Dissecting the Interactions between Chlorin e6 and Human Serum Albumin
Abstract
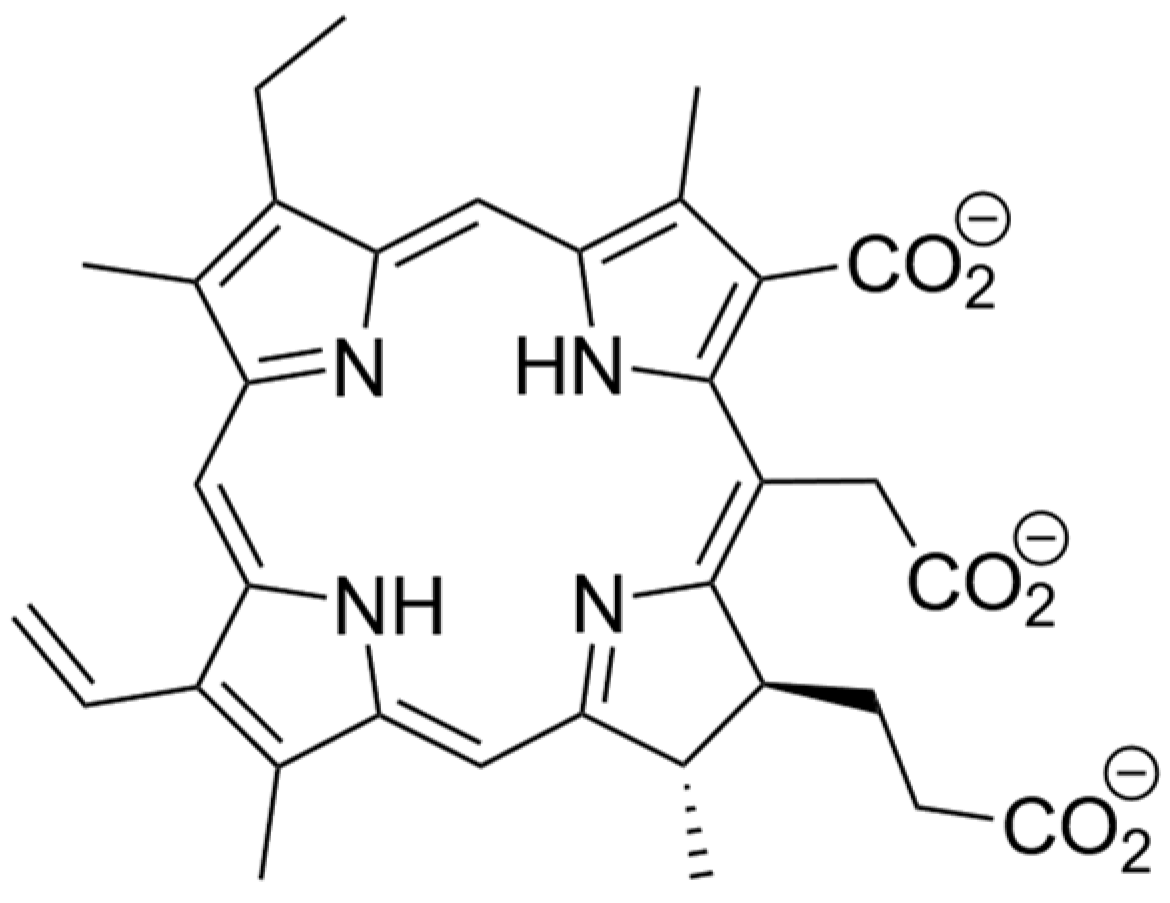
1. Introduction
2. Results and Discussion
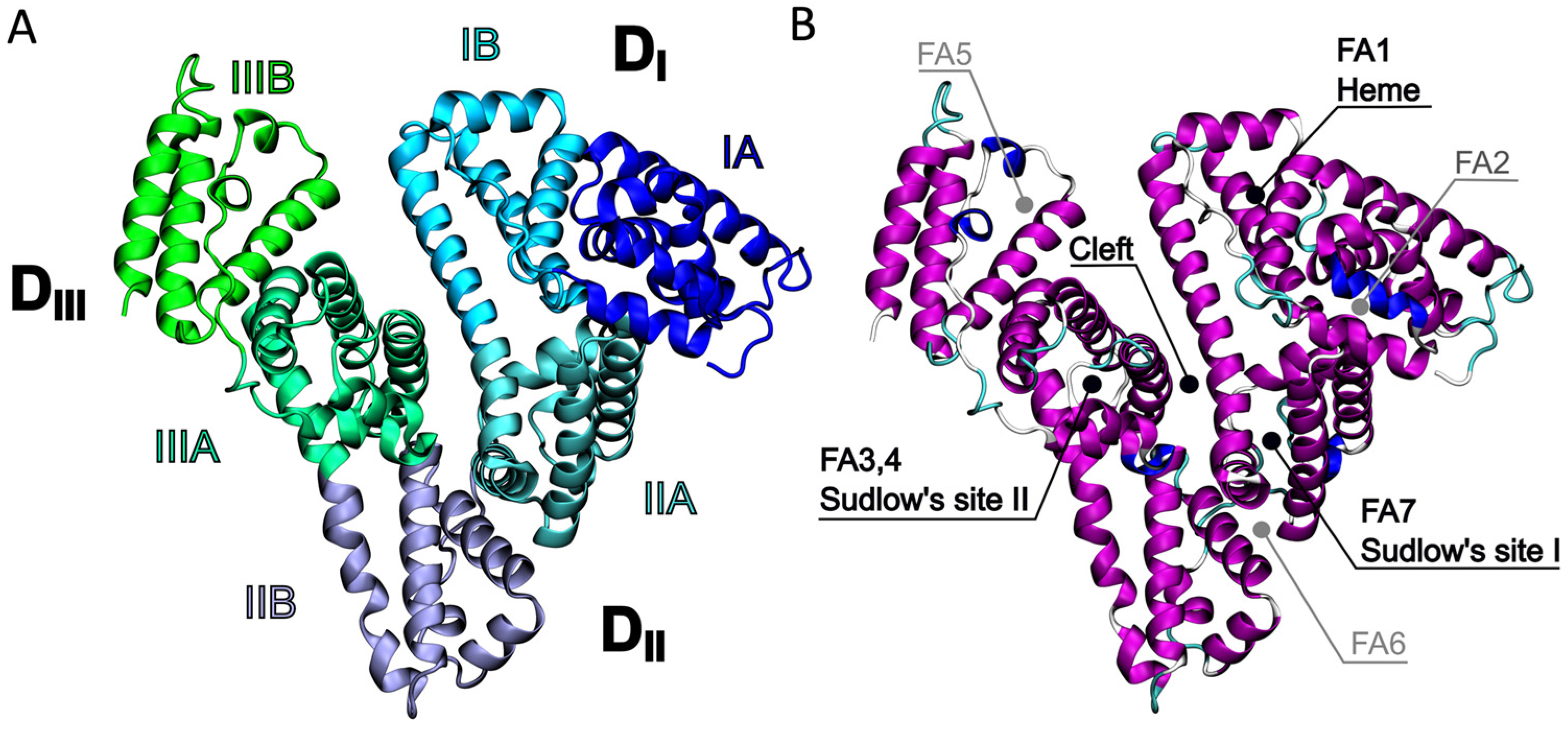
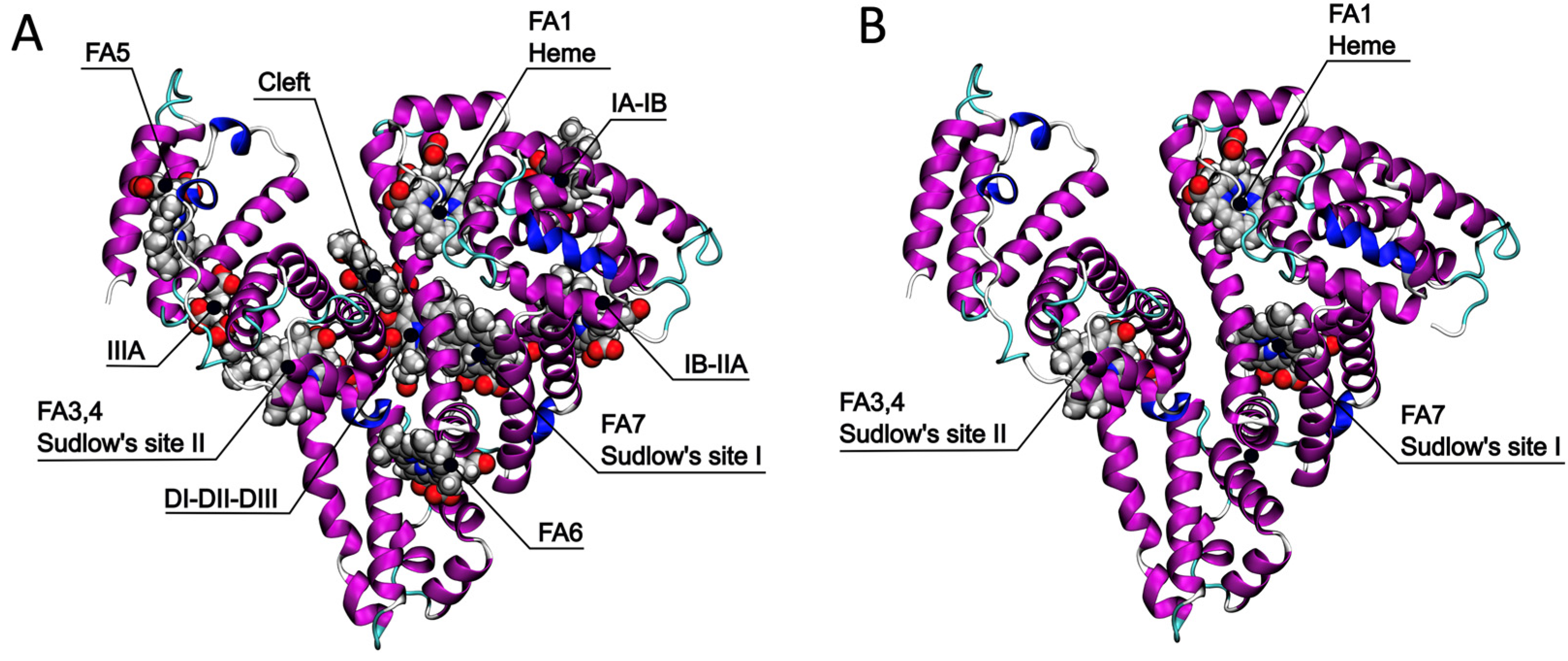
2.1. Identification of the Ce6 Binding Pockets
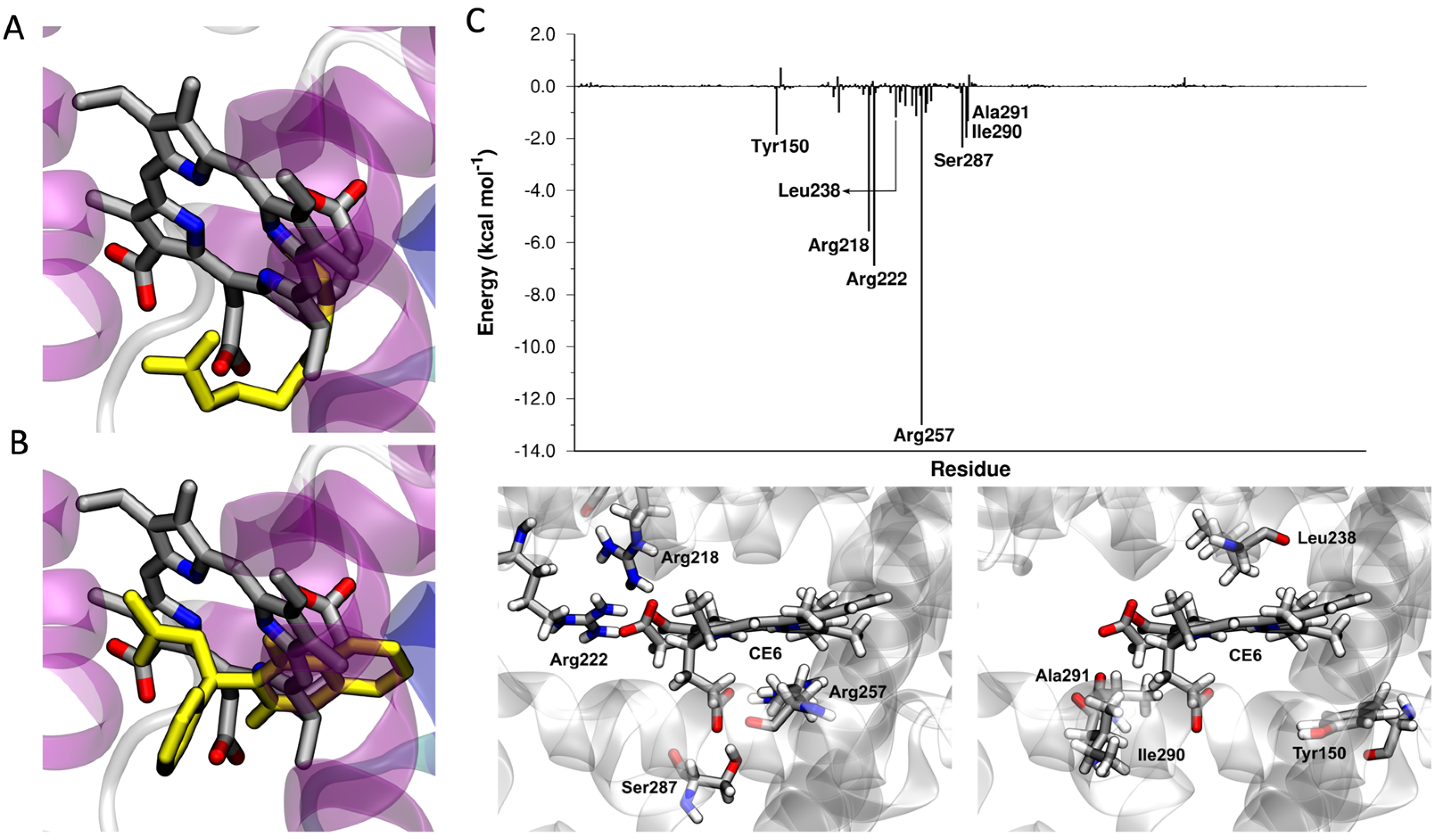
2.2. Ce6 in the Sudlow I Binding Site
2.3. Ce6 in the Heme Binding Site
2.4. Effect of the Binding of Ce6 on the Structure of HSA
2.5. Comparison between Experimental and Computational Results about Ce6 Binding
2.6. Synthesis and Characterization of Ce6@HSA
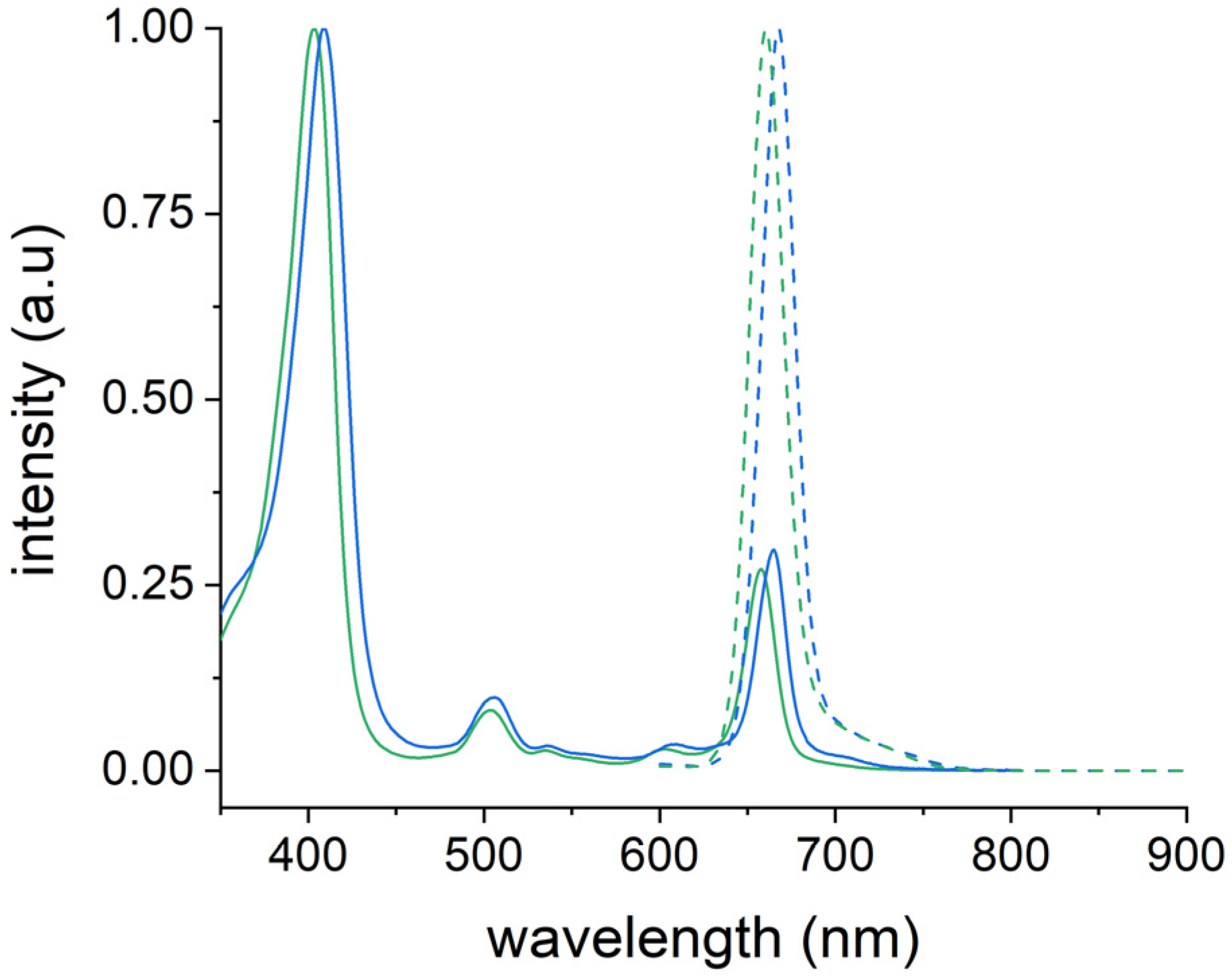
2.7. Photophysical Properties of Ce6@HSA
2.8. Generation of ROS by Ce6@HSA
3. Materials and Methods
3.1. Computational Analysis of the Ce6@HSA Complex
3.1.1. Human Serum Albumin Structural Database
3.1.2. Docking
3.1.3. Minimization and MD Simulations
3.1.4. Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) Analysis
3.2. Synthesis and Characterization of the Ce6@HSA Complex
3.2.1. Materials
3.2.2. Synthesis and Purification of the Ce6@HSA Complex
3.2.3. Characterization of the Ce6@HSA Complex
3.2.4. Detection of Reactive Oxygen Species
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Juzeniene, A. Chlorin E6-Based Photosensitizers for Photodynamic Therapy and Photodiagnosis. Photodiagn. Photodyn. Ther. 2009, 6, 94–96. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Cai, M.; Zhu, R.; Fu, T.; Du, Y.; Kong, J.; Zhang, Y.; Qu, C.; Dong, X.; Ni, J.; et al. Antitumor Effect of Photodynamic Therapy/Sonodynamic Therapy/Sono-Photodynamic Therapy of Chlorin E6 and Other Applications. Mol. Pharm. 2023, 20, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, J.M.; Bilgin, M.D.; Grossweiner, L.I. Singlet Oxygen Generation by Photodynamic Agents. J. Photochem. Photobiol. B 1997, 37, 131–140. [Google Scholar] [CrossRef]
- Shi, H.; Liu, Q.; Qin, X.; Wang, P.; Wang, X. Pharmacokinetic Study of a Novel Sonosensitizer Chlorin-E6 and Its Sonodynamic Anti-Cancer Activity in Hepatoma-22 Tumor-Bearing Mice. Biopharm. Drug Dispos. 2011, 32, 319–332. [Google Scholar] [CrossRef]
