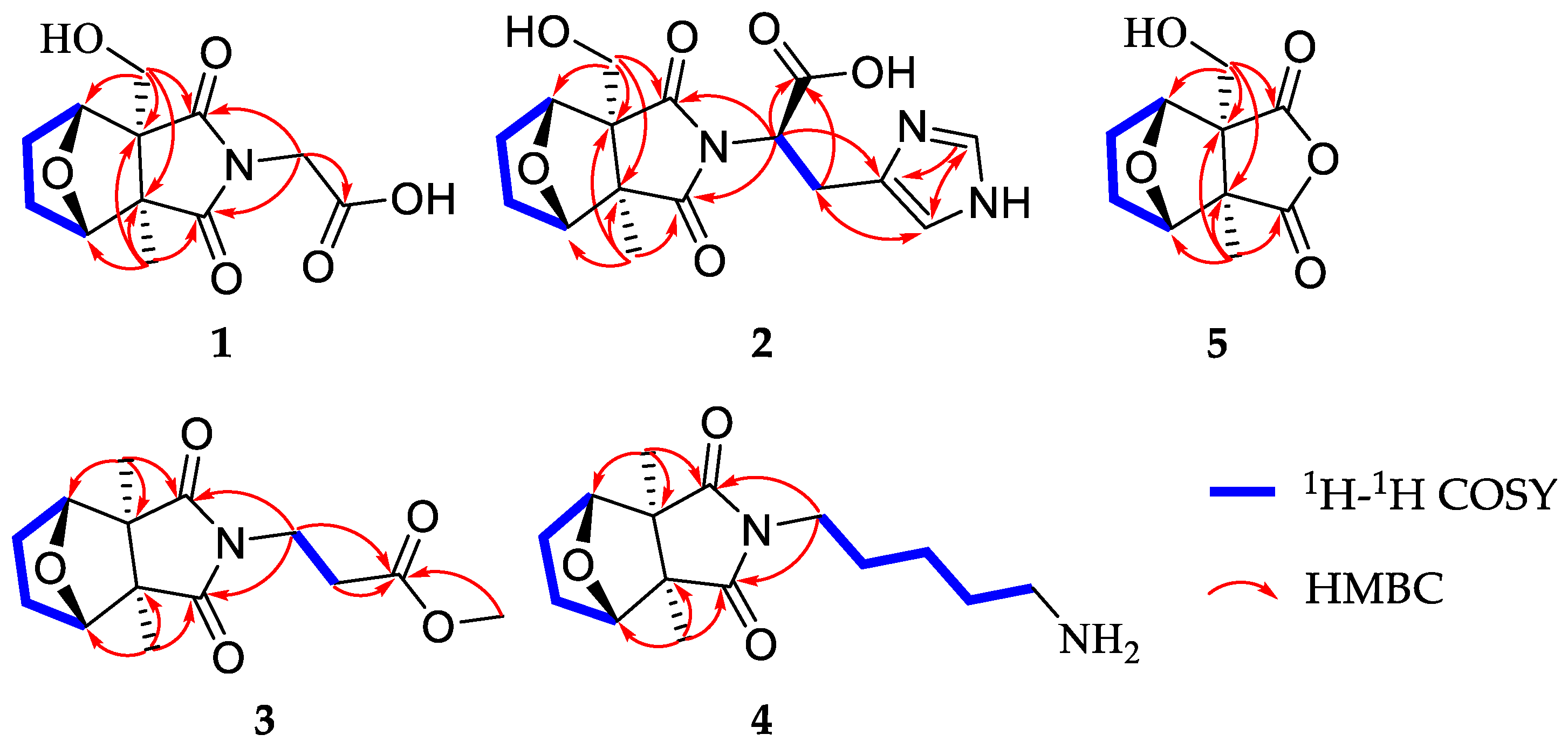
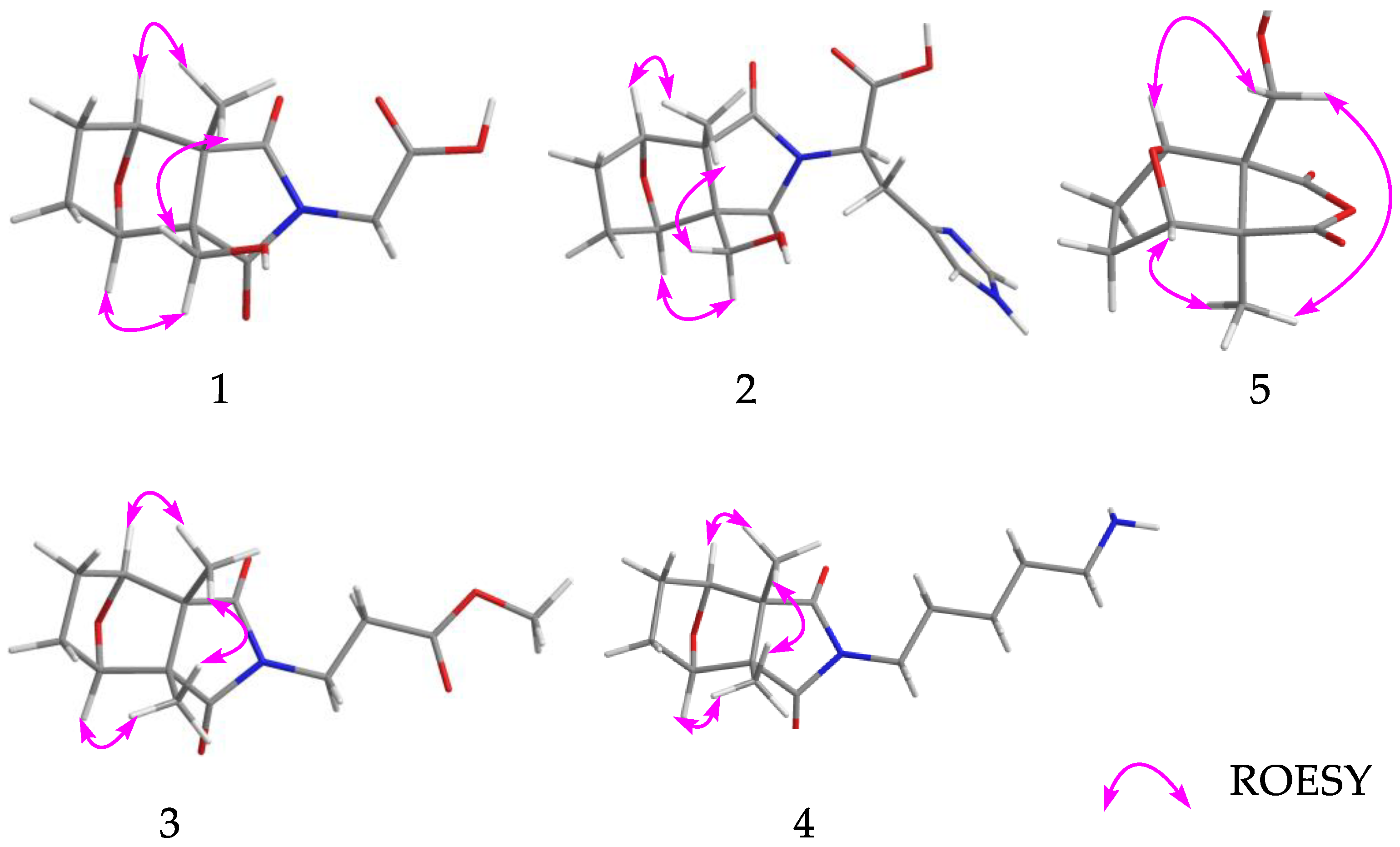
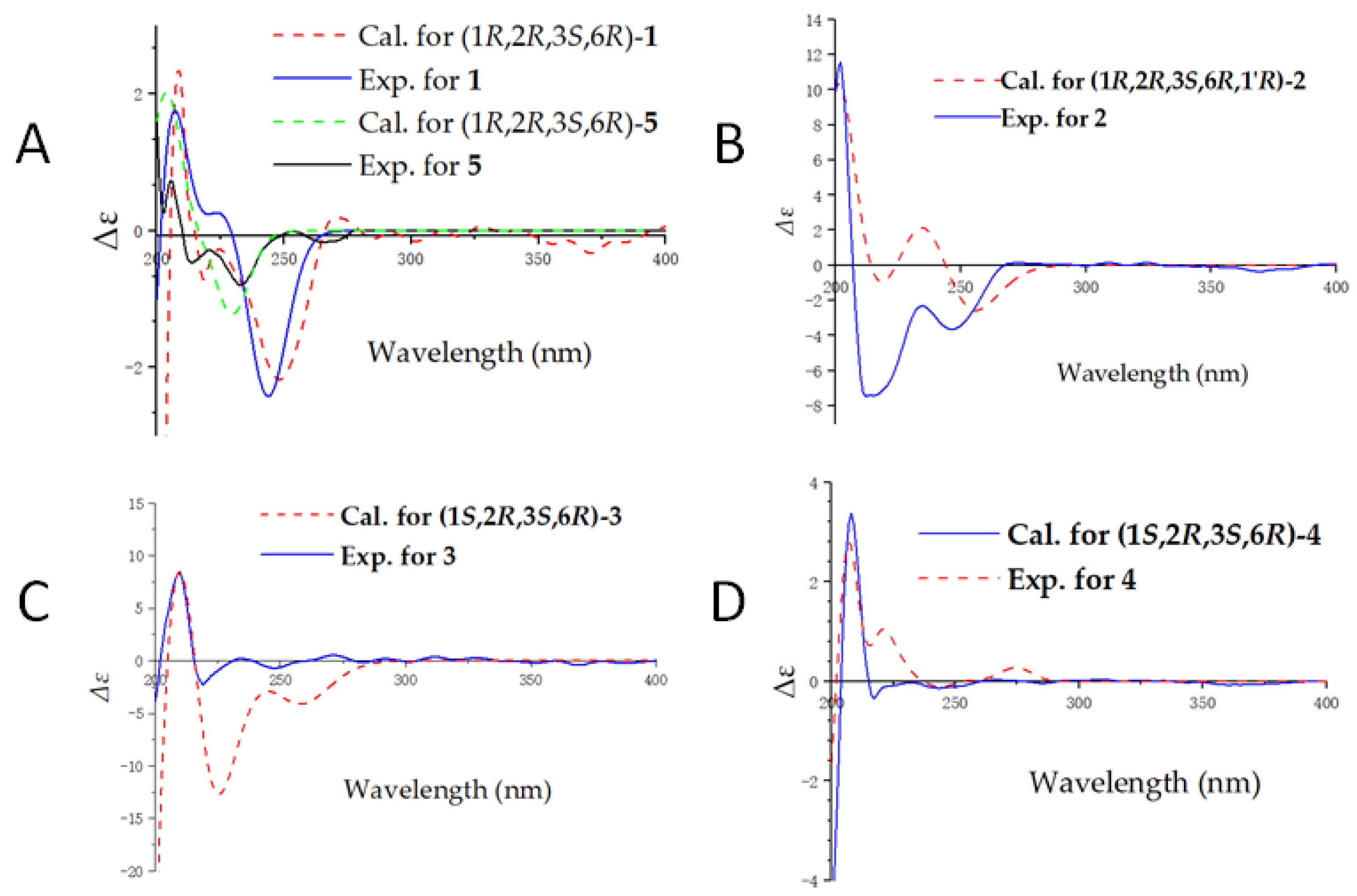
2.1. Structure Elucidation of the Compounds
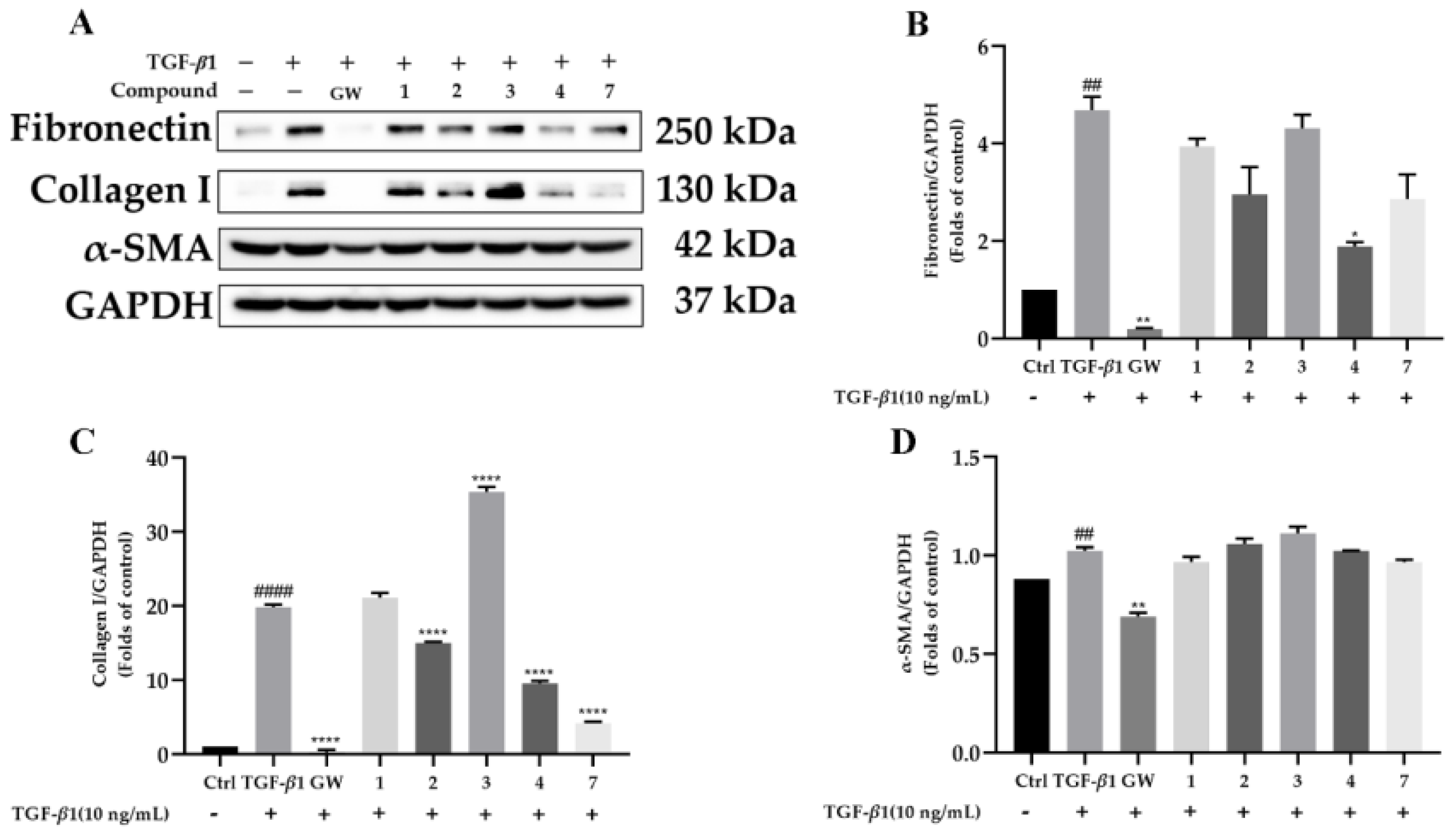
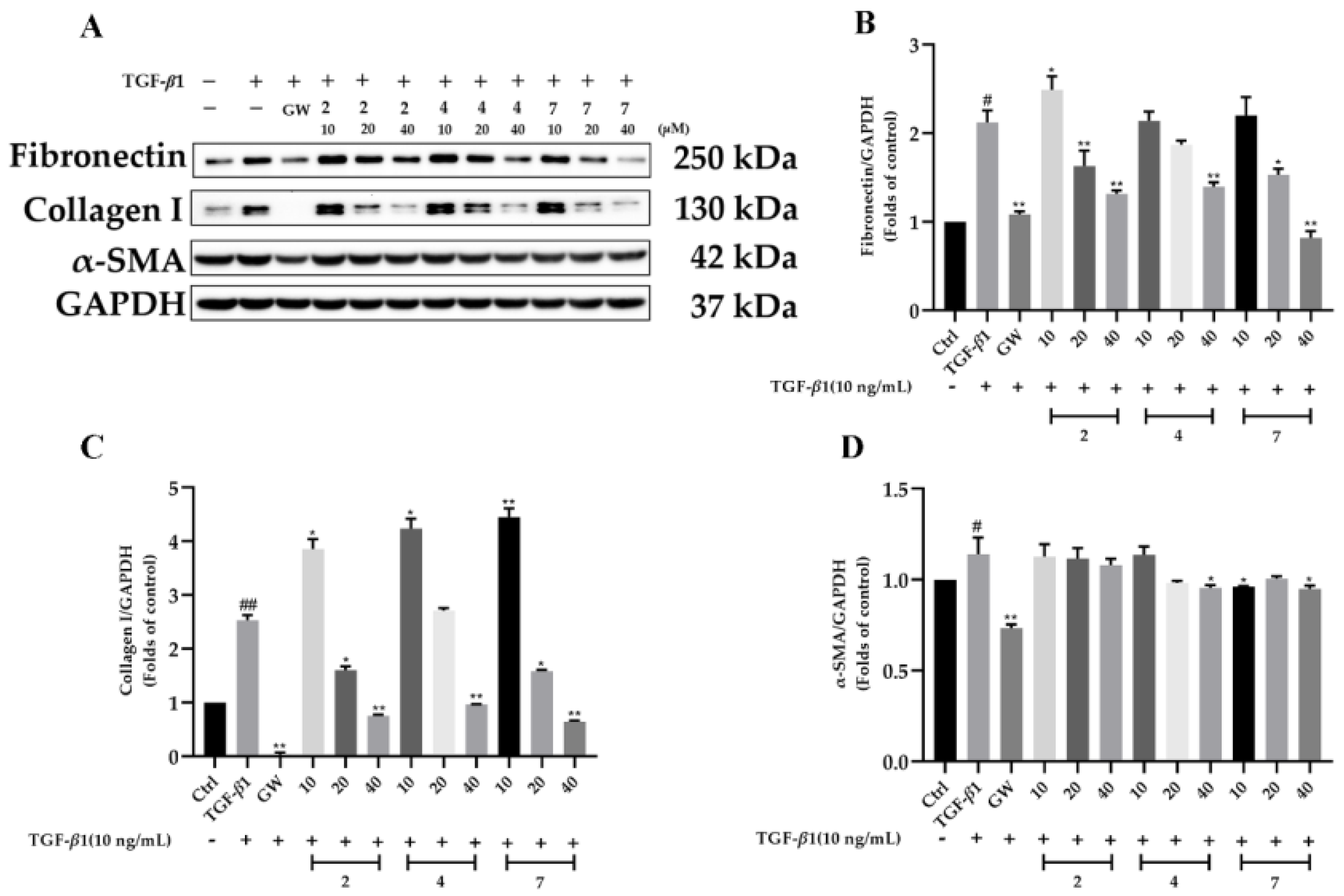
2.2. Biological Evaluation
3. Experimental Section
3.1. General Procedures
3.2. Insect Material
3.3. Extraction and Isolation
3.4. Compound Characterization Data
3.5. Computational Methods
3.6. Kidney Fibrosis Activity
3.6.1. Cell Culture
3.6.2. Cell Viability Assay
3.6.3. Western Blot
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Yan, Y.M.; Meng, X.H.; Bai, H.F.; Cheng, Y.X. Nonpeptide small molecules with a ten-membered macrolactam or a morpholine motif from the insect American cockroach and their antiangiogenic activity. Org. Chem. Front. 2021, 8, 1401–1408. [Google Scholar] [CrossRef]
- Ding, W.Y.; Yan, Y.M.; Meng, X.H.; Nafie, L.A.; Xu, T.; Dukor, R.K.; Qin, H.B.; Cheng, Y.X. Isolation, total synthesis, and absolute configuration determination of renoprotective dimeric N-acetyldopamine–adenine hybrids from the insect Aspongopus chinensis. Org. Lett. 2020, 22, 5726–5730. