Click Triazole as a Linker for Pretargeting Strategies: Synthesis, Docking Investigations, Fluorescence Diagnosis, and Antibacterial Action Studies
Abstract
1. Introduction
2. Results and Discussion
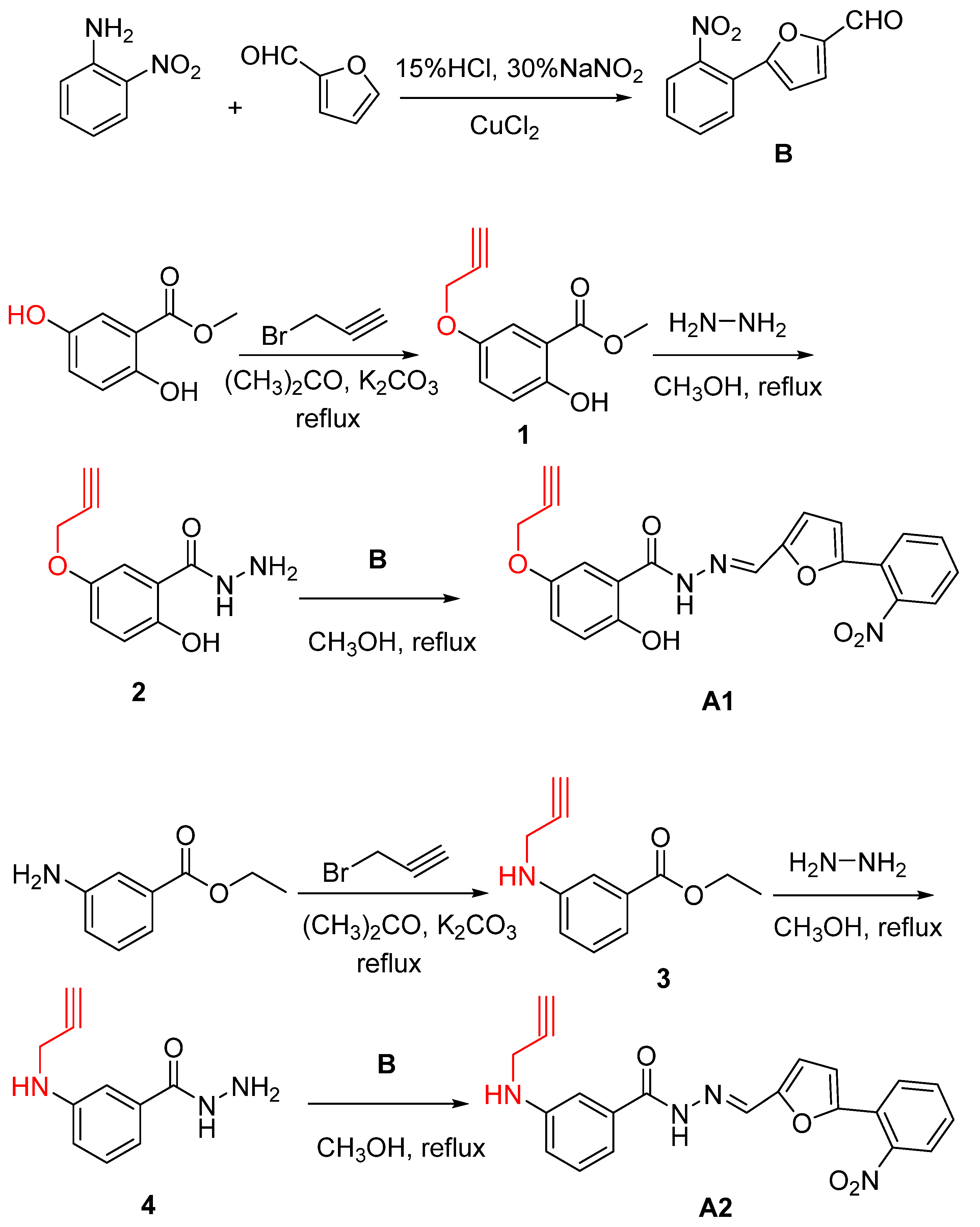
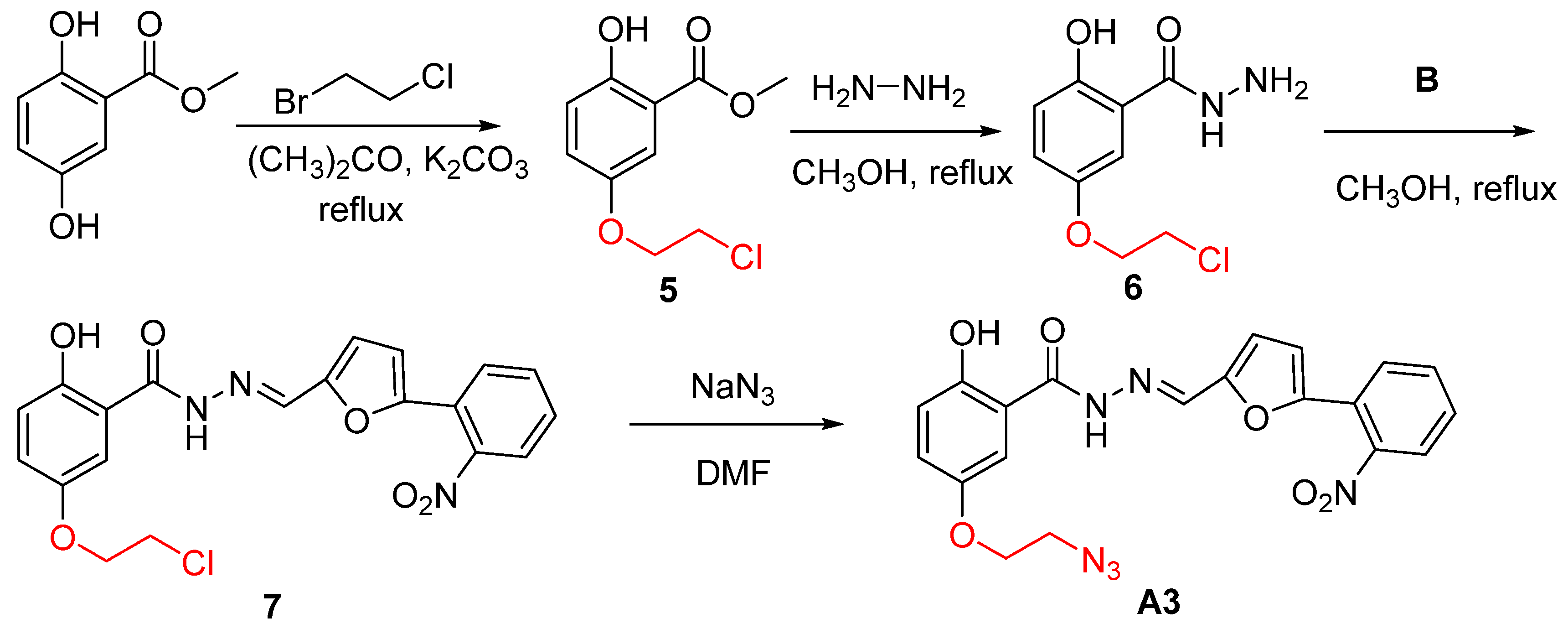
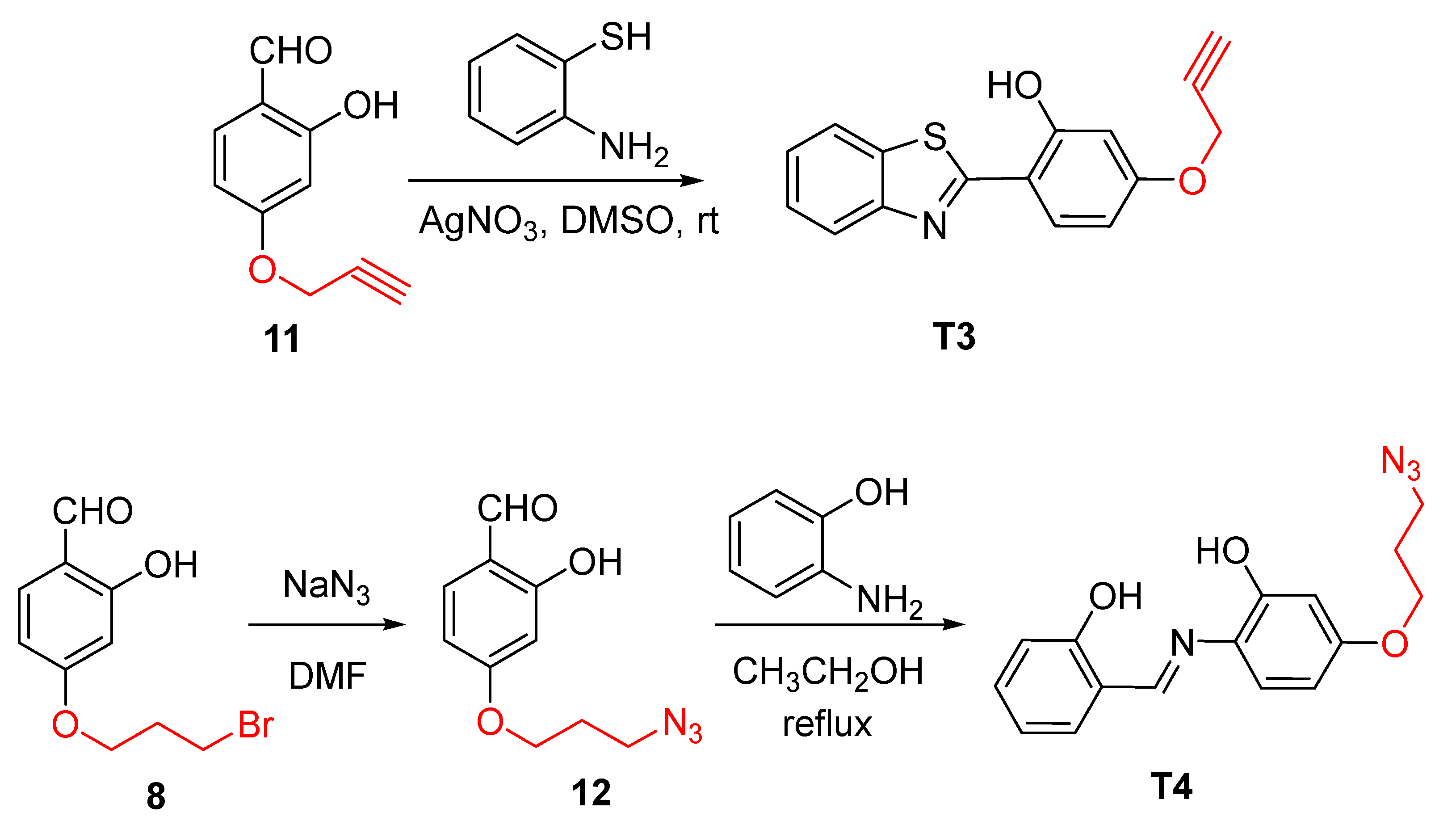
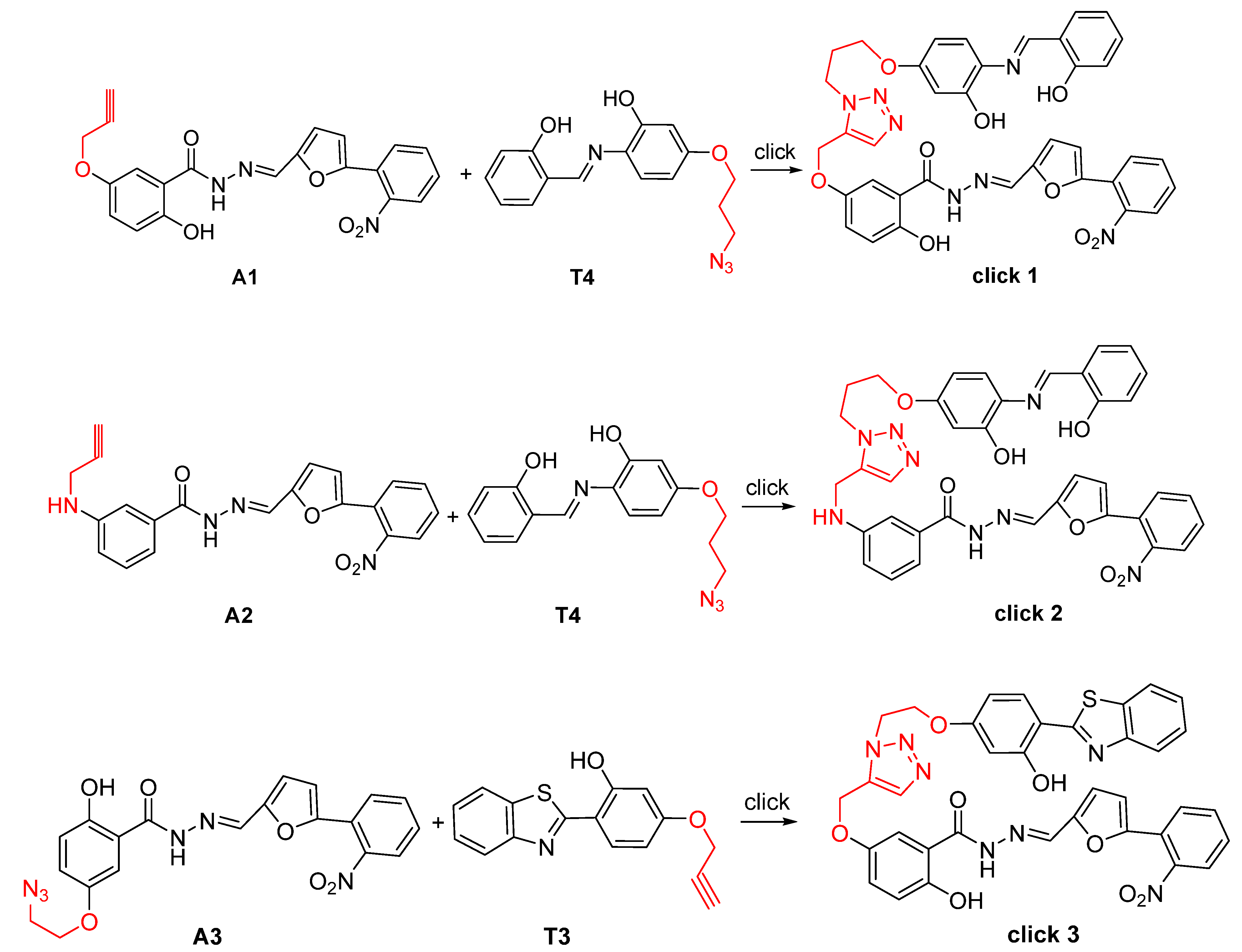
2.1. Design and Synthesis of B8I-2 Derivatives and Fluorescent Probes
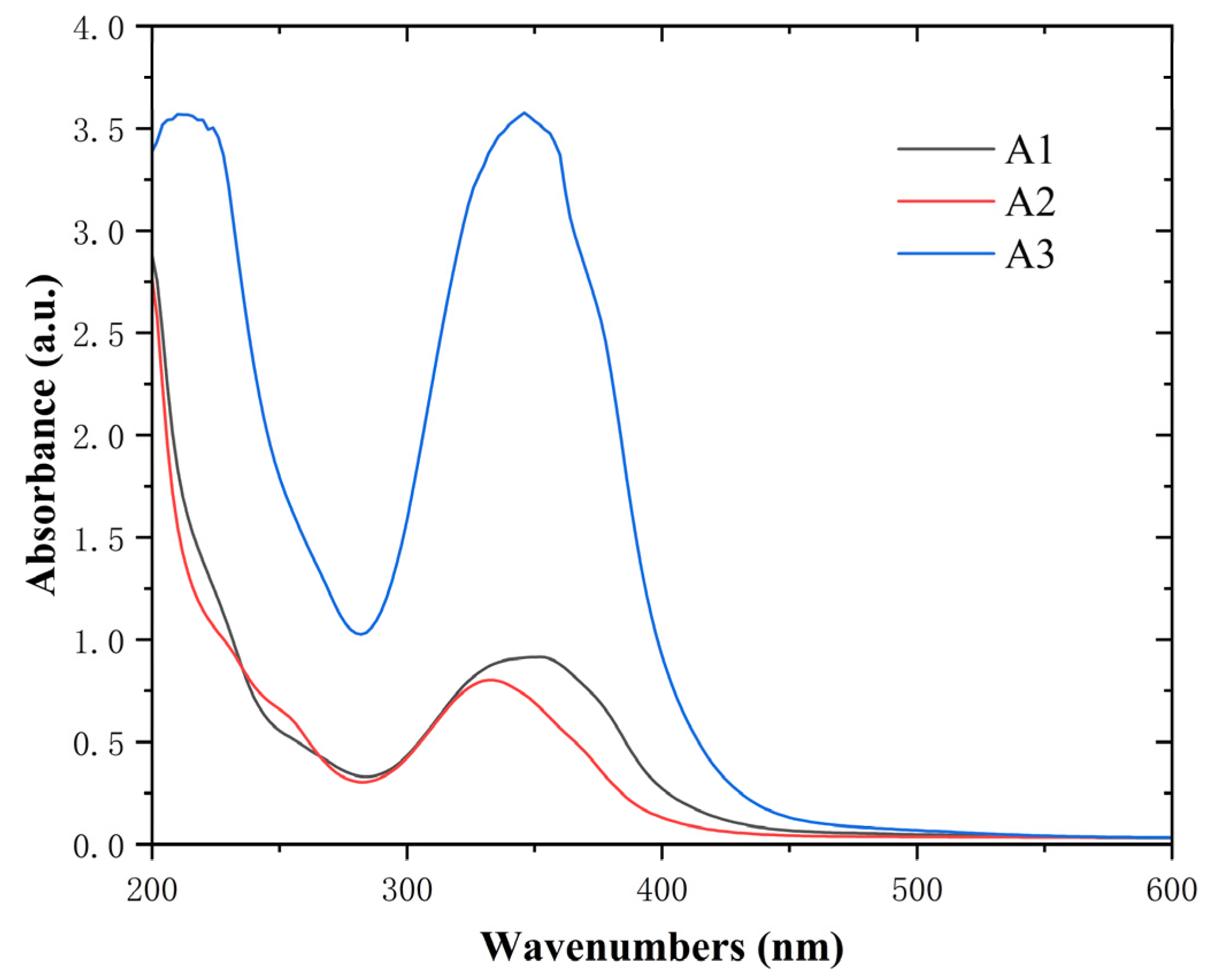
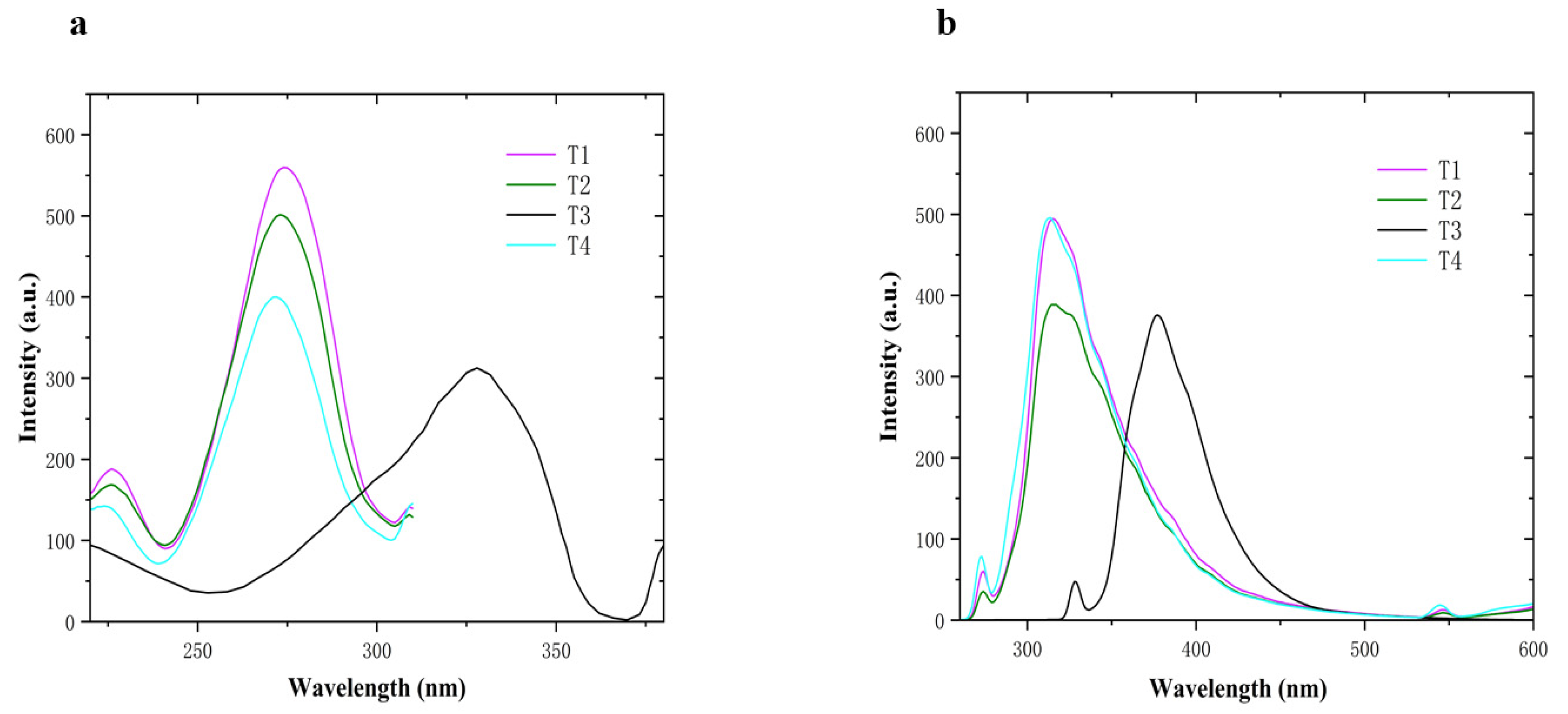
2.2. UV-Visible Absorption Spectroscopy and Fluorescence Spectroscopic
2.3. The HPLC-MS Results for the “Click” Reactions
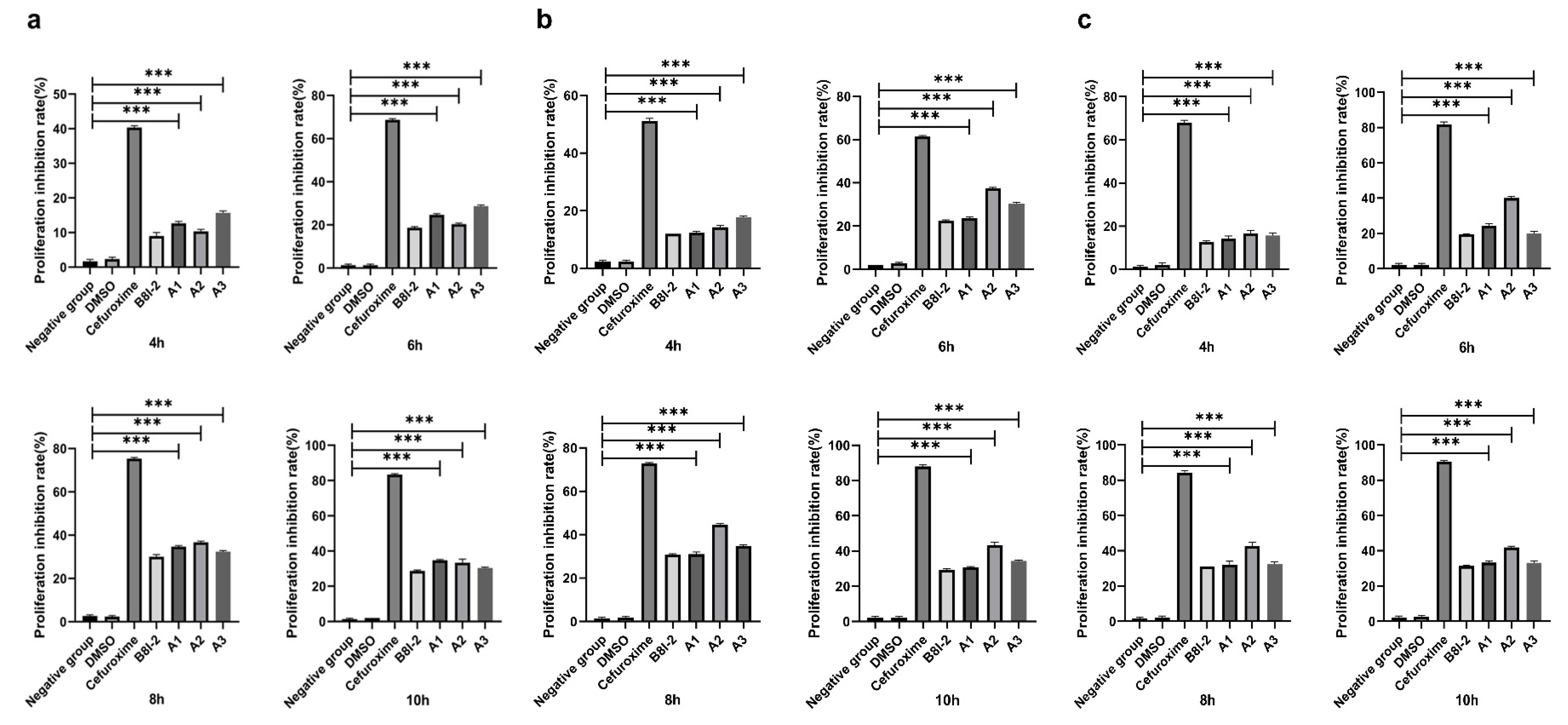
2.4. Determination of the Maximum Safe Concentration of the Drugs
2.5. Antibacterial Activity on Agrobacterium Tumefaciens
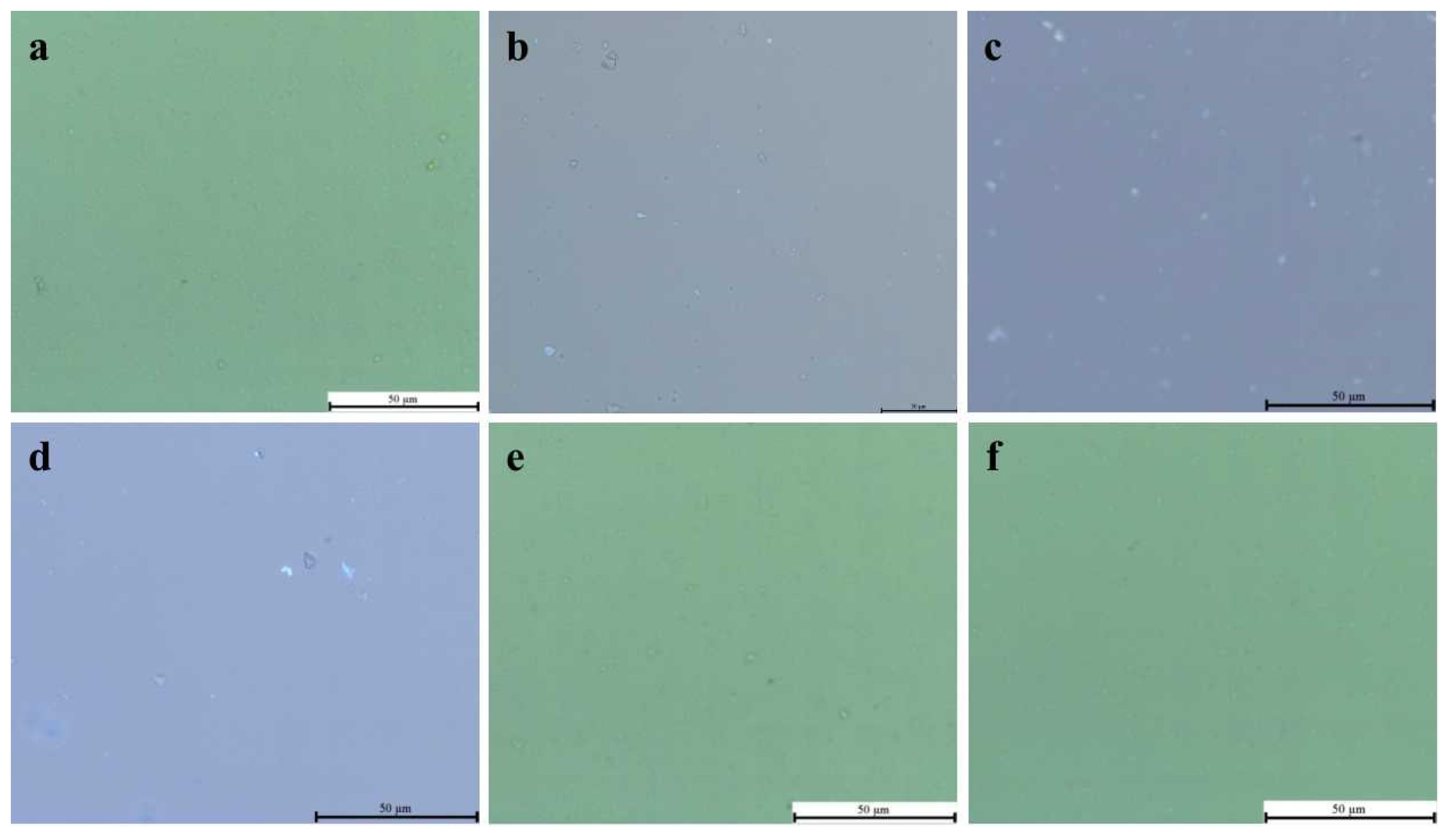
2.6. Fluorescence Detection of Agrobacterium tumefaciens by Diagnostic Molecules
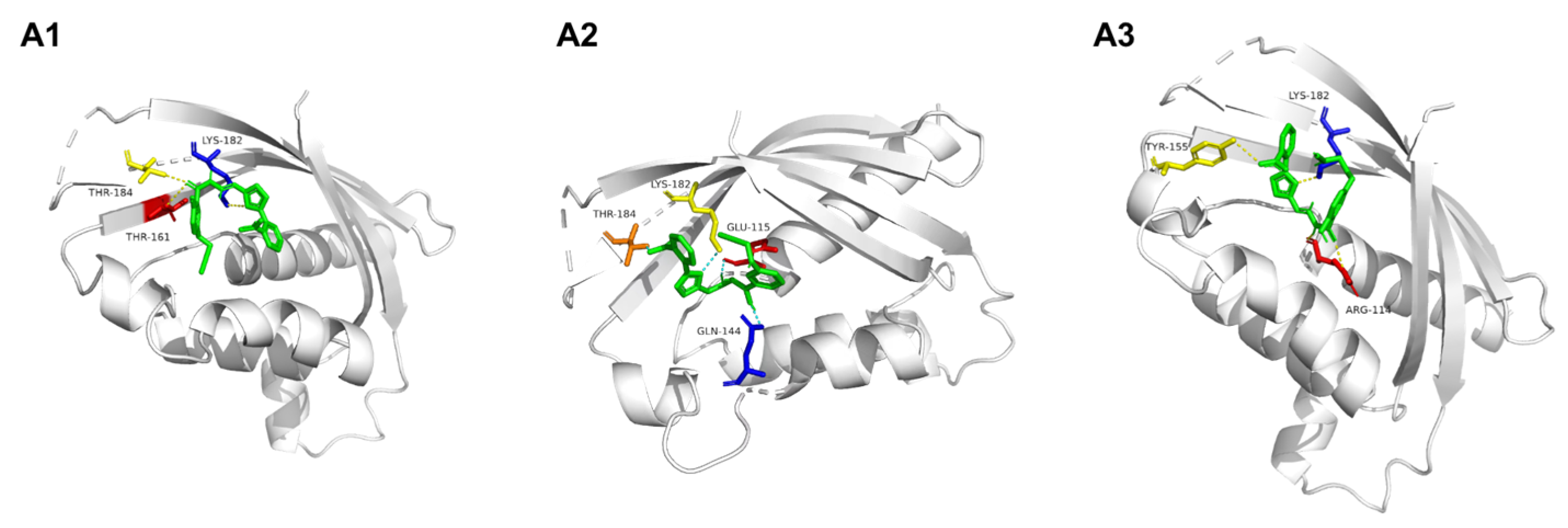
2.7. Docking of A1–A3 to VirB8
3. Materials and Methods
3.1. Apparatus and Characterization
3.2. Synthesis
3.2.1. General Procedure for Synthesis of the Compounds (A1–A3)
(E)-2-hydroxy-N’-((5-(2-nitrophenyl)furan-2-yl)methylene)-5-(prop-2-ynyloxy)benzohydrazide (A1)
(E)-N’-((5-(2-nitrophenyl)furan-2-yl)methylene)-3-(prop-2-ynylamino)benzohydrazide (A2)
(E)-5-(2-azidoethoxy)-2-hydroxy-N’-((5-(2-nitrophenyl)furan-2-yl)methylene)benzohydrazide (A3)
3.2.2. General Procedure for the Synthesis of the Probes (T1–T4)
(E)-2-((2-(2-nitrobenzyloxy)phenylimino)methyl)-5-(2-azidoethoxy)phenol (T1)
(E)-2-((2-hydroxyphenylimino)methyl)-5-(prop-2-ynyloxy)phenol (T2)
2-(benzo[d]thiazol-2-yl)-5-(prop-2-ynyloxy)phenol (T3)
(E)-5-(3-azidopropoxy)-2-((2-hydroxyphenylimino)methyl)phenol (T4)
3.3. Determination of the UV Absorption Wavelength of Inhibitors