- Li, Y.; Wang, P.; Wang, X.; Su, X.; Liu, Q. Involvement of Mitochondrial and Reactive Oxygen Species in the Sonodynamic Toxicity of Chlorin E6 in Human Leukemia K562 Cells. Ultrasound Med. Biol. 2014, 40, 990–1000. [Google Scholar] [CrossRef]
- Mocanu, M.N.; Yan, F. Ultrasound-Assisted Interaction between Chlorin-E6 and Human Serum Albumin: PH Dependence, Singlet Oxygen Production, and Formulation Effect. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 190, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Chin, L.; Wan, P.; Heng, S.; Soo, P.; Thong, P.; Bhuvaneswari, R.; Hirt, W.; Kuenzel, S.; Chee, K.; et al. Improved Formulation of Photosensitizer Chlorin E6 Polyvinylpyrrolidone for Fluorescence Diagnostic Imaging and Photodynamic Therapy of Human Cancer. Eur. J. Pharm. Biopharm. 2008, 69, 1083–1093. [Google Scholar] [CrossRef]
- Park, H.; Na, K. Conjugation of the Photosensitizer Chlorin E6 to Pluronic F127 for Enhanced Cellular Internalization for Photodynamic Therapy. Biomaterials 2013, 34, 6992–7000. [Google Scholar] [CrossRef]
- Jinadasa, R.G.W.; Hu, X.; Vicente, M.G.H.; Smith, K.M. Syntheses and Cellular Investigations of 17 3-, 15 2-, and 13 1-Amino Acid Derivatives of Chlorin e 6. J. Med. Chem. 2011, 54, 7464–7476. [Google Scholar] [CrossRef]
- Shinoda, Y.; Kujirai, K.; Aoki, K.; Morita, M.; Masuda, M.; Zhang, L.; Kaixin, Z.; Nomoto, A.; Takahashi, T.; Tsuneoka, Y.; et al. Novel Photosensitizer β-Mannose-Conjugated Chlorin E6 as a Potent Anticancer Agent for Human Glioblastoma U251 Cells. Pharmaceuticals 2020, 13, 316. [Google Scholar] [CrossRef]
- Liu, W.; Ma, X.; Jin, Y.; Zhang, J.; Li, Y.; Tang, Y.; Song, Y.; Wang, S. Chlorin E6-Biotin Conjugates for Tumor-Targeting Photodynamic Therapy. Molecules 2021, 26, 7342. [Google Scholar] [CrossRef] [PubMed]
- Hamblin, M.R.; Miller, J.L.; Rizvi, I.; Ortel, B.; Maytin, E.V.; Hasan, T. Pegylation of a Chlorin E6 Polymer Conjugate Increases Tumor Targeting of Photosensitizer. Cancer Res. 2001, 61, 7155–7162. [Google Scholar] [PubMed]
- Ulfo, L.; Costantini, P.E.; di Giosia, M.; Danielli, A.; Calvaresi, M. EGFR-Targeted Photodynamic Therapy. Pharmaceutics 2022, 14, 241. [Google Scholar] [CrossRef] [PubMed]
