MYB24 Negatively Regulates the Biosynthesis of Lignin and Capsaicin by Affecting the Expression of Key Genes in the Phenylpropanoid Metabolism Pathway in Capsicum chinense
Abstract
1. Introduction
2. Results
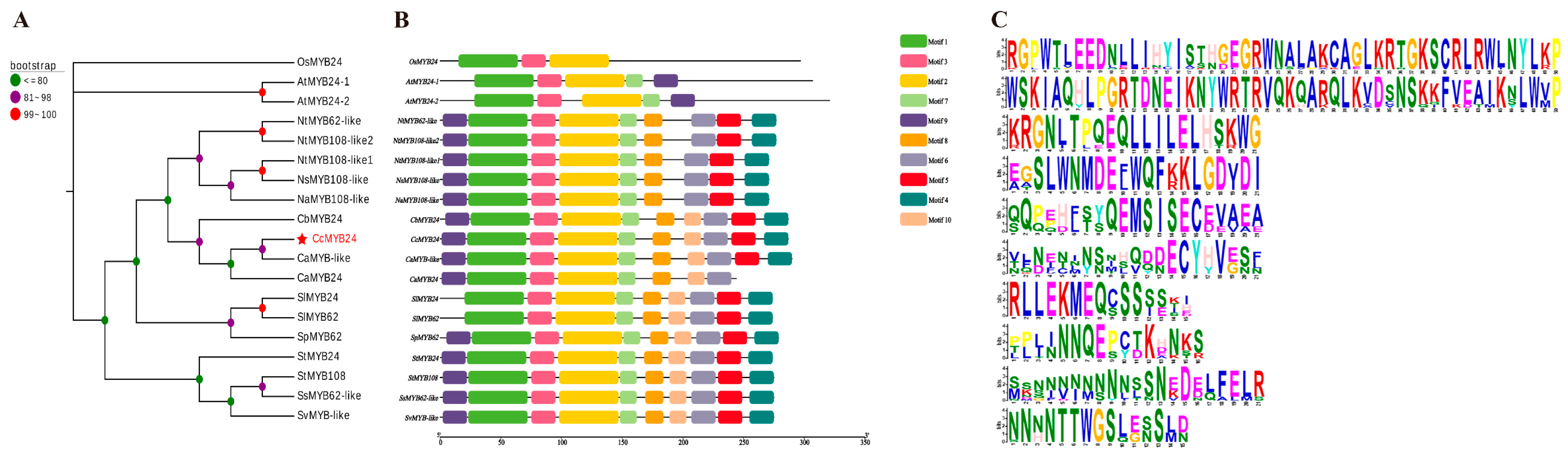
2.1. Cloning and Sequence Analysis of CcMYB24 Transcription Factors
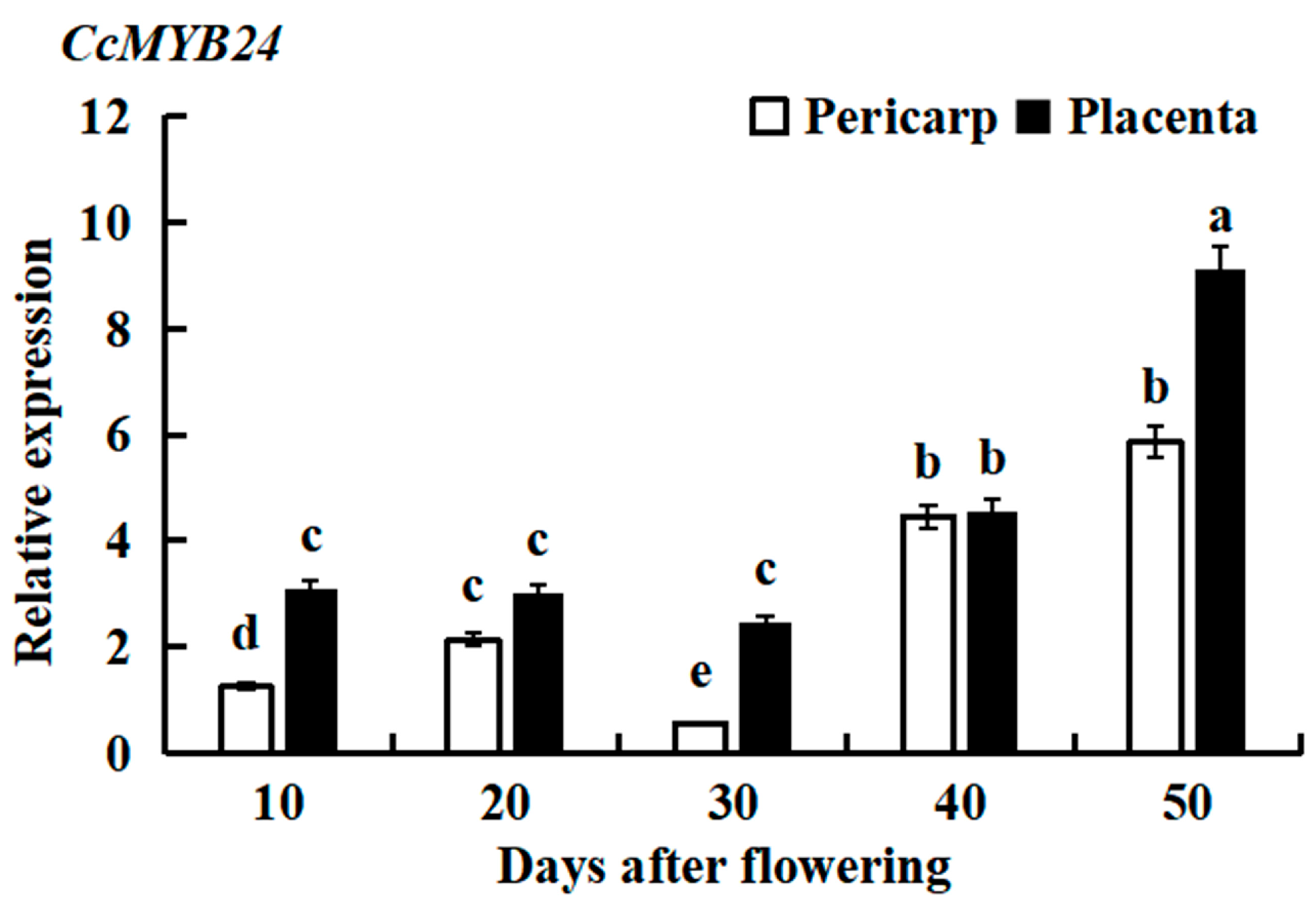
2.2. Differential Expression of CcMYB24 in Different Growth and Development Stages of Pepper
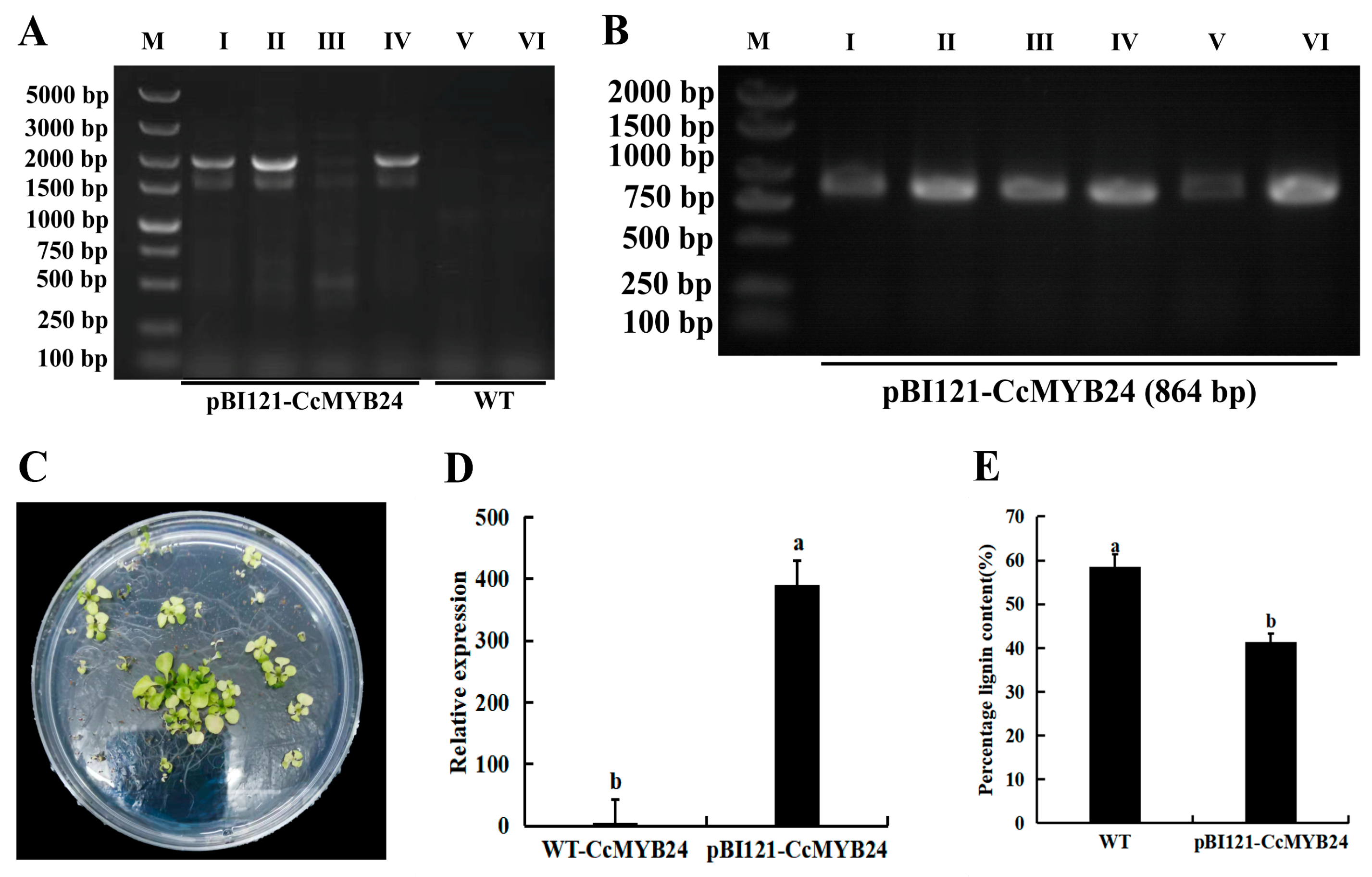
2.3. Heterologous Expression of the CcMYB24 Gene in A. thaliana
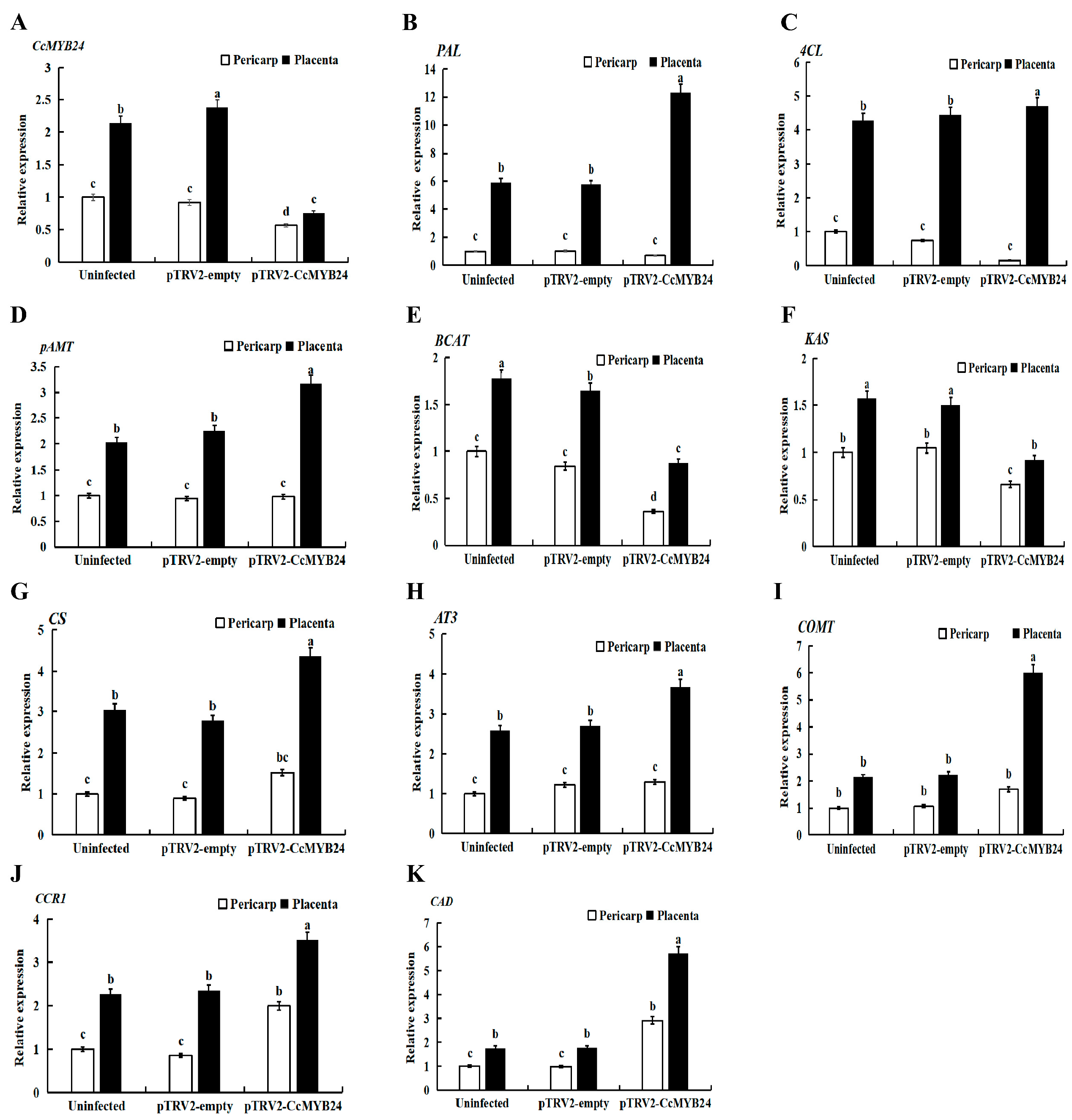
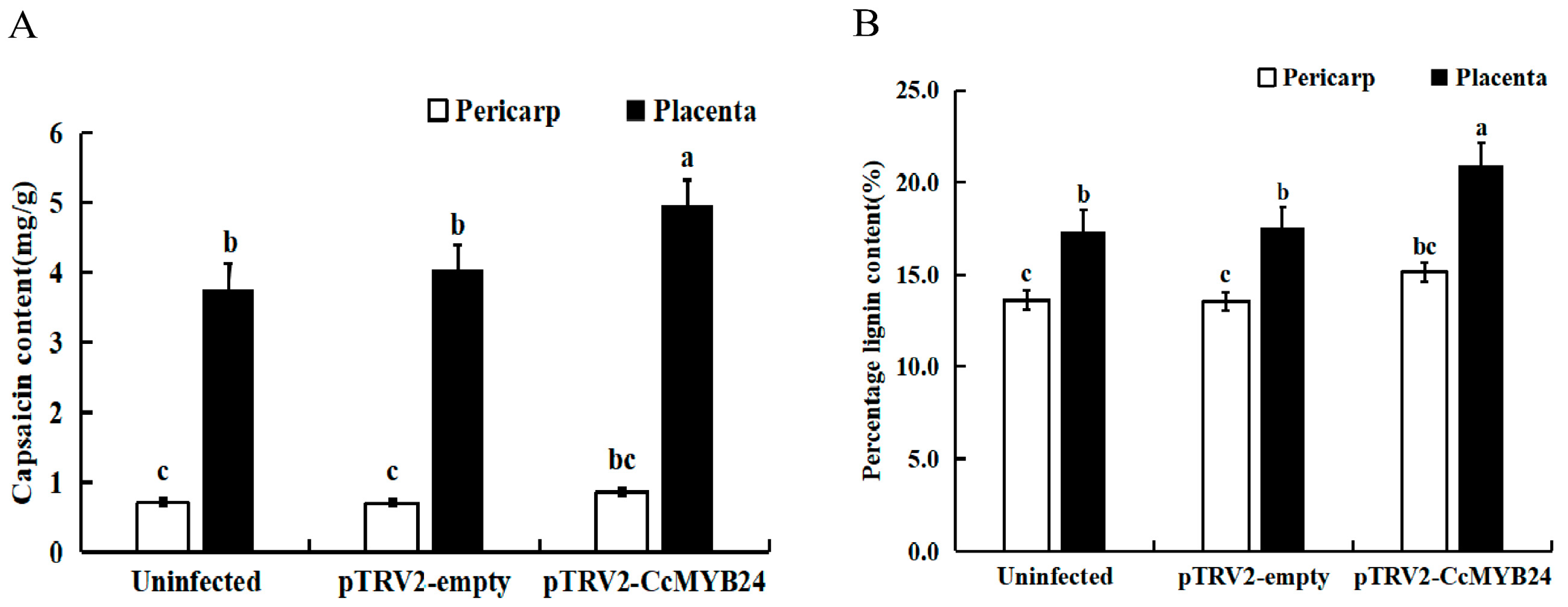
2.4. VIGS Identification of CcMYB24 in Hainan Huangdenglong Pepper
3. Discussion
4. Materials and Methods
4.1. Experimental Materials
4.2. Main Reagents
4.3. Extraction of Total RNA and Synthesis of the First Strand of cDNA
4.4. Cloning of the CcMYB24 Transcription Factor
4.5. Bioinformatics Analysis of the CcMYB24 Gene
4.6. qRT-PCR Analysis of CcMYB24 in Different Growth and Development Stages of Pepper
4.7. Heterologous Expression of CcMYB24 in A. thaliana
4.8. VIGS of CcMYB24 in Hainan Huangdenglong Pepper
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Appendix A. Primer Sequences
| Primer | Sequence (5′–3′) | Use |
|---|---|---|
| CcMYB24-F | ATGAGTAGTAATAATAATAATAATAATTTATCATC | Clone |
| CcMYB24-R | TCAAATATCTACATCTCCTAGC | Clone |
| pB-CcMYB24-F | gagaacacgggggactctagaATGAGTAGTAATAATAATAATAATAATTTATCATC | Overexpression |
| pB-CcMYB24-R | cgatcggggaaattcgagctcTCAAATATCTACATCTCCTAGC | Overexpression |
| pT-CcMYB24-F | gtgagtaaggttaccgaattcGAAGCAATAAAGAATCTATGGGTA | VIGS |
| pT-CcMYB24-R | cgtgagctcggtaccggatccACAATGATCCTTCATTAGTAAACC | VIGS |
| QMYB24-F | AATAGCAAGAAGTTTGTTGAAGCA | qRT-PCR |
| QMYB24-R | TAAGGGTAAATTTGGAAATGAAAG | qRT-PCR |
| Actin-F | GTCCATCTGCTCTCTGTTG | qRT-PCR |
| Actin-R | CACCCCAAGCACAATAAGAC | qRT-PCR |
| PAL-F | TGTCCCGTTGTCCTACATTGCT | qRT-PCR |