3.4. Fluorescence Intensity Measurement by Fluorescence Spectrophotometer
3.5. In Vitro Validation of the “Click” Reaction and Optimization of Conditions
3.6. Cytotoxicity: MTT Cell Proliferation Assay
3.7. Antibacterial Activity on Agrobacterium tumefaciens
3.8. Fluorescent Detection of Agrobacterium tumefaciens by Diagnostic Molecules
3.9. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Wang, C.; Liu, M.; Wang, Z.; Li, S.; Deng, Y.; He, N. Point-of-care diagnostics for infectious diseases: From methods to devices. Nano Today 2021, 37, 101092. [Google Scholar]
- Cascella, M.; Rajnik, M.; Aleem, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation, and Treatment of Coronavirus (COVID-19); Statpearls: Orlando, FL, USA, 2022. [Google Scholar]
- Kinimi, E.; Odongo, S.; Muyldermans, S.; Kock, R.; Misinzo, G. Paradigm shift in the diagnosis of peste des petits ruminants: Scoping review. Acta Vet. Scand. 2020, 62, 7. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Y.; Wang, C.; Feng, Q.; Fan, F.; Zhang, G.; Kang, X.; Qin, X.; Sun, J.; Li, Y. Integrated microcapillary for sample-to-answer nucleic acid pretreatment, amplification, and detection. Anal. Chem. 2014, 86, 10461–10466. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhu, C.; Deng, G.; Zhang, L.; Zhao, S.; Lin, J.; Li, L.; Jiao, P.; Liao, M.; Liu, Y. Rapid identification of H5 avian influenza virus in chicken throat swab specimens using microfluidic real-time RT-PCR. Anal. Methods 2014, 6, 2628–2632. [Google Scholar] [CrossRef]
- Chekli, Y.; Peron-Cane, C.; Dell’Arciprete, D.; Allemand, J.-F.; Li, C.; Ghigo, J.-M.; Gautier, A.; Lebreton, A.; Desprat, N.; Beloin, C. Visualizing the dynamics of exported bacterial proteins with the chemogenetic fluorescent reporter FAST. Sci. Rep. 2020, 10, 15791. [Google Scholar] [CrossRef] [PubMed]