- Akhlynina, T.V.; Jans, D.A.; Rosenkranz, A.A.; Statsyuk, N.V.; Balashova, I.Y.; Toth, G.; Pavo, I.; Rubin, A.B.; Sobolev, A.S. Nuclear Targeting of Chlorin E6 Enhances Its Photosensitizing Activity. J. Biol. Chem. 1997, 272, 20328–20331. [Google Scholar] [CrossRef]
- Abramova, O.B.; Demyashkin, G.A.; Drozhzhina, V.V.; Yakovleva, N.D.; Kozlovtseva, E.A.; Sivovolova, T.P.; Shegay, P.V.; Ivanov, S.A.; Kaprin, A.D. On the Pathomorphological Pattern of the Efficiency of Photodynamic Therapy of Murine Melanoma B16 Using a New Photosensitizer Based on Chlorin E6 Conjugate with a Prostate-Specific Membrane Antigen. Molecules 2022, 27, 3445. [Google Scholar] [CrossRef]
- Li, Z.; Qiu, L. The Current Status of Chlorin E6-Based Nanoscale Delivery Systems for Cancer Therapy. Oncologie 2021, 23, 515–531. [Google Scholar] [CrossRef]
- Čunderlíková, B.; Gangeskar, L.; Moan, J. Acid-Base Properties of Chlorin E6: Relation to Cellular Uptake. J. Photochem. Photobiol. B 1999, 53, 81–90. [Google Scholar] [CrossRef]
- Khludeev, I.I.; Kozyr’, L.A.; Zorina, T.E.; Zorin, V.P. PH-Dependent Changes in the Mechanisms of Transport of Chlorine E6 and Its Derivatives in the Blood. Bull. Exp. Biol. Med. 2015, 160, 208–212. [Google Scholar] [CrossRef]
- Mojzisova, H.; Bonneau, S.; Vever-Bizet, C.; Brault, D. The PH-Dependent Distribution of the Photosensitizer Chlorin E6 among Plasma Proteins and Membranes: A Physico-Chemical Approach. Biochim. Biophys. Acta Biomembr. 2007, 1768, 366–374. [Google Scholar] [CrossRef]
- Čunderlíková, B.; Kongshaug, M.; Gangeskar, L.; Moan, J. Increased Binding of Chlorin E6 to Lipoproteins at Low PH Values. Int. J. Biochem. Cell Biol. 2000, 32, 759–768. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, Z. Albumin Carriers for Cancer Theranostics: A Conventional Platform with New Promise. Adv. Mater. 2016, 28, 10557–10566. [Google Scholar] [CrossRef]
- Cantelli, A.; Malferrari, M.; Soldà, A.; Simonetti, G.; Forni, S.; Toscanella, E.; Mattioli, E.J.; Zerbetto, F.; Zanelli, A.; di Giosia, M.; et al. Human Serum Albumin–Oligothiophene Bioconjugate: A Phototheranostic Platform for Localized Killing of Cancer Cells by Precise Light Activation. JACS Au 2021, 1, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Parodi, A.; Miao, J.; Soond, S.M.; Rudzińska, M.; Zamyatnin, A.A. Albumin Nanovectors in Cancer Therapy and Imaging. Biomolecules 2019, 9, 218. [Google Scholar] [CrossRef] [PubMed]
