Abstract
The Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) virus has been causing the COVID-19 pandemic since December 2019, with over 600 million infected persons worldwide and over six million deaths. We investigated the anti-viral effects of polyphenolic green tea ingredients and the synthetic resveratrol analogue 3,3′,4,4′,5,5′-hexahydroxy-trans-stilbene (HHS), a compound with antioxidant, antitumor and anti-HIV properties. In the TCID50 assay, four out of nine green tea constituents showed minor to modest cell protective effects, whereas HHS demonstrated the highest reduction (1103-fold) of the TCID50, indicating pronounced inhibition of virus replication. HHS was also a highly effective inhibitor of SARS-CoV-2 proliferation in VeroE6 cells with an IC50 value of 31.1 µM. HSS also inhibited the binding of the receptor-binding domain (RBD) of the spike protein to the human angiotensin-converting enzyme 2 (ACE2) receptor (RBD-ACE2) binding with 29% at 100 µM and with 9.2% at 50 µM indicating that the SARS-CoV-2 inhibitory effect might at least in part be attributed to the inhibition of virus binding to ACE2. Based on the chemical similarity to other polyphenols, the oral bioavailability of HHS is likely also very low, resulting in blood levels far below the inhibitory concentration of EGCG against SARS-CoV-2 observed in vitro. However, administration of HHS topically as a nose or throat spray would increase concentrations several-fold above the minimal inhibitory concentration (MIC) in the mucosa and might reduce virus load when administered soon after infection. Due to these promising tissue culture results, further preclinical and clinical studies are warranted to develop HHS as an additional treatment option for SARS-CoV-2 infection to complement vaccines, which is and will be the main pillar to combat the COVID-19 pandemic.
1. Introduction
In December 2019, starting in Wuhan, China, a novel coronavirus (SARS-CoV-2) caused a pandemic that will continue until effective and safe vaccines can be provided to the world or causal drug treatment of the disease can help to avoid severe and fatal cases of the illness. Over 600 million persons have been infected worldwide, with increasing numbers who are awaiting additional effective treatment options. Although various vaccines and antiviral compounds were introduced, additional options to treat or prevent SARS-CoV-2 infections are warranted.
Green tea polyphenols might be promising candidates as they exhibit antiviral activity against a wide range of diseases, including viral diseases. Epigallocatechin 3-gallate (EGCG) is the major ingredient in green tea and accounts for 50–80% of a brewed cup [1]. EGCG showed antiviral activity not only against influenza, hepatitis B and C, herpes simplex and HIV but also against SARS-CoV-2 [1]. It was also observed that EGCG had lowered the production of coronavirus-associated proteins in infected cells [1] and decreased the level of detected virus particles in a mouse model [2]. So far, no in vitro or in vivo data about the activity of other polyphenols in green tea against SARS-CoV-2 exist. However, molecular visualization and molecular docking experiments demonstrated the binding of EGCG and other green tea polyphenols to the central channel of the SARS-CoV-2 spike protein [3] and the SARS-CoV-2 main protease (Mpro) [4] as well as interactions with papain-like protease (PLpro), RNA-dependent RNA polymerase (RdRP), Helicase (nsp13) and the endonuclease Nsp15 (NendoU) [5].
We therefore investigated the effects of the main green tea catechins, namely catechin (C), catechin gallate (CG), EGCG, epicatechin (EC), epicatechin gallate (ECG), epigallocatechin gallate (EGC), and gallocatechin gallate (GCG) regarding their ability to inhibit or lower virus replication in vitro compared to the potent and clinically used anti-COVID-19 drug remdesivir. As Maiti and Banerjee [3] showed that the number of galloyl groups favors greater attachment to the receptor ACE2, we also included gallic acid (GA) and ellagic acid (ELG), a dimeric gallic acid derivative, in our study—both compounds also present in considerable amounts in green tea. Previous investigations in our lab demonstrated that the anticancer and anti-inflammatory effects of resveratrol analogues are increased by the number of OH groups [6,7,8]. In order to investigate whether the numbers and position of phenolic groups of green tea compounds may also correlate with activity, we included 3,3′,4,4′,5,5′-hexahydroxy-trans-stilbene (HHS), a synthetic resveratrol analogue. The chemical structures of all compounds used are shown in Figure 1.
Figure 1.
Chemical structures of the nine green tea constituents GA, C, CG, EGCG, EC, ECG, EGC, GCG, and ELG, as well as the synthetic hexahydroxy resveratrol analog HHS as a control compound, for structure–activity relationships.
As mentioned previously, HHS is an excellent free radical scavenger, thus inhibiting various enzymes crucial for nucleoside and nucleotide synthesis [9]. It inhibits ribonucleotide reductase, a key enzyme of nucleic acid synthesis [10]. In addition, HHS was shown to be a selective cyclooxygenase-2 inhibitor [7]. In vivo activity could be proved in a mouse melanoma model, showing anti-tumor activity and even inhibition of the development of melanoma metastasis [11]. Later HHS was shown by Han and coworkers to be an effective HIV-1 inhibitor [12]. They investigated this compound in different cell lines after infection with different strains of HIV-1. They found that HHS is acting at a very early stage of virus infection. It might inhibit a step prior to reverse transcription, possibly interacting with the entry of the virus into the target cells. The observed mechanism of action is different from the effects seen with resveratrol. Resveratrol competes with the substrates deoxynucleoside triphosphates (dNTPs), thus blocking chain polymerization [13].