- Tabb, J.; Rapoport, E.; Han, I.; Lombardi, J.; Green, O. An antigen-targeting assay for Lyme disease: Combining aptamers and SERS to detect the OspA protein. Nanomedicine 2022, 41, 102528. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Li, N.; Xiao, Y. Assessing chromatin condensation for epigenetics with a DNA-targeting sensor by FRET and FLIM techniques. Chin. Chem. Lett. 2021, 32, 2395–2399. [Google Scholar] [CrossRef]
- Radkov, A.D.; Hsu, Y.-P.; Booher, G.; VanNieuwenhze, M.S. Imaging bacterial cell wall biosynthesis. Annu. Rev. Biochem. 2018, 87, 991. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.R.; Wodzanowski, K.A.; Santiago, C.C.; Hyland, S.N.; Follmar, J.L.; Asare-Okai, P.; Grimes, C.L. Protected N-acetyl muramic acid probes improve bacterial peptidoglycan incorporation via metabolic labeling. ACS Chem. Biol. 2021, 16, 1908–1916. [Google Scholar] [CrossRef]
- Liu, C.; Zhou, L.; Xie, L.; Zheng, Y.; Man, H.; Xiao, Y. Forthrightly monitoring ferroptosis induced by endoplasmic reticulum stresses through fluorescence lifetime imaging of microviscosity increases with a specific rotor. Chin. Chem. Lett. 2022, 33, 2537–2540. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, Q.; Wang, T.; Shi, D.; Fu, Z.; Si, Z.; Xu, Z.; Cheng, Y.; Shi, H.; Cheng, D. Targeting Infiltrating Myeloid Cells in Gastric Cancer Using a Pretargeted Imaging Strategy Based on Bio-Orthogonal Diels-Alder Click Chemistry and Comparison with Zr-Labeled Anti-CD11b Positron Emission Tomography Imaging. Mol. Pharm. 2022, 19, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xing, B. Small-molecule fluorescent probes: Big future for specific bacterial labeling and infection detection. Chem. Commun. 2022, 58, 155–170. [Google Scholar] [CrossRef]