- di Giosia, M.; Zerbetto, F.; Calvaresi, M. Incorporation of Molecular Nanoparticles Inside Proteins: The Trojan Horse Approach in Theranostics. Acc. Mater. Res. 2021, 2, 594–605. [Google Scholar] [CrossRef]
- Kratz, F. A Clinical Update of Using Albumin as a Drug Vehicle—A Commentary. J. Control. Release 2014, 190, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Hoogenboezem, E.N.; Duvall, C.L. Harnessing Albumin as a Carrier for Cancer Therapies. Adv. Drug Deliv. Rev. 2018, 130, 73–89. [Google Scholar] [CrossRef]
- Rubio-Camacho, M.; Encinar, J.A.; Martínez-Tomé, M.J.; Esquembre, R.; Mateo, C.R. The Interaction of Temozolomide with Blood Components Suggests the Potential Use of Human Serum Albumin as a Biomimetic Carrier for the Drug. Biomolecules 2020, 10, 1015. [Google Scholar] [CrossRef]
- Rapozzi, V.; Moret, F.; Menilli, L.; Guerrini, A.; Tedesco, D.; Naldi, M.; Bartolini, M.; Gani, M.; Zorzet, S.; Columbaro, M.; et al. HSA-Binding Prodrugs-Based Nanoparticles Endowed with Chemo and Photo-Toxicity against Breast Cancer. Cancers 2022, 14, 877. [Google Scholar] [CrossRef] [PubMed]
- Gradishar, W.J. Albumin-Bound Paclitaxel: A next-Generation Taxane. Expert Opin. Pharmacother. 2006, 7, 1041–1053. [Google Scholar] [CrossRef]
- Cantelli, A.; Malferrari, M.; Mattioli, E.J.; Marconi, A.; Mirra, G.; Soldà, A.; Marforio, T.D.; Zerbetto, F.; Rapino, S.; di Giosia, M.; et al. Enhanced Uptake and Phototoxicity of C60@albumin Hybrids by Folate Bioconjugation. Nanomaterials 2022, 12, 3501. [Google Scholar] [CrossRef]
- Mattioli, E.J.; Ulfo, L.; Marconi, A.; Pellicioni, V.; Costantini, P.E.; Marforio, T.D.; di Giosia, M.; Danielli, A.; Fimognari, C.; Turrini, E.; et al. Carrying Temoporfin with Human Serum Albumin: A New Perspective for Photodynamic Application in Head and Neck Cancer. Biomolecules 2023, 13, 68. [Google Scholar] [CrossRef]