As there are no in vitro data on whether HHS is able to inhibit virus uptake into cells, we further investigated the interaction of HHS with the angiotensin-converting enzyme 2 (ACE2) receptor, which is located on cell membranes and used by SARS-CoV-2 to gain entry into human cells. As host cell invasion is initiated through direct binding of the viral spike protein to ACE2, disrupting the spike-ACE2 interaction would be a potential therapeutic target for treating COVID-19.
2. Results
2.1. Median Tissue Culture Infective Dose (TCID50)
To investigate in a preliminary screen whether green tea constituents or HHS inhibit SARS-CoV-2 infection, VeroCCL81 cells were pretreated with the various compounds at a concentration of 50 µM, and the mixtures were subjected to TCID50 in order to determine virus titers. GA, EGCG, EGC, GCG and HHS showed cell protective effects and reduced the TCID50 by at least 1 Log scale. TCID50 was significantly lowered by 50 µM of GA (9.62 fold reduction), EGCG (451 fold reduction), EGC (4.09 fold reduction), GCG (52.4 fold reduction) and HHS (1103 fold reduction), indicating pronounced inhibition of virus replication. TCID50 -values for compounds C, CG, EC, EGC and ELG were below the detection limit and in the range of DMSO (Figure 2). In additional experiments, GA, EGCG, EGC, GCG and HHS were also incubated with non-infected Vero cells showing that the compounds did not interfere with cell growth, at least in concentrations up to 50 µM.
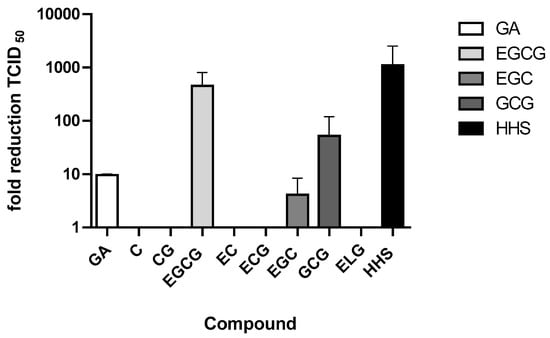
Figure 2.
Effect of treatment of Vero cells with the respective substances compound (50 µM) after infection of the cells with SARS-CoV-2. Vero cells were preincubated for 45 min with either DMSO or the compound and infected with a SARS-CoV-2 stock. After five to seven days TCID50 dose was calculated. Values represent the average + half range of two determinations.
2.2. Reduction of Virus Infection Efficiency
As HHS showed the most pronounced inhibitor effects against SARS-CoV-2 in VeroCL81 cells, only this compound was used in further experiments but not the other minor potent green tea polyphenols. Figure 3 shows the effect of treatment with HHS on virus-infected VeroE6 cells. We observed a clear reduction of the amount of detected viral particles in the supernatant after 48 h in the presence of HHS; remdesivir, a clinically used anti-COVID-19 drug, served as a control. At 50 µM, HHS was highly effective, leading to a cycle threshold (CT) value of 31.9, indicating a 106-fold reduction of infection and viral replication, a value significantly different from the untreated control (no compound). The CT value, therefore, shows the number of cycles necessary to spot the virus (the lower the CT value, the higher the viral load). Samples treated with remdesivir also showed a significant reduction of SARS-CoV-2 with a CT value of 26.9 at 10 µM. DMSO samples had no effect on the virus infection efficiency and were not significantly different from samples of the untreated control, indicating that DMSO (1%) does not interfere with the assay. This was further confirmed by a metabolic interference assay using non-infected VeroE6 cells resulting again in no significant changes in the metabolic activity of DMSO (1%) compared with untreated controls (the slopes k of linear regression analysis resulted in 89% metabolic activity; see Figure S1). We also investigated the possible cytotoxic properties of HHS against non-infected VeroE6 cells covering all conditions in which the virus neutralization assays were performed. As shown in Supplementary Materials Figure S2, HHS does not interfere with normal cell growth at 40 µM, 60 µM and even at 80 µM compared to DMSO (Figure S2).
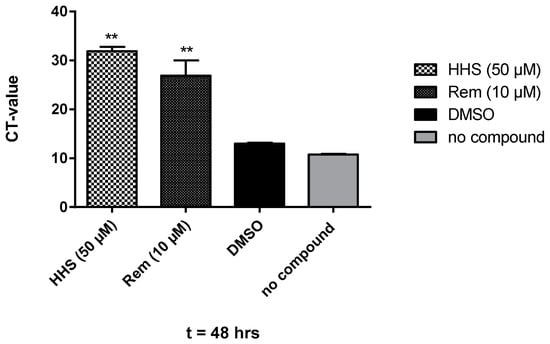
Figure 3.