| PAL-R | CTCGGGCTTTCCATTCATCAC | qRT-PCR |
| 4CL-F | CTTCTTCTCAACCATCCCAACA | qRT-PCR |
| 4CL-R | ACGAAATCCTTGACTTCATCCTC | qRT-PCR |
| pAMT-F | TGGATTTGGAAGACTTGGGACA | qRT-PCR |
| pAMT-R | GCTTACAAGGACAGCGGCA | qRT-PCR |
| COMT-F | TAGCACATAACCCAGGAGGCA | qRT-PCR |
| COMT-R | CACAGCACACCTTACG GAATCT | qRT-PCR |
| KAS-F | TCGGCTATTGGTGTTGGTGGT | qRT-PCR |
| KAS-R | CACCTCCCAAGTATTCCGCTA | qRT-PCR |
| AT3-F | TCAAATGGCAGTTTCCCTTCTC | qRT-PCR |
| AT3-R | TCAAATGGCAGTTTCCCTTCTC | qRT-PCR |
| CAD-F | TATGGCACCAGAACAAGCAG | qRT-PCR |
| CAD-R | CTCCAAGTCCCAATATTCCAC | qRT-PCR |
| CCR1-F | GATCCAGAACAAATGGTGGAGC | qRT-PCR |
| CCR1-R | CCAGCAAGTCTCGTCCA CAAC | qRT-PCR |
| CS-F | TTGGCTCGCGTATAATGACTT | qRT-PCR |
| CS-R | TGCCGCTGGAATAACACCTC | qRT-PCR |
References
- Zhang, D.; Sun, X.; Battino, M.; Wei, X.; Shi, J.; Zhao, L.; Liu, S.; Xiao, J.; Shi, B.; Zou, X. A comparative overview on chili pepper (capsicum genus) and sichuan pepper (zanthoxylum genus): From pungent spices to pharma-foods. Trends Food Sci. Technol. 2021, 117, 148–162. [Google Scholar] [CrossRef]
- Saleh, B.K.; Omer, A.; Teweldemedhin, B. Medicinal uses and health benefits of chili pepper (Capsicum spp.): A review. MOJ Food Process. Technol. 2018, 6, 325–328. [Google Scholar] [CrossRef]
- Baenas, N.; Belović, M.; Ilic, N.; Moreno, D.A.; García, V.C. Industrial use of pepper (Capsicum annum L.) derived products: Technological benefits and biological advantages. Food Chem. 2019, 274, 872–885. [Google Scholar] [CrossRef]
- Pyo, M.Y.; Park, B.K.; Choi, J.J.; Yang, M.; Yang, H.O.; Cha, J.W.; Kim, J.-C.; Kim, I.S.; Lee, H.B.; Jin, M. Pheophytin a and chlorophyll a identified from environmentally friendly cultivation of green pepper enhance interleukin-2 and interferon-γ in Peyer’s patches ex vivo. Biol. Pharm. Bull. 2013, 36, 1747–1753. [Google Scholar] [CrossRef] [PubMed]
- Gudeva, L.K.; Mitrev, S.; Maksimova, V.; Spasov, D. Content of capsaicin extracted from hot pepper (Capsicum annuum ssp. microcarpum L.) and its use as an ecopesticide. Hem. Ind. 2013, 67, 671–675. [Google Scholar] [CrossRef]