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Y.P.; Qin, F.Y.; Yan, Y.M.; Zhang, H.X.; Cheng, Y.X. Racemic xanthine and dihydroxydopamine conjugates from Cyclopelta parva and their COX-2 inhibitory activity. Fitoterapia 2020, 142, 104534. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.K.; Prasad, S.B. Bioactive component, cantharidin from Mylabris cichorii and its antitumor activity against ehrlich ascites carcinoma. Cell Biol. Toxicol. 2012, 28, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Kadioglu, O.; Kermani, N.S.; Kelter, G.; Schumacher, U.; Fiebig, H.H.; Greten, H.J.; Efferth, T. Pharmacogenomics of cantharidin in tumor cells. Biochem. Pharmacol. 2014, 87, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chen, Z.; Zong, Y.; Gong, F.R.; Zhu, Y.; Zhu, Y.X.; Lv, J.H.; Zhang, J.J.; Xie, L.; Sun, Y.J.; et al. PP2A inhibitors induce apoptosis in pancreatic cancer cell line PANC-1 through persistent phosphorylation of IKKα and sustained activation of the NF-κB pathway. Cancer Lett. 2011, 304, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.S. Medical uses of mylabris in ancient China and recent studies. J. Ethnopharmacol. 1989, 26, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.B.; Liu, X.L.; Zhang, Y.; Li, C.J.; Zhang, D.M.; Peng, Y.Z.; Zhou, X.; Du, H.F.; Tan, C.B.; Zhang, Y.Y.; et al. Cantharimide and its derivatives from the blister beetle Mylabris phalerata Palla. J. Nat. Prod. 2016, 79, 2032–2038. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, T.; Konishi, T.; Miyahara, K.; Noda, N. Three novel cantharidin-related compounds from the Chinese blister beetle, Mylabris phalerata PALL. Chem. Pharm. Bull. 2004, 52, 807–809. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.B.; Guo, Y.L.; Zhang, Y.; Wang, X.Y.; Jiang, Y.; Yang, D.J. Rapid profiling of cantharidin analogues in Mylabris phalerata Pallas by ultra-performance liquid chromatography-quadrupole time-of-flight-tandem mass spectrometry. Biomed. Chromatogr. 2020, 34, e4801. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.Y.; Zhang, W.; Li, N.P.; Lei, X.P.; Gong, X.Y.; Zhang, D.M.; Wang, L.; Ye, W.C. Cantharidin derivatives from the medicinal insect Mylabris phalerata. Tetrahedron 2017, 73, 5932–5939. [Google Scholar] [CrossRef]
- Zhang, X.J.; Cao, Y.; Pan, D.B.; Yao, X.J.; Wang, F.; Zhang, G.L.; Luo, Y.G. Antifibrotic pyridine-containing monoterpene alkaloids from Caryopteris glutinosa. Phytochemistry 2022, 203, 113378. [Google Scholar] [CrossRef] [PubMed]