Treatment of Vero E6 cells with HHS (50 µM), remdesivir (10 µM) and DMSO (1% in medium). Compounds were present pre- and during infection with SARS-CoV-2 and during incubation. After 48 h, RNA was extracted, and the viral copy numbers were quantified via qRT-PCR. Values represent the CT values (means ± SD) of five determinations. Asterisks (**) indicate significantly different (p < 0.01) in comparison to no compound (non-parametric unpaired t-test).
2.3. Inhibitory Concentrations of HHS
In the next set of experiments, VeroE6 cells were seeded in 48 well plates, infected and incubated for 24 h with different concentrations of HHS, which showed the most pronounced reduction in virus replication in the previous experiments. HHS inhibited with an IC50 of 31.1 µM, and remdesivir inhibited 50% of virus proliferation at 4.2 µM (Figure 4). Inhibition of virus proliferation could also be confirmed by immunohistochemical (IHC) staining with an anti-SARS-CoV-2 nucleocapsid-specific antibody, as depicted in Figure 5. Infected cells appeared red, and a clear dose dependency of viral inactivation was visible. Treatment of the cells with 7.5 µM HHS and 0.25 µM remdesivir still led to the infection of numerous cells, whereas 50 µM HHS and 10 µM remdesivir showed a clear viral inhibiting effect; only single cells were stained positive in a confluent cellular monolayer.
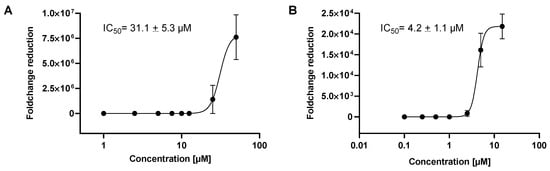
Figure 4.
Inhibition of SARS-CoV-2 viral replication by HHS and remdesivir for 24 h, (A) HHS, (B) remdesivir. IC50 values are the concentrations where the viral infection was inhibited by 50%. Values were calculated by GraphPad Prism 9 and represent the means ± SD of three determinations.
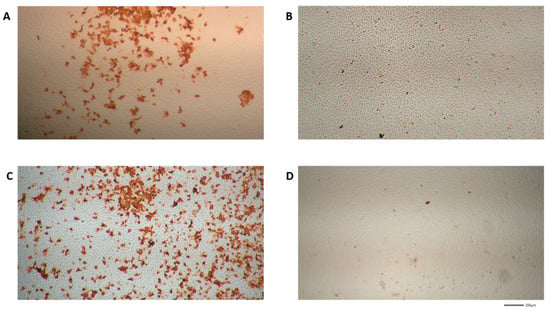
Figure 5.
SARS-CoV-2 specific immunohistochemical staining (IHC) of VeroE6 cells treated with different concentrations of HHS and remdesivir as described in the methods part (images were made from IC50 experiments). Virus-infected cells appeared red. The microscopy images are shown in four-fold magnification. (A,B) HHS 7.5 µM and 50 µM; (C,D) remdesivir 0.25 µM and 10 µM.
2.4. Inhibition of Virus Uptake
In order to determine whether inhibition of virus replication was due to alterations of virus uptake into the host cells and/or inactivation of the virus, we incubated the cells with HHS for one hour in the presence of the virus. HHS could inhibit virus replication in a concentration-dependent manner after short-term incubation for one hour, as evidenced by significantly increased CT after incubation with 25 or 50 µM compared to no compound control (results are shown in Figure 6). These results indicate that virus uptake into the host cell or viral integrity could be inhibited by HHS. Remdesivir, which acts as a nucleoside analogue, showed no inhibitory effect during the one-hour infection period (CT values: 11.7 and 11.4 for 10 µM and 5 µM).
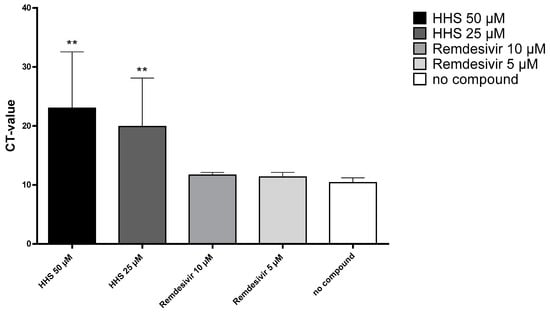
Figure 6.
Incubation of VeroE6 cells with HHS and remdesivir for one hour in parallel to virus infection. Compounds were then washed away and the cells were further incubated in medium (MEM 2% FCS) only. After 24 h, the cell supernatant was removed and tested by qPCR as described above. Values for the viral uptake inhibition represent the CT values (means ± SD) of five determinations. Asterisks (**) indicate significantly different (p < 0.01) in comparison to no compound (Kruskal–Wallis multiple comparisons).