- Pezzuoli, D.; Cozzolino, M.; Montali, C.; Brancaleon, L.; Bianchini, P.; Zantedeschi, M.; Bonardi, S.; Viappiani, C.; Abbruzzetti, S. Serum Albumins Are Efficient Delivery Systems for the Photosensitizer Hypericin in Photosensitization-Based Treatments against Staphylococcus Aureus. Food Control 2018, 94, 254–262. [Google Scholar] [CrossRef]
- Bhattacharya, A.A.; Grüne, T.; Curry, S. Crystallographic Analysis Reveals Common Modes of Binding of Medium and Long-Chain Fatty Acids to Human Serum Albumin. J. Mol. Biol. 2000, 303, 721–732. [Google Scholar] [CrossRef]
- Ghuman, J.; Zunszain, P.A.; Petitpas, I.; Bhattacharya, A.A.; Otagiri, M.; Curry, S. Structural Basis of the Drug-Binding Specificity of Human Serum Albumin. J. Mol. Biol. 2005, 353, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Zsila, F. Subdomain IB Is the Third Major Drug Binding Region of Human Serum Albumin: Toward the Three-Sites Model. Mol. Pharm. 2013, 10, 1668–1682. [Google Scholar] [CrossRef] [PubMed]
- Zunszain, P.A.; Ghuman, J.; Komatsu, T.; Tsuchida, E.; Curry, S. Crystal Structural Analysis of Human Serum Albumin Complexed with Hemin and Fatty Acid. BMC Struct. Biol. 2003, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Wardell, M.; Wang, Z.; Ho, J.X.; Robert, J.; Ruker, F.; Ruble, J.; Carter, D.C. The Atomic Structure of Human Methemalbumin at 1.9 Å. Biochem. Biophys. Res. Commun. 2002, 291, 813–819. [Google Scholar] [CrossRef]
- Sułkowski, L.; Pawełczak, B.; Chudzik, M.; Maciazek-Jurczyk, M. Characteristics of the Protoporphyrin IX Binding Sites on Human Serum Albumin Using Molecular Docking. Molecules 2016, 21, 1519. [Google Scholar] [CrossRef]
- Li, X.; Yu, S.; Lee, Y.; Guo, T.; Kwon, N.; Lee, D.; Yeom, S.C.; Cho, Y.; Kim, G.; Huang, J.D.; et al. In Vivo Albumin Traps Photosensitizer Monomers from Self-Assembled Phthalocyanine Nanovesicles: A Facile and Switchable Theranostic Approach. J. Am. Chem. Soc. 2019, 141, 1366–1372. [Google Scholar] [CrossRef]
- Li, X.; Jeong, K.; Lee, Y.; Guo, T.; Lee, D.; Park, J.; Kwon, N.; Na, J.H.; Hong, S.K.; Cha, S.S.; et al. Water-Soluble Phthalocyanines Selectively Bind to Albumin Dimers: A Green Approach toward Enhancing Tumor-Targeted Photodynamic Therapy. Theranostics 2019, 9, 6412–6423. [Google Scholar] [CrossRef]