- Siegrist, M.S.; Swarts, B.M.; Fox, D.M.; Lim, S.A.; Bertozzi, C.R. Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface. FEMS Microbiol. Rev. 2015, 39, 184–202. [Google Scholar] [CrossRef]
- Banahene, N.; Kavunja, H.W.; Swarts, B.M. Chemical reporters for bacterial glycans: Development and applications. Chem. Rev. 2021, 122, 3336–3413. [Google Scholar] [CrossRef]
- Nibin Joy, M.; Bodke, Y.D.; Telkar, S.; Bakulev, V.A. Synthesis of coumarins linked with 1, 2, 3-triazoles under microwave irradiation and evaluation of their antimicrobial and antioxidant activity. J. Mex. Chem. Soc. 2020, 64, 53–73. [Google Scholar]
- Tornøe, C.W.; Christensen, C.; Meldal, M. Peptidotriazoles on solid phase: [1, 2, 3]-triazoles by regiospecific copper (I)-catalyzed 1, 3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002, 67, 3057–3064. [Google Scholar] [CrossRef]
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A stepwise huisgen cycloaddition process: Copper (I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. 2002, 114, 2708–2711. [Google Scholar] [CrossRef]
- Pachón, L.D.; van Maarseveen, J.H.; Rothenberg, G. Click chemistry: Copper clusters catalyse the cycloaddition of azides with terminal alkynes. Adv. Synth. Catal. 2005, 347, 811–815. [Google Scholar] [CrossRef]
- Shankaraiah, N.; Sakla, A.; Laxmikeshav, K.; Tokala, R. Reliability of Click Chemistry on Drug Discovery: A Personal Account. Chem. Rec. 2020, 20, 253–272. [Google Scholar] [CrossRef]
- Kolb, H.C.; Sharpless, K.B. The growing impact of click chemistry on drug discovery. Drug Discov. Today 2003, 8, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Giffin, M.J.; Heaslet, H.; Brik, A.; Lin, Y.-C.; Cauvi, G.; Wong, C.