2.5. Inhibition of ACE2 Binding
We then tested if remdesivir or HHS were able to inhibit RBD-ACE2 binding. As shown in Figure 7, remdesivir had no inhibitory effect on the binding to the receptor. However, HHS could inhibit binding to ACE 2 in a concentration-dependent manner (Figure 7). At 100 µM, HHS significantly inhibited ACE2 binding with 29% and at 50 µM with 9.2% compared to the lowest HHS concentration (6 µM). Even at low concentrations of 25 µM, 12.5 µM and 6 µM, inhibitions rates of 4.9, 5.2 and 3.1 percent were seen. However, based on the high standard deviations at 12.5 µM and 25 µM, these concentrations did not show significance to 6 µM. We, therefore, conclude that HHS can also inhibit virus uptake into the cell at a very early stage, which, at least in part, could be attributed to the blocking of the RBD-ACE2 binding.
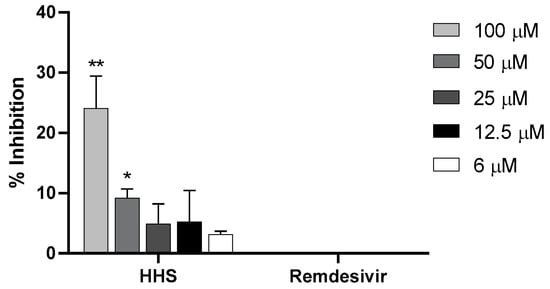
Figure 7.
Percentages of inhibition of RBD-ACE2 binding (y-axes) after pre-incubation for 3 h at RT with various concentrations of HHS and remdesivir (x-axes) followed by a 3 h overlay onto plate-bound ACE2. Bound RBD was detected with a mouse monoclonal anti-His antibody followed by a HRP-labelled anti-mouse IgG1 antibody and detected with ABTS. Values represent the means ± SD of three determinations. Asterisks (*, **) indicate significantly different (p < 0.5 and p < 0.01, respectively) in comparison to 6 µM HHS (non-parametric unpaired t-test).
3. Discussion
In the present study, we screened nine green tea polyphenols against SARS-CoV-2. Up to now, only the main constituent EGCG was shown to be active against infected Vero cells. For minor green tea constituents, only modeling and data, e.g., against the central channel of the SARS-CoV-2 spike protein, the proteases Mpro, Plpro, RdRP, and nsp13 and Nsp15 are available so far [3,4,5].
Calculating the median tissue culture infective dose of TCID50, four out of nine green tea compounds were active (Figure 2). The structure–activity relationships revealed that EGCG, EGC and GCG contain a pyrogallol group in their chemical structure (pyrogallol: benzene 1,2,3, triol is a decarboxylation product of gallic acid). Compounds with only one galloyl group (CG and ECG), however, showed no virus inhibition. Interestingly, GA was slightly active, possibly because of the additional carboxyl group. One galloyl and one pyrogallol group further increased the potency (13-110-fold reduction), as shown for GCG and ECGC. HHS, a synthetic resveratrol analog containing two pyrogallol groups, showed the highest fold reduction of viral replication (1103-fold), indicating that pyrogallol groups are more important for activity than galloyl groups. Our data are in contrast to Maiti and coworker [3], who did not discriminate between the galloyl and pyrogallol groups and, therefore, wrongly stated that the numbers of galloyl groups of catechins favor binding in the central channel of the spike protein. However, we are well aware that in addition to galloyl and pyrogallol groups, other structural features of green tea constituents, e.g., lipophilicity, molecular weight, and binding affinities to SARS-CoV-2 proteins, may also contribute to the activity.
Next, we elucidated the effect of HHS (the compound which proved most effective in a previous experiment) on the amount of detected viral particles of infected VeroE6 cells. Again, HHS at 50 µM was very effective, with a CT value above 30 (Figure 3). As expected, remdesivir was more potent than HHS, showing a CT value of 26.9 already at 10 µM leading to an IC50 value of 4.2 µM compared to that of 31.1 µM for HHS. The IC50 values of HHS in the Vero cells are much higher than in HL-60 leukemia cells, as HHS is inhibits key enzymes of DNA synthesis, thus specifically targeting rapidly proliferating malignant cells [9]. The IC50 for remdesivir in our experiments was 4.2 and only slightly higher than what was determined by Pruijssers et al. with a value of 1.65 µM [14]. This difference might be due to different assay parameters and the use of infected lung cells instead of Vero cells.
HHS incubation for only one hour could also effectively inhibit virus replication, as shown in Figure 6. This suggests that HHS might block virus entry into the cells, at least to a certain extent. An interaction between HHS and the viral envelope may also be a possible mode of action, as inhibitory effects in this setup likely occur before or during viral attachment to the cell surface or in a very early uptake state. As HHS acts through different mechanisms, mutations of the virus could be sensitive toward treatment with HHS. We hypothesize that due to its unique mechanisms of action, HHS can also be used successfully in combination with other antiviral compounds to target certain steps of virus proliferation for early treatment of SARS-CoV-2 infections.
In a final experiment, we tested whether HHS might inhibit RBD-ACE2 binding. Indeed, HSS showed a partly inhibitory effect on the binding to the receptor (29% inhibition at 50 µM and 9.2% at 25 µM). In addition to binding to the ACE2 receptor, further mechanisms inhibiting nucleic acid synthesis or inhibition of proteases [1,15], such as 3C-like protease, the main protease found in coronaviruses, might also contribute to the observed inhibition of virus entry into VeroE6 cells. Remdesivir showed no binding to ACE2 as it exerts its antiviral activity inside the host cell, where it is metabolized into nucleotide triphosphate (NTP). NTP then suppresses viral replication by targeting the RNA-dependent RNA polymerase [16].