- Srinivasan, K. Biological Activities of Red Pepper (Capsicum annuum) and Its Pungent Principle Capsaicin: A Review. Crit. Rev. Food Sci. Nutr. 2016, 56, 1488–1500. [Google Scholar] [CrossRef] [PubMed]
- Crapnell, R.D.; Banks, C.E. Electroanalytical overview: The pungency of chile and chilli products determined via the sensing of capsaicinoids. Analyst 2021, 146, 2769–2783. [Google Scholar] [CrossRef]
- Hideshi, F.; Tetsuya, S.; Kazuo, I. Capsaicinoid Formation in the Protoplast from the Placenta of Capsicum Fruits. Agric. Biol. Chem. 2014, 46, 2591–2592. [Google Scholar] [CrossRef]
- Liu, S.; Chen, C.; Chen, G.; Cao, B.; Chen, Q.; Lei, J. RNA-sequencing tag profiling of the placenta and pericarp of pungent pepper provides robust candidates contributing to capsaicinoid biosynthesis. Plant Cell Tissue Organ Cult. 2012, 110, 111–121. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; Alseekh, S.; Tohge, T.; Rallapalli, G.; Luo, J.; Kawar, P.G.; Hill, L.; Santino, A.; Fernie, A.R.; et al. Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 2015, 6, 8635. [Google Scholar] [CrossRef]
- Han, K.; Lee, H.Y.; Ro, N.Y.; Hur, O.S.; Lee, J.H.; Kwon, J.K.; Kang, B.C. QTL mapping and GWAS reveal candidate genes controlling capsaicinoid content in Capsicum. Plant Biotechnol. J. 2018, 16, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Mazourek, M.; Pujar, A.; Borovsky, Y.; Paran, I.; Mueller, L.; Jahn, M.M. A dynamic interface for capsaicinoid systems biology. Plant Physiol. 2009, 150, 1806–1821. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Yu, C.S.; Shen, Y.; Fang, X.D.; Chen, L.; Min, J.M.; Cheng, J.W.; Zhao, S.C.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, M.; Yeom, S.; Kim, Y.M.; Lee, J.M.; Lee, H.A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef]