-H.; McRee, D.E.; Elder, J.H.; Stout, C.D.; Torbett, B.E. A copper (I)-catalyzed 1, 2, 3-triazole azide− alkyne click compound is a potent inhibitor of a multidrug-resistant HIV-1 protease variant. J. Med. Chem. 2008, 51, 6263–6270. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Tang, Z.; Sun, Z.; Meng, X.; Song, S.; Quan, Z. Supported copper (I) catalyst from fish bone waste: An efficient, green and reusable catalyst for the click reaction toward N-substituted 1, 2, 3-TRIAZOLES. Appl. Organomet. Chem. 2018, 32, e3946. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, M.; Zhou, K.; Rao, J. Pre-targeted imaging of protease activity through in situ assembly of nanoparticles. Angew. Chem. 2020, 132, 7938–7944. [Google Scholar] [CrossRef]
- Poulie, C.; Sporer, E.; Hvass, L.; Jørgensen, J.; Kempen, P.; Lopes van den Broek, S.; Shalgunov, V.; Kjaer, A.; Jensen, A.; Herth, M. Bioorthogonal Click of Colloidal Gold Nanoparticles to Antibodies In vivo. Chemistry 2022, 28, e202201847. [Google Scholar] [CrossRef]
- Taiariol, L.; Chaix, C.; Farre, C.; Moreau, E. Click and bioorthogonal chemistry: The future of active targeting of nanoparticles for nanomedicines? Chem. Rev. 2022, 122, 340–384. [Google Scholar] [CrossRef] [PubMed]
- Summer, D.; Petrik, M.; Mayr, S.; Hermann, M.; Kaeopookum, P.; Pfister, J.; Klingler, M.; Rangger, C.; Haas, H.; Decristoforo, C. Hybrid Imaging Agents for Pretargeting Applications Based on Fusarinine C—Proof of Concept. Molecules 2020, 25, 2123. [Google Scholar] [CrossRef]
- Teng, Y.; Zhang, R.; Yang, B.; Yang, H.; Li, X.; Yin, D.; Feng, X.; Tian, Y. Bio-orthogonally activated tetraphenylene-tetrazine aggregation-induced emission fluorogenic probes. J. Mater. Chem. B 2022, 10, 8642–8649. [Google Scholar] [CrossRef]