Until now, there are no data about peroral absorption, distribution, and metabolism of HHS in animal models or humans. However, based on two pyrogallol groups, oral bioavailability should be low and comparable to the green tea polyphenol EGCG. Even at a high dose of EGCG (375 to 1200 mg), maximum blood levels were only 4.3–5.6 μM and far below the inhibitory concentration of EGCG against SARS-CoV-2 observed in vitro [17]. However, since SARS-CoV-2 particles accumulate in epithelial cells of the saliva glands and oral mucosa, HHS could be applied as a throat spray, which should lead to concentrations several-fold above the minimal inhibitory concentration (MIC). Indeed, in a very recent study, human volunteers applied two puffs of green tea throat spray (8% green tea extract), and throat swaps were taken from the pharyngeal region of each participant before and after spray application [18]. The median ECGC amount in the saliva 30 min after spray application was very high and ranged between 344 and 407 µg/mL saliva (740 µM and 888 µM).
In conclusion, our data demonstrated that the synthetic polyphenolic compound HSS is an effective inhibitor of SARS-CoV-2 infection in vitro. HHS also inhibited ACE2-RBD binding, indicating that its SARS-CoV-2 inhibitory effect could be at least in part attributed to the ACE2-RBD binding. As polyphenols are extensively metabolized in the gut and liver after peroral administration, HHS might therefore be locally used as a throat spray or be inhaled to prevent bronchial virus proliferation and thus prevent severe illness after SARS-CoV-2 infection. Animal models and patient data would have to confirm our in vitro results. Further preclinical and clinical studies are therefore warranted to develop HSS as a promising treatment option for SARS-CoV-2 infection.
4. Materials and Methods
4.1. Chemicals
GA, C, CG, EGCG, EC, ECG, EGC, GCG and ELG with purities of ≥95% were obtained from Sigma-Aldrich, Munich, Germany. DMSO for molecular biology (99.9%) was purchased from Merck KgaA (Darmstadt, Germany). Remdesivir (MCE HY-104007) was obtained from THP Medical Product, Vienna, Austria. HSS was provided by the Department of Pharmaceutical Sciences, University of Vienna, Austria.
4.2. Median Tissue Culture Infective Dose (TCID50) Assay
The human SARS-CoV-2 isolate BetaCoV/Munich/BavPat1/2020 (referred to as BavPat1) was kindly provided by Christian Drosten, Charité, Berlin, and proliferated in VeroCCL81 cells. Infected VeroCCL81 cells were then plated in growth medium (DMEM containing 10% fetal calf serum (FCS), 100 µM non-essential amino acids, 1 mM sodium pyruvate and penicillin/streptomycin) into a 96-well plate at a density of 1.5–2 × 104/well on the evening before the experiment. On the next day in the morning, the growth medium was removed, and 95% confluent Vero cells were preincubated for 45 min with either DMSO or GA, C, CG, EGCG, EC, ECG, EGC, GCG, ELG and HHS at a dose of 50 µM and infected with 100 µL of serial dilutions of a SARS-CoV-2 virus stock (serial ten-fold dilutions; range from 110–108). Preincubation and virus infection was carried out in a medium containing only 2% FCS instead of 10% FCS, as previous data showed interference with the assay by the containing scavenging substances. In contrast to a more precise plaque assay, the number of plaque-forming units was not counted, but wells were only classified as infected (dead cells) or uninfected (healthy cells) for each dilution step. After five to seven days (no differences in the results between these two days were seen) all wells were checked for cell viability by light microscopy, and the TCID50 dose in the presence or absence of indicated compounds was calculated according to the Reed and Munch method [19]. Wells with viable and infected cells were identified using an inverse light microscope showing either an intact monolayer of Vero cells (viable) or wells with a massive CPE (cytopathic effect), characterized by a large number of rounded or detached cells and the absence of a monolayer (infected).