- Anandakumar, P.; Kamaraj, S.; Vanitha, M.K. D-limonene: A multifunctional compound with potent therapeutic effects. J. Food Biochem. 2021, 45, e13566. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Peng, L.Y.; Liu, P.; Yang, M.X.; Zhou, H.; Ding, Y.R.; Wang, J.J.; Huang, W.; Tan, Q.; Wang, Y.L.; et al. Paeoniflorin ameliorates chronic hypoxia/SU5416-induced pulmonary arterial hypertension by inhibiting endothelial-to-mesenchymal transition. Drug Des. Dev. Ther. 2020, 14, 1191–1202. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, T.; Jinpo, K.; Noda, N. Cantharimide dimers from the Chinese blister beetle, Mylabris phalerate Pallas. Chem. Pharm. Bull. 2007, 55, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.B.; Liu, X.L.; Zhou, X.; Zhang, X.M.; Yang, Y.; Du, H.F.; Tan, C.B.; Zhang, Y.Y.; Zhang, Y.; Yang, D.J. Chemical constituents from Mylabris phalerata and their cytotoxic activity in vitro. China J. Chin. Mater. Med. 2016, 41, 859–863. [Google Scholar] [CrossRef]
- Dauben, W.G.; Lam, J.Y.L.; Guo, Z.R. Total synthesis of (−)-palasonin and (+)-palasonin and related chemistry. J. Org. Chem. 1996, 61, 4816–4819. [Google Scholar] [CrossRef] [PubMed]