- Paschos, A.; den Hartigh, A.; Smith, M.A.; Atluri, V.L.; Sivanesan, D.; Tsolis, R.M.; Baron, C. An in vivo high-throughput screening approach targeting the type IV secretion system component VirB8 identified inhibitors of Brucella abortus 2308 proliferation. Infect. Immun. 2011, 79, 1033–1043. [Google Scholar] [CrossRef]
- Smith, M.A.; Coinçon, M.; Paschos, A.; Jolicoeur, B.; Lavallée, P.; Sygusch, J.; Baron, C. Identification of the binding site of Brucella VirB8 interaction inhibitors. Chem. Biol. 2012, 19, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Ogawara, H. Possible drugs for the treatment of bacterial infections in the future: Anti-virulence drugs. J. Antibiot. 2021, 74, 24–41. [Google Scholar] [CrossRef]
- Altamirano-Silva, P.; Cordero-Serrano, M.; Méndez-Montoya, J.; Chacón-Díaz, C.; Guzmán-Verri, C.; Moreno, E.; Chaves-Olarte, E. Intracellular passage triggers a molecular response in Brucella abortus that increases its infectiousness. Infect. Immun. 2021, 89, e00004-21. [Google Scholar] [CrossRef]
- Blasey, N.; Rehrmann, D.; Riebisch, A.K.; Mühlen, S. Targeting bacterial pathogenesis by inhibiting virulence-associated Type III and Type IV secretion systems. Front. Cell. Infect. Microbiol. 2023, 12, 1955. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.B.; Whitlock, H.W., Jr. Syntheses and Synthetic Studies of 2-Hydroxy-5-(propargyloxy) benzoic Acid. Synth. Commun. 1993, 23, 23–34. [Google Scholar] [CrossRef]
- Pringle, W.; Sharpless, K.B. The osmium-catalyzed aminohydroxylation of Baylis-Hillman olefins. Tetrahedron Lett. 1999, 40, 5151–5154. [Google Scholar] [CrossRef]