4.3. Virus Neutralization Assay
The human 2019-nCoV isolate (Ref-SKU: 026V-03883 (Wuhan Strain) obtained from Charité Berlin was proliferated in VeroE6 cells (African green monkey kidney cells purchased from Biomedica, Vienna, Austria; VC-FTV6), TCID50 titers were determined according to the Reed Munch method [19]. Plaque-forming units (PFU) were calculated with a conversion factor of 0.7, based on the ATCC-LGC Standards (www.atcc.org/support/technical-support/faqs/converting-tcid-50-to-plaque-forming-units-pfu). For virus neutralization experiments, the working stocks were diluted to a calculated multiplicity of infection (MOI) 0.002 in minimal essential medium (MEM) supplemented with 2% FCS. Experiments with active SARS-CoV-2 virus samples were performed under BSL-3 conditions. VeroE6 cells were seeded at a density of 3.0E + 04 cells per well in a 48-well plate (Corning Costar (Corning, NY, USA), cell culture treated) in 300 µL (MEM) + 2% FCS, 24 h prior to the virus neutralization assay. On the infection day, the seeding medium was removed, a medium change was made, and the pre-diluted test compounds (60 mM to 20 mM) were diluted 1:100 directly in the well containing medium and mixed properly. The final compound concentration in the well ranged from 60 µM to 20 µM. This was made 30 min prior to the viral infection. During the one-hour infection with the virus diluted to a calculated MOI of 0.002, cells were kept at 37 °C and 5% CO2. After that, the infection medium was removed, and the cells were washed twice with MEM w/o supplements to remove the unbound virus. Subsequently, fresh MEM + 2% FCS was added, and HHS was diluted 1:100 in the well and mixed properly. The cells were incubated for an additional 48 h at 5% CO2 and 37 °C until 140 µL supernatant was harvested and inactivated with AVL (viral lysis) buffer to extract RNA and quantify the viral copy numbers via quantitative real-time polymerase chain reaction (qRT-PCR). Untreated infected cells served as positive controls in the assay; no additional compounds to lower or inhibit the viral replication were used in these controls. Non-infected cells served as the background reference of the assay; no compounds and no viruses were used in the negative controls. Remdesivir was used as a control with known antiviral activity [14]. In addition, the 48-well plate was fixed in 4% formaldehyde for SARS-CoV-2-specific immunohistochemical staining (IHC).
4.4. Metabolic Activity (MA) Assay
VeroE6 Cells were seeded in 48-well plates with a 3.0E + 04 cells per well in 300 µL MEM supplemented with 2% FCS at 37 °C and 5% CO2. After 24 h, the medium was changed, and cells in MEM + 2% FCS were exposed to 1% DMSO or HHS (40, 60, and 80 µM final concentration in 1% DMSO) and further incubated for 1, 24, or 48 h. Then cells were washed twice with MEM without supplements, and the metabolic activity was measured over 120 min using a resazurin-based assay (BioTek Plate Reader Synergy 4, Thermo Fisher Scientific, Waltham, MA, USA). After the addition of resazurin (10 µM), the increase of relative fluorescent unit (RFU) was measured at 485/590 nm and a linear regression analysis was performed. The slopes (k) from 1% DMSO-treated cells were compared and normalized to the untreated controls or to HHS-treated cells (40, 60 and 80 µM), respectively. From the slopes of the linear regression, the relative metabolic activity (RMA) was calculated.
4.5. RNA Isolation, qRT-PCR and Determination of Viral Copy Numbers
Viral RNA was isolated using the QIAamp viral RNA Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s protocol. The viral RNA was then amplified with the CDC-recommended primers and probe set of N1 and N2 from 2019-Novel Coronavirus (2019-nCoV). A real-time qRT-PCR Panel was conducted (Table 1) using a QuantiTect Multiplex RT-PCR Kit (QIAGEN, Hilden, Germany) with a Rotor Gene Q cycler. The reaction volume was reduced to 25 µL with amplification for 30 min. at 50 °C and 15 min. at 95 °C; the steps were followed by 45 cycles (95 °C for 3 min. and 55 °C for 30 s). Table 1 lists the sequences of the used primers and probes. The calculation of viral copy numbers was performed identically to the protocol described by Kicker et al. [18]. Briefly, a commercially available copy number standard (ATCC VR-1986D genomic RNA from 2019 Novel Coronavirus, Lot: 70035624) was serially diluted and analyzed via qRT-PCR. The resulting CT values were plotted against ln[copy numbers], and the equation received from linear regression analysis was used to calculate the viral copy numbers from the CT values of the samples for Primer and Probe N1 and N2, respectively. Regression equations obtained for N1: y = −1.442 x + 35.079 and N2: y = −1.5 x + 38.357. These calculated copy numbers and measured CT values refer to a volume of 5 µL RNA eluate, which was used as a template in qRT-PCR.

Table 1.
qRT-PCR primers and probes.
4.6. Immunohistochemistry (IHC)
Immunohistochemistry was performed as described previously [18]. Briefly, after fixation of the cells with 4% formaldehyde, cells were permeabilized using 0,1% Triton X 100 in PBS for 10 min, and the endogen peroxidases were blocked with 3% H2O2 in methanol for 30 min. After three washing steps with PBS, the cells were incubated for 1 h with a 1:1000 dilution of primary antibody (SARS-CoV-2 (2019-nCoV) Nucleocapsid Antibody (Rabbit Mab, Sinobiological Cat: 40143-R019) in antibody diluent (REAL Antibody diluent, Agilent Technologies, Santa Clara, CA, USA, Dako Cat: S202230_2) followed by the treatment with the secondary antibody (EnVision™ + Dual Link System HRP, Agilent Technologies, Dako Cat: K5007). After washing (PBS 3x), the substrate (AEC substrate-Chromogen, Agilent Technologies, Dako, Cat: K346430-2, 2 drops) was dropped on the cells and incubated until viral infected cells were stained red. The reaction was then stopped with washing in PBS (3x) and wells were kept humid until photo documentation. For photo documentation, fourfold magnification with a Nikon Eclipse TS100 microscope (Nikon, Japan, Tokyo) was used with the JENOPTIK GRYPHAX® camera and software (Jena, Germany) were used.