- Rinco, O.; Brenton, J.; Douglas, A.; Maxwell, A.; Henderson, M.; Indrelie, K.; Wessels, J.; Widin, J. The Effect of Porphyrin Structure on Binding to Human Serum Albumin by Fluorescence Spectroscopy. J. Photochem. Photobiol. A Chem. 2009, 208, 91–96. [Google Scholar] [CrossRef]
- Patel, S.; Datta, A. Steady State and Time-Resolved Fluorescence Investigation of the Specific Binding of Two Chlorin Derivatives with Human Serum Albumin. J. Phys. Chem. B 2007, 111, 10557–10562. [Google Scholar] [CrossRef]
- Sarbadhikary, P.; Dube, A. Spectroscopic Investigations on the Binding of an Iodinated Chlorin: P 6-Copper Complex to Human Serum Albumin. Photochem. Photobiol. Sci. 2017, 16, 1762. [Google Scholar] [CrossRef]
- Patel, S.; Sharma, K.K.; Datta, A. Competitive Binding of Chlorin P6and Dansyl-L-Proline to Sudlow’s Site II of Human Serum Albumin. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015, 138, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.Y.; Zou, X. Ensemble Docking of Multiple Protein Structures: Considering Protein Structural Variations in Molecular Docking. Proteins Struct. Funct. Genet. 2007, 66, 399–421. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Castañeda, T.R.; Bader, E.; Elshorst, B.; Cummings, S.; Scherer, P.; Bangari, D.S.; Loewe, C.; Schreuder, H.; Pöverlein, C.; et al. Triantennary GalNAc Molecular Imaging Probes for Monitoring Hepatocyte Function in a Rat Model of Nonalcoholic Steatohepatitis. Adv. Sci. 2020, 7, 2002997. [Google Scholar] [CrossRef] [PubMed]
- Sugio, S.; Kashima, A.; Mochizuki, S.; Noda, M.; Kobayashi, K. Crystal Structure of Human Serum Albumin at 2.5 Å Resolution. Protein Eng. 1999, 12, 439–446. [Google Scholar] [CrossRef]
- Greco, G.; Ulfo, L.; Turrini, E.; Marconi, A.; Costantini, P.E.; Marforio, T.D.; Mattioli, E.J.; di Giosia, M.; Danielli, A.; Fimognari, C.; et al. Light-Enhanced Cytotoxicity of Doxorubicin by Photoactivation. Cells 2023, 12, 392. [Google Scholar] [CrossRef]
- di Giosia, M.; Nicolini, F.; Ferrazzano, L.; Soldà, A.; Valle, F.; Cantelli, A.; Marforio, T.D.; Bottoni, A.; Zerbetto, F.; Montalti, M.; et al. Stable and Biocompatible Monodispersion of C60 in Water by Peptides. Bioconj. Chem. 2019, 30, 808–814. [Google Scholar] [CrossRef]