- Zhou, Y.; Sun, W.; Peng, J.; Yan, H.; Zhang, L.; Liu, X.; Zuo, Z. Design, synthesis and biological evaluation of novel copper-chelating acetylcholinesterase inhibitors with pyridine and N-benzylpiperidine fragments. Bioorg. Chem. 2019, 93, 103322. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Komatsu, T.; Kamiya, M.; Campos, C.U.; González-Gaitán, M.; Terai, T.; Hanaoka, K.; Nagano, T.; Urano, Y. Highly activatable and environment-insensitive optical highlighters for selective spatiotemporal imaging of target proteins. J. Am. Chem. Soc. 2012, 134, 11153–11160. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Prakash, S.; Prakash, D. Synthesis and characterization of some hetero binuclear Ni-II Schiff base complexes. J. Indian Chem. Soc. 2012, 89, 19–23. [Google Scholar]
- Ma, X.; Cheng, J.; Liu, J.; Zhou, X.; Xiang, H. Ratiometric fluorescent pH probes based on aggregation-induced emission-active salicylaldehyde azines. New J. Chem. 2015, 39, 492–500. [Google Scholar] [CrossRef]
- Nehra, P.; Khungar, B.; Singh, R.P.; Sivasubramanian, S.; Jha, P.N.; Saini, V. Synthesis, characterization and applications of imidazolium ionic liquid-tagged zinc (II) complex. Inorg. Chim. Acta 2018, 478, 260–267. [Google Scholar] [CrossRef]
- Al-Jorani, K.R.; Atia, A.J.K.; Lafta, S.J.; Al-Bayti, R.I.; Kadhem, S.A.; Baqer, S.M. Antibacterial activity of new benzimidazole moiety synthesis via a acid chloride and related heterocyclic chalcones. J. Pharm. Sci. Res. 2019, 11, 1195–1203. [Google Scholar]
- Dias, G.G.; Paz, E.R.; Kadooca, J.Y.; Sabino, A.A.; Cury, L.A.; Torikai, K.; De Simone, C.A.; Fantuzzi, F.; da Silva Junior, E.N. Rhodium (III)-Catalyzed C–H/N–H Alkyne Annulation of Nonsymmetric 2-Aryl (Benz) imidazole Derivatives: Photophysical and Mechanistic Insights. J. Org. Chem. 2020, 86, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zheng, N.; Li, J.; Zheng, Y.; Song, W. Iridium-catalyzed orthogonal and regioselective synthesis of triazole disulfides in aqueous media under mild conditions. Green Chem. 2020, 22, 2394–2398. [Google Scholar] [CrossRef]