4.7. Inhibition of ACE2 Binding
In addition to classical virus neutralization tests, molecular interaction assay (MIAs) allow to test components for their specific ability to inhibit the binding of RBD from SARS-CoV-2 to its cognate receptor ACE2 [20]. The MIA to detect the inhibition of RBD to ACE2 receptor binding was performed as described [21,22] with the following alterations. An amount of 200 ng His-tagged RBD was incubated with various doses (100, 50, 25, 12.2, and 6 mM) of HHS, EGCG, GCG, and remdesivir for three hours at RT followed by a 3 h overlay onto plate-bound human ACE2 receptor protein (2 µg/mL). Bound RBD was then detected with a mouse monoclonal anti-His antibody followed by a HRP-labelled anti-mouse IgG1 antibody and detected with (ABTS). All measurements were performed in duplicate with a variation of <5%.
4.8. Statistical Analysis
Statistical analyses were performed using GraphPad Prism software for Windows, version 9.0 (San Diego, CA, USA). For the calculation of p-values, either one-way ANOVA (non-parametric) with multiple comparison test (Kruskal–Wallis test) when n = 5 or unpaired test (non-parametric: Mann–Whitney test) when n = 3. Differences with p < 0.05 are considered statistically significant. Unless stated otherwise, all data are shown as means ± standard deviation (SD). The IC50 values were calculated as nonlinear regression: Log (inhibitor) vs. response (two parameters) constrain equal to 0.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28062612/s1; Figure S1: Metabolic activity of VeroE6 cells incubated with DMSO (1%) for 48 h; Figure S2: VeroE6 cells were incubated with HHS (40–80 µM) for 1 h (washed and incubated in MEM + 2 % FCS for additional 24 h), 24 h or 48 h.
Author Contributions
Conceptualization, W.J. and T.S.; methodology, S.K., E.K., M.H. and R.V.; formal analysis E.K., M.H., R.G. and M.B.; resources, W.J., S.K., K.Z. and R.V.; writing—original draft, W.J. and T.S.; writing—review and editing, W.J., T.S., E.K., M.H., R.G. and P.G.; supervision: S.K., K.Z., R.V. and W.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
K.Z. is the CEO of Zatloukal Innovations, Austria; T.S.; W.J. are applicants of the international patent (No. WO2022/200086 A1). R.V. has received research grants from Viravaxx Inc, Vienna, Austria, HVD Biotech, Vienna, Austria and Worg Pharmaceuticals, Hangzhou, China. He serves as a consultant for Viravaxx and Worg. The other authors have no conflict of interest to declare.
References
- Jang, M.; Park, Y.I.; Cha, Y.E.; Park, R.; Namkoong, S.; Lee, J.I.; Park, J. Tea polyphenols EGCG and theaflavin inhibit the activity of SARS-CoV-2 3CL-protease in vitro. Evid.-Based Complement. Alternat. Med. 2020, 2020, 5630838. [Google Scholar] [CrossRef] [PubMed]
- Park, R.; Jang, M.; Park, Y.I.; Park, Y.; Jung, W.; Park, J.; Park, J. Epigallocatechin gallate (EGCG), a green tea polyphenol, reduces coronavirus replication in a mouse model. Viruses 2021, 13, 2533. [Google Scholar] [CrossRef] [PubMed]
- Maiti, S.; Banerjee, A. Epigallocatechin gallate and theaflavin gallate interaction in SARS-CoV-2 spike-protein central channel with reference to the hydroxychloroquine interaction: Bioinformatics and molecular docking study. Drug Dev. Res. 2021, 82, 86–96. [Google Scholar] [CrossRef]
- Ghosh, R.; Chakraborty, A.; Biswas, A.; Chowdhuri, S. Evaluation of green tea polyphenols as novel corona virus (SARS-CoV 2) main protease (Mpro) inhibitors—An in silico docking and molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2021, 39, 4362–4374. [Google Scholar] [CrossRef] [PubMed]
- Umar, H.I.; Siraj, B.; Ajayi, A.; Jimoh, T.O.; Chukwuemeka, P.O. Molecular docking studies of some selected gallic acid derivatives against five non-structural proteins of novel coronavirus. J. Genet. Eng. Biotechnol. 2021, 19, 16. [Google Scholar] [CrossRef] [PubMed]
- Szekeres, T.; Fritzer-Szekeres, M.; Saiko, P.; Jäger, W. Resveratrol and resveratrol analogues—Structure-activity relationship. Pharm. Res. 2010, 27, 1042–1048. [Google Scholar] [CrossRef]
- Murias, M.; Handler, N.; Erker, T.; Pleban, K.; Ecker, G.; Saiko, P.; Szekeres, T.; Jäger, W. Resveratrol analogues as selective cyclooxygenase-2 inhibitors: Synthesis and structure-activity relationship. Bioorg. Med. Chem. 2004, 12, 5571–5578. [Google Scholar] [CrossRef]