- Arce-Rodríguez, M.L.; Ochoa-Alejo, N. Biochemistry and molecular biology of capsaicinoid biosynthesis: Recent advances and perspectives. Plant Cell Rep. 2019, 38, 1017–1030. [Google Scholar] [CrossRef]
- Stewart, C.J.; Kang, B.C.; Liu, K.; Mazourek, M.; Moore, S.L.; Yoo, E.Y.; Kim, B.D.; Paran, I.; Jahn, M.M. The Pun1 gene for pungency in pepper encodes a putative acyltransferase. Plant J. 2005, 42, 675–688. [Google Scholar] [CrossRef]
- Kabita, K.C.; Sharma, S.K.; Sanatombi, K. Analysis of capsaicinoid biosynthesis pathway genes expression in callus cultures of Capsicum chinense Jacq. cv. ‘Umorok’. Plant Cell Tissue Organ Cult. 2019, 137, 565–573. [Google Scholar] [CrossRef]
- Arce-Rodriguez, M.L.; Ochoa-Alejo, N. Silencing AT3 gene reduces the expression of pAmt, BCAT, Kas, and Acl genes involved in capsaicinoid biosynthesis in chili pepper fruits. Biol. Plant. 2015, 59, 477–484. [Google Scholar] [CrossRef]
- Abraham-Juárez, M.; Rocha-Granados, M.; López, M.G.; Rivera-Bustamante, R.F.; Ochoa-Alejo, N. Virus-induced silencing of Comt, pAmt and Kas genes results in a reduction of capsaicinoid accumulation in chili pepper fruits. Planta 2008, 227, 681–695. [Google Scholar] [CrossRef]
- Koeda, S.; Sato, K.; Saito, H.; Nagano, A.J.; Yasugi, M.; Kudoh, H.; Tanaka, Y. Mutation in the putative ketoacyl-ACP reductase CaKR1 induces loss of pungency in Capsicum. Theor. Appl. Genet. 2019, 132, 65–80. [Google Scholar] [CrossRef]
- Christian, D.; Stracke, R.; Erich, G.; Bernd, W.; Cathie, M.; Loïc, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Ma, D.; Constabel, C.P. MYB respressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Sarpras, M.; Rashmi, G.; Vineet, S.; Chhapekar, S.S.; Jharna, D.; Ajay, K.; Satish, K.Y.; Mukesh, N.; Vijaya, B.; Suresh, K.A.; et al. Comparative analysis of fruit metabolites and pungency candidate genes expression between bhut jolokia and other Capsicum species. PLoS ONE 2016, 11, e0167791. [Google Scholar] [CrossRef]