- Chu, N.; Wang, Y.; Jia, H.; Han, J.; Wang, X.; Hou, Z. Design, Synthesis and Biological Evaluation of New Carbohydrate-Based Coumarin Derivatives as Selective Carbonic Anhydrase IX Inhibitors via “Click” Reaction. Molecules 2022, 27, 5464. [Google Scholar] [CrossRef] [PubMed]


| Comp | A1 | A2 | A3 | T1 | T2 | T3 | T4 |
|---|---|---|---|---|---|---|---|
| IC50/μmol·L−1 | >98 | >96 | >100 | >100 | >92 | >100 | >87 |
| Comp | A1 | A2 | A3 |
|---|---|---|---|
| 4AKY | 6.4 kcal/mol | 7.3 kcal/mol | 7.1 kcal/mol |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Zhao, M.; Song, C.; Sun, J.; Tao, J.; Sun, B.; Jiang, J. Click Triazole as a Linker for Pretargeting Strategies: Synthesis, Docking Investigations, Fluorescence Diagnosis, and Antibacterial Action Studies. Molecules 2023, 28, 2758. https://doi.org/10.3390/molecules28062758
Liu Q, Zhao M, Song C, Sun J, Tao J, Sun B, Jiang J. Click Triazole as a Linker for Pretargeting Strategies: Synthesis, Docking Investigations, Fluorescence Diagnosis, and Antibacterial Action Studies. Molecules. 2023; 28(6):2758. https://doi.org/10.3390/molecules28062758
Chicago/Turabian StyleLiu, Qian, Mingxia Zhao, Cairong Song, Jiankang Sun, Jiali Tao, Bin Sun, and Junbing Jiang. 2023. "Click Triazole as a Linker for Pretargeting Strategies: Synthesis, Docking Investigations, Fluorescence Diagnosis, and Antibacterial Action Studies" Molecules 28, no. 6: 2758. https://doi.org/10.3390/molecules28062758
APA StyleLiu, Q., Zhao, M., Song, C., Sun, J., Tao, J., Sun, B., & Jiang, J. (2023). Click Triazole as a Linker for Pretargeting Strategies: Synthesis, Docking Investigations, Fluorescence Diagnosis, and Antibacterial Action Studies. Molecules, 28(6), 2758. https://doi.org/10.3390/molecules28062758