- König, W.A. Collection of enantiomeric separation factors obtained by capillary gas chromatography on chiral stationary phases. J. High Res. Chrom. 1993, 16, 569–586. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision B.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions, and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions, or products referred to in the content. |

| Position | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 3 | 4.52 (br d, 4.5) | 4.46 (br d, 4.6) | 4.47 (br d, 2.6) | 4.47 (br d, 2.2) | 4.67 (br d, 2.7) |
| 4 | Ha: 1.95 (m) | Ha: 1.91 (m) | Ha: 1.89 (overlap) a | Ha: 1.91 (overlap) a | Ha: 1.97 (m) |
| Hb: 1.67 (overlap) a | Hb: 1.61 (overlap) a | Hb: 1.64 (overlap) a | Hb: 1.65 (overlap) a | Hb: 1.68 (overlap) a | |
| 5 | Ha: 1.75 (m) | Ha: 1.67 (m) | Ha: 1.89 (overlap) a | Ha: 1.91 (overlap) a | Ha: 1.68 (overlap) a |
| Hb: 1.67 (overlap) a | Hb: 1.61 (overlap) a | Hb: 1.64 (overlap) a | Hb: 1.65 (overlap) a | Hb: 1.68 (overlap) a | |
| 6 | 4.56 (br d, 4.7) | 4.36 (br d, 4.8) | 4.48 (br d, 2.6) | 4.48 (br d, 2.2) | 4.66 (br d, 2.7) |
| 10 | Ha: 3.97 (d, 11.3) | Ha: 3.94 (d, 11.2) | 1.15 (overlap)a | 1.15 (overlap)a | Ha: 3.98 (d, 11.1) |