- Tanaka, Y.; Nakashima, F.; Kirii, E.; Goto, T.; Yoshida, Y.; Yasuba, K.I. Difference in capsaicinoid biosynthesis gene expression in the pericarp reveals elevation of capsaicinoid contents in chili peppers (Capsicum chinense). Plant Cell Rep. 2017, 36, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Arce-Rodríguez, M.L.; Ochoa-Alejo, N. An R2R3-MYB transcription factor regulates capsaicinoid biosynthesis. Plant Physiol. 2017, 174, 1359–1370. [Google Scholar] [CrossRef]
- Sun, B.; Zhu, Z.; Chen, C.; Chen, G.; Cao, B.; Chen, C.; Lei, J. Jasmonate-inducible R2R3-MYB transcription factor regulates capsaicinoid biosynthesis and stamen development in Capsicum. J. Agric. Food Chem. 2019, 67, 10891–10903. [Google Scholar] [CrossRef]
- Sun, B.M.; Zhou, X.; Chen, C.M.; Chen, C.J.; Chen, K.H.; Chen, M.X.; Liu, S.Q.; Chen, G.J.; Cao, B.H.; Cao, F.R.; et al. Coexpression network analysis reveals an MYB transcriptional activator involved in capsaicinoid biosynthesis in hot peppers. Hortic. Res. 2020, 7, 816–824. [Google Scholar] [CrossRef]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Marie, P.; Meike, B. Regulation of MYB and bHLH transcription factors: A glance at the protein level. Mol. Plant 2015, 8, 378–388. [Google Scholar] [CrossRef]
- Wang, S.; Shi, M.; Zhang, Y.; Xie, X.; Sun, P.; Fang, C.; Zhao, J. FvMYB24, a strawberry R2R3-MYB transcription factor, improved salt stress tolerance in transgenic Arabidopsis. Biochem. Biophys. Res. Commun. 2021, 569, 93–99. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, W.; Jiang, H.; Mao, Z.; Wang, N.; Jiang, S.; Xu, H.; Yang, G.; Zhang, Z.; Chen, X. The R2R3-MYB transcription factor MdMYB24-like is involved in methyl jasmonate-induced anthocyanin biosynthesis in apple. Plant Physiol. Biochem. 2019, 139, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Yu, Y.N.; Jia, J.H.; Li, Q.H.; Gong, Z.H. The pepper MYB transcription factor CaMYB306 accelerates fruit coloration and negatively regulates cold resistance. Sci. Hortic. 2022, 295, 0304–4238. [Google Scholar] [CrossRef]