- Cantelli, A.; Piro, F.; Pecchini, P.; di Giosia, M.; Danielli, A.; Calvaresi, M. Concanavalin A-Rose Bengal Bioconjugate for Targeted Gram-Negative Antimicrobial Photodynamic Therapy. J. Photochem. Photobiol. B 2020, 206, 111852. [Google Scholar] [CrossRef]
- Ding, H.; Yu, H.; Dong, Y.; Tian, R.; Huang, G.; Boothman, D.A.; Sumer, B.D.; Gao, J. Photoactivation Switch from Type II to Type I Reactions by Electron-Rich Micelles for Improved Photodynamic Therapy of Cancer Cells under Hypoxia. J. Control. Release 2011, 156, 276–280. [Google Scholar] [CrossRef]
- di Giosia, M.; Bomans, P.H.H.; Bottoni, A.; Cantelli, A.; Falini, G.; Franchi, P.; Guarracino, G.; Friedrich, H.; Lucarini, M.; Paolucci, F.; et al. Proteins as Supramolecular Hosts for C60: A True Solution of C60 in Water. Nanoscale 2018, 10, 9908–9916. [Google Scholar] [CrossRef] [PubMed]
- di Giosia, M.; Genovese, D.; Cantelli, A.; Cingolani, M.; Rampazzo, E.; Strever, G.; Tavoni, M.; Zaccheroni, N.; Calvaresi, M.; Prodi, L. Synthesis and Characterization of a Reconstituted Myoglobin-Chlorin E6 Adduct for Theranostic Applications. J. Porphyr. Phthalocyanines 2020, 24, 887–893. [Google Scholar] [CrossRef]
- Soldà, A.; Cantelli, A.; di Giosia, M.; Montalti, M.; Zerbetto, F.; Rapino, S.; Calvaresi, M. C60@lysozyme: A New Photosensitizing Agent for Photodynamic Therapy. J. Mater. Chem. B 2017, 5, 6608–6615. [Google Scholar] [CrossRef] [PubMed]
- Ulfo, L.; Cantelli, A.; Petrosino, A.; Costantini, P.E.; Nigro, M.; Starinieri, F.; Turrini, E.; Zadran, S.K.; Zuccheri, G.; Saporetti, R.; et al. Orthogonal Nanoarchitectonics of M13 Phage for Receptor Targeted Anticancer Photodynamic Therapy. Nanoscale 2022, 14, 632–641. [Google Scholar] [CrossRef]
- di Giosia, M.; Soldà, A.; Seeger, M.; Cantelli, A.; Arnesano, F.; Nardella, M.I.; Mangini, V.; Valle, F.; Montalti, M.; Zerbetto, F.; et al. A Bio-Conjugated Fullerene as a Subcellular-Targeted and Multifaceted Phototheranostic Agent. Adv. Funct. Mater. 2021, 31, 2101527. [Google Scholar] [CrossRef]