| Hb: 3.73 (d, 11.3) | Hb: 3.69 (d, 11.2) | Hb: 3.72 (d, 11.1) | |||
| 11 | 1.33 (s) | 1.24 (s) | 1.15 (overlap)a | 1.15 (overlap)a | 1.41 (s) |
| 1′ | Ha: 4.23 (d, 17.2) | 4.97 (br t, 7.4) | 3.74 (t, 7.0) | 3.51 (t, 7.0) | |
| Hb: 4.18 (d, 17.2) | |||||
| 2′ | 3.49 (br d, 8.0) | 2.58 (t, 7.1) | 1.61 (m) | ||
| 3′ | 1.36 (m) | ||||
| 4′ | 7.26 (s) | 3.64 (s) | 1.66 (m) | ||
| 5′ | 2.89 (t, 7.7) | ||||
| 7′ | 8.71 (s) |
| Position | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | 61.9, C | 61.6, C | 55.3, C | 55.2, C | 64.3, C |
| 2 | 55.3, C | 55.0, C | 55.3, C | 55.2, C | 55.9, C |
| 3 | 85.3, CH | 85.3, CH | 85.1, CH | 85.2, CH | 86.7, CH |
| 4 | 24.6 CH2 | 24.5, CH2 | 24.6, CH2 | 24.6, CH2 | 24.7, CH2 |
| 5 | 25.1, CH2 | 24.9, CH2 | 24.6, CH2 | 24.6, CH2 | 24.6, CH2 |
| 6 | 83.2, CH | 83.2, CH | 85.1, CH | 85.2, CH | 84.2, CH |
| 7 | 180.9, C | 180.7, C | 183.1, C | 183.6, C | 176.0, C |
| 9 | 182.5, C | 182.7, C | 183.1, C | 183.6, C | 177.9, C |
| 10 | 59.5, CH2 | 59.4, CH2 | 12.6, CH3 | 12.5, CH3 | 59.1, CH2 |
| 11 | 12.2, CH3 | 12.2, CH3 | 12.6, CH3 | 12.5, CH3 | 12.7, CH3 |
| 1′ | 40.5, CH2 | 53.2, CH | 35.8, CH2 | 39.4, CH2 | |
| 2′ | 169.7, C | 25.1, CH2 | 32.7, CH2 | 27.89, CH2 | |