- Xu, Q.; Yin, X.-R.; Zeng, J.-K.; Ge, H.; Song, M.; Xu, C.-J.; Li, X.; Ferguson, I.B.; Chen, K.-S. Activator- and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J. Exp. Bot. 2014, 65, 4349–4359. [Google Scholar] [CrossRef]
- Diao, W.P.; Snyder, J.C.; Wang, S.B.; Liu, J.B.; Pan, B.G.; Guo, G.J.; Wei, J. Genome-Wide Identification and Expression Analysis of WRKY Gene Family in Capsicum annuum L. Front. Plant Sci. 2016, 7, 211. [Google Scholar] [CrossRef]
- Song, J.; Chen, C.; Zhang, S.; Wang, J.; Huang, Z.; Chen, M.; Cao, B.; Zhu, Z.; Lei, J. Systematic analysis of the Capsicum ERF transcription factor family: Identification of regulatory factors involved in the regulation of species-specific metabolites. BMC Genom. 2020, 21, 573. [Google Scholar] [CrossRef]
- Yang, L.; Zhao, X.; Ran, L.; Li, C.; Fan, D.; Luo, K. PtoMYB156 is involved in negative regulation of phenylpropanoid metabolism and secondary cell wall biosynthesis during wood formation in poplar. Sci. Rep. 2017, 7, 41209. [Google Scholar] [CrossRef]
- Zhu, Z.; Sun, B.; Cai, W.; Zhou, X.; Mao, Y.; Chen, C.; Wei, J.; Cao, B.; Chen, C.; Chen, G.; et al. Natural variations in the MYB transcription factor MYB31 determine the evolution of extremely pungent peppers. New Phytol. 2019, 223, 1111. [Google Scholar] [CrossRef]
- Chen, L.; Wu, F.; Zhang, J. NAC and MYB Families and Lignin Biosynthesis-Related Members Identification and Expression Analysis in Melilotus albus. Plants 2021, 10, 303. [Google Scholar] [CrossRef]
- Wang, X.C.; Wu, J.; Guan, M.L.; Zhao, C.H.; Geng, P.; Zhao, Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020, 101, 637–652. [Google Scholar] [CrossRef]
- Chen, L.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Biochem. 2019, 136, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Surbhi, S.; Sathi, P.; Sampa, D. Overexpression of CaMYB78 transcription factor enhances resistance response in chickpea against Fusarium oxysporum and negatively regulates anthocyanin biosynthetic pathway. Protoplasma 2023, 260, 589–605. [Google Scholar] [CrossRef]