- Bortot, B.; Apollonio, M.; Baj, G.; Andolfi, L.; Zupin, L.; Crovella, S.; di Giosia, M.; Cantelli, A.; Saporetti, R.; Ulfo, L.; et al. Advanced Photodynamic Therapy with an Engineered M13 Phage Targeting EGFR: Mitochondrial Localization and Autophagy Induction in Ovarian Cancer Cell Lines. Free Radic. Biol. Med. 2022, 179, 242–251. [Google Scholar] [CrossRef]
- Zhou, Z.; Song, J.; Nie, L.; Chen, X. Reactive Oxygen Species Generating Systems Meeting Challenges of Photodynamic Cancer Therapy. Chem. Soc. Rev. 2016, 45, 6597–6626. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; He, P.; Wang, Z.; Tang, B.Z. A Feasible Strategy of Fabricating Type i Photosensitizer for Photodynamic Therapy in Cancer Cells and Pathogens. ACS Nano 2021, 15, 7735–7743. [Google Scholar] [CrossRef]
- Mroz, P.; Pawlak, A.; Satti, M.; Lee, H.; Wharton, T.; Gali, H.; Sarna, T.; Hamblin, M.R. Functionalized Fullerenes Mediate Photodynamic Killing of Cancer Cells: Type I versus Type II Photochemical Mechanism. Free Radic. Biol. Med. 2007, 43, 711–719. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Schneidman-Duhovny, D.; Inbar, Y.; Polak, V.; Shatsky, M.; Halperin, I.; Benyamini, H.; Barzilai, A.; Dror, O.; Haspel, N.; Nussinov, R.; et al. Taking Geometry to Its Edge: Fast Unbound Rigid (and Hinge-Bent) Docking. Proteins 2003, 52, 107–112. [Google Scholar] [CrossRef]
- Calvaresi, M.; Zerbetto, F. Baiting proteins with C60. ACS Nano 2010, 4, 2283–2299. [Google Scholar] [CrossRef] [PubMed]
- Calvaresi, M.; Zerbetto, F. Fullerene sorting proteins. Nanoscale 2011, 3, 2873–2881. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, L.; Rasulev, B.; Kar, S.; Krupa, P.; Mozolewska, M.A.; Leszczynski, J. Inhibitors or Toxins? Large Library Target-Specific Screening of Fullerene-Based Nanoparticles for Drug Design Purpose. Nanoscale 2017, 9, 10263–10276. [Google Scholar] [CrossRef]
- di Giosia, M.; Valle, F.; Cantelli, A.; Bottoni, A.; Zerbetto, F.; Calvaresi, M. C60 Bioconjugation with Proteins: Towards a Palette of Carriers for All PH Ranges. Materials 2018, 11, 691. [Google Scholar] [CrossRef]
- Calvaresi, M.; Bottoni, A.; Zerbetto, F. Thermodynamics of Binding between Proteins and Carbon Nanoparticles: The Case of C60@Lysozyme. J. Phys. Chem. C 2015, 119, 28077–28082. [Google Scholar] [CrossRef]
- Calvaresi, M.; Furini, S.; Domene