| 3′ | 130.8, C | 172.9, C | 24.3, CH2 | ||
| 4′ | 119.1, CH | 52.4, CH3 | 27.9, CH2 | ||
| 5′ | 40.5, CH2 | ||||
| 6′ | 134.7, CH | ||||
| 8′ | 171.1, C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.-M.; Li, J.-J.; Wan, L.; Cheng, Y.-X. Five New Cantharidin Derivatives from the Insect Mylabris cichorii L. and Their Potential against Kidney Fibrosis In Vitro. Molecules 2023, 28, 2822. https://doi.org/10.3390/molecules28062822
Li K-M, Li J-J, Wan L, Cheng Y-X. Five New Cantharidin Derivatives from the Insect Mylabris cichorii L. and Their Potential against Kidney Fibrosis In Vitro. Molecules. 2023; 28(6):2822. https://doi.org/10.3390/molecules28062822
Chicago/Turabian StyleLi, Ke-Ming, Ji-Jun Li, Li Wan, and Yong-Xian Cheng. 2023. "Five New Cantharidin Derivatives from the Insect Mylabris cichorii L. and Their Potential against Kidney Fibrosis In Vitro" Molecules 28, no. 6: 2822. https://doi.org/10.3390/molecules28062822
APA StyleLi, K.-M., Li, J.-J., Wan, L., & Cheng, Y.-X. (2023). Five New Cantharidin Derivatives from the Insect Mylabris cichorii L. and Their Potential against Kidney Fibrosis In Vitro. Molecules, 28(6), 2822. https://doi.org/10.3390/molecules28062822