- Zhao, W.; Wu, W.; Liu, Y.; Fan, L.; Meng, H.; Wang, L.; Wu, G. The TT2-type MYB transcription factor JrMYB12 positively regulates proanthocyanidin biosynthesis in red walnut. Sci. Hortic. 2022, 307, 111515. [Google Scholar] [CrossRef]
- Kang, W.H.; Sim, Y.M.; Koo, N.; Nam, J.Y.; Lee, J.; Kim, N.; Jang, H.; Kim, Y.M.; Yeom, S.I. Transcriptome profiling of abiotic responses to heat, cold, salt, and osmotic stress of Capsicum annuum L. Sci. Data 2020, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Yu, X.; Hori, C.; Demura, T.; Ohtani, M.; Zhu, Q. Heterologous Overexpression of Poplar SnRK2 Genes Enhanced Salt Stress Tolerance in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 612. [Google Scholar] [CrossRef]
- Li, M.; Yang, Y.; Raza, A.; Yin, S.; Wang, H.; Zhang, Y.; Dong, J.; Wang, G.; Zhong, C.; Zhang, H.; et al. Heterologous expression of Arabidopsis thaliana rty gene in strawberry (Fragaria × ananassa Duch.) improves drought tolerance. BMC Plant Biol. 2021, 21, 57. [Google Scholar] [CrossRef]
- Prasad, B.C.; Kumar, V.; Gururaj, H.B.; Parimalan, R.; Giridhar, P.; Ravishankar, G.A. Characterization of capsaicin synthase and identification of its gene (csy1) for pungency factor capsaicin in pepper (Capsicum sp.). Proc. Natl. Acad. Sci. USA 2006, 103, 13315–13320. [Google Scholar] [CrossRef]
- Zhang, Z.X.; Zhao, S.N.; Liu, G.F.; Huang, Z.M.; Cao, Z.M.; Cheng, S.H.; Lin, S.S. Discovery of putative capsaicin biosynthetic genes by RNA-Seq and digital gene expression analysis of pepper. Sci. Rep. 2016, 6, 34121. [Google Scholar] [CrossRef]
- Jeanne, C.; Maneesha, A.; Marcus, M.; Jacob, N.; Martin, M.; Mary, A.O. Transcripts for possible capsaicinoid biosynthetic genes are differentially accumulated in pungent and non-pungent Capsicum spp. Plant Sci. 1999, 148, 47–57. [Google Scholar] [CrossRef]