- Murias, M.; Jäger, W.; Handler, N.; Erker, T.; Horvath, Z.; Szekeres, T.; Nohl, H.; Gille, L. Antioxidant, prooxidant and cytotoxic activity of hydroxylated resveratrol analogues: Structure-activity relationship. Biochem. Pharmacol. 2005, 15, 903–912. [Google Scholar] [CrossRef]
- Saiko, P.; Horvath, Z.; Murias, M.; Handler, N.; Jaeger, W.; Erker, T.; Fritzer-Szekeres, M.; Szekeres, T. Antitumor effects of 3,3′,4,4′,5,5′-hexahydroxystilbene in HL-60 human promyelocytic leukemia cells. Nucleosides Nucleotides Nucleic Acids 2006, 25, 1013–1017. [Google Scholar] [CrossRef]
- Saiko, P.; Pemberger, M.; Horvath, Z.; Savinc, I.; Grusch, M.; Handler, N.; Erker, T.; Jaeger, W.; Fritzer-Szekeres, M.; Szekeres, T. Novel resveratrol analogs induce apoptosis and cause cell cycle arrest in HT29 human colon cancer cells: Inhibition of ribonucleotide reductase activity. Oncol. Rep. 2008, 19, 1621–1626. [Google Scholar]
- Paulitschke, V.; Schicher, N.; Szekeres, T.; Jäger, W.; Elbling, L.; Riemer, A.B.; Scheiner, O.; Trimurtulu, G.; Venkateswarlu, S.; Mikula, M.; et al. 3,3′,4,4′,5,5′-Hexahydroxystilbene impairs melanoma progression in a metastatic mouse model. J. Investig. Dermatol. 2010, 130, 1668–1679. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.S.; Quashie, P.K.; Mesplède, T.; Xu, H.; Quan, Y.; Jaeger, W.; Szekeres, T.; Wainberg, M.A. A resveratrol analog termed 3,3′,4,4′,5,5′-hexahydroxy-trans-stilbene is a potent HIV-1 inhibitor. J. Med. Virol. 2015, 87, 2054–2060. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Hsieh, T.C.; Wu, J.M.; Wang, X.; Christopher, J.S.; Pham, A.H.; Swaby, J.D.; Lou, L.; Xie, Z.R. Elucidating the inhibitory effect of resveratrol and its structural analogs on selected nucleotide-related enzymes. Biomolecules 2020, 10, 1223. [Google Scholar] [CrossRef] [PubMed]
- Pruijssers, A.J.; George, A.S.; Schäfer, A.; Leist, S.R.; Gralinksi, L.E.; Dinnon, K.H., 3rd; Yount, B.L.; Agostini, M.L.; Stevens, L.J.; Chappell, J.D.; et al. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 2020, 32, 107940. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.H.; Woo, H.J.; Kang, H.K.; Nguyen, V.D.; Kim, Y.M.; Kim, D.W.; Ahn, S.A.; Xia, Y.; Kim, D. Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol. Lett. 2012, 34, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Kokic, G.; Hillen, H.S.; Tegunov, D.; Dienemann, C.; Seitz, F.; Schmitzova, J.; Farnung, L.; Siewert, A.; Höbartner, C.; Cramer, P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021, 12, 279. [Google Scholar] [CrossRef]
- Chow, H.H.; Hakim, I.A.; Vining, D.R.; Crowell, J.A.; Ranger-Moore, J.; Chew, W.M.; Celaya, C.A.; Rodney, S.R.; Hara, Y.; Alberts, D.S. Effects of dosing condition on the oral bioavailability of green tea catechins after single-dose administration of Polyphenon E in healthy individuals. Clin. Cancer Res. 2005, 11, 4627–4633. [Google Scholar] [CrossRef]
- Kicker, E.; Tittel, G.; Schaller, T.; Pferschy-Wenzig, E.M.; Zatloukal, K.; Bauer, R. SARS-CoV-2 neutralizing activity of polyphenols in a special green tea extract preparation. Phytomedicine 2022, 98, 153970. [Google Scholar] [CrossRef]
- Ramakrishnan, M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016, 5, 85–86. [Google Scholar] [CrossRef]
- Gattinger, P.; Ohradanova-Reic, A.; Valenta, R. Importance, applications and features of assays measuring SARS-CoV-2 neutralizing antibodies. Int. J. Mol. Sci. 2023, 24, 5352. [Google Scholar] [CrossRef]
- Gattinger, P.; Borochova, K.; Dorofeeva, Y.; Henning, R.; Kiss, R.; Kratzer, B.; Mühl, B.; Perkmann, T.; Trapin, D.; Trella, M.; et al. Antibodies in serum of convalescent patients following mild COVID-19 do not always prevent virus-receptor binding. Allergy 2021, 76, 878–883. [Google Scholar] [CrossRef] [PubMed]
- Gattinger, P.; Niespodziana, K.; Stiasny, K.; Sahanic, S.; Tulaeva, I.; Borochova, K.; Dorofeeva, Y.; Schlederer, T.; Sonnweber, T.; Hofer, G.; et al. Neutralization of SARS-CoV-2 requires antibodies against conformational receptor-binding domain epitopes. Allergy 2022, 77, 230–242. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).