- Yoel, S.; Tzvi, T. Agrobacterium-mediated plant genetic transformation. Plant Biotechnol. Agric. 2012, 12, 99–116. [Google Scholar] [CrossRef]
- Ramkumar, T.R.; Lenka, S.K.; Arya, S.S.; Bansal, K.C. A Short History and Perspectives on Plant Genetic Transformation. Methods Mol. Biol. 2020, 2124, 39–68. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, A. Genetic Transformation of Plants: Introduction and Recent Advances. Res. J. Pharmacogn. Phytochem. 2017, 9, 125–127. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Zhang, W.; Zhang, L.; Wu, D.; Sun, P.; Huang, C.; Fu, G.; Deng, Q.; Wang, Z.; Cheng, S. MYB24 Negatively Regulates the Biosynthesis of Lignin and Capsaicin by Affecting the Expression of Key Genes in the Phenylpropanoid Metabolism Pathway in Capsicum chinense. Molecules 2023, 28, 2644. https://doi.org/10.3390/molecules28062644
Yu S, Zhang W, Zhang L, Wu D, Sun P, Huang C, Fu G, Deng Q, Wang Z, Cheng S. MYB24 Negatively Regulates the Biosynthesis of Lignin and Capsaicin by Affecting the Expression of Key Genes in the Phenylpropanoid Metabolism Pathway in Capsicum chinense. Molecules. 2023; 28(6):2644. https://doi.org/10.3390/molecules28062644
Chicago/Turabian StyleYu, Shuang, Wei Zhang, Liping Zhang, Dan Wu, Peixia Sun, Chuang Huang, Genying Fu, Qin Deng, Zhiwei Wang, and Shanhan Cheng. 2023. "MYB24 Negatively Regulates the Biosynthesis of Lignin and Capsaicin by Affecting the Expression of Key Genes in the Phenylpropanoid Metabolism Pathway in Capsicum chinense" Molecules 28, no. 6: 2644. https://doi.org/10.3390/molecules28062644
APA StyleYu, S., Zhang, W., Zhang, L., Wu, D., Sun, P., Huang, C., Fu, G., Deng, Q., Wang, Z., & Cheng, S. (2023). MYB24 Negatively Regulates the Biosynthesis of Lignin and Capsaicin by Affecting the Expression of Key Genes in the Phenylpropanoid Metabolism Pathway in Capsicum chinense. Molecules, 28(6), 2644. https://doi.org/10.3390/molecules28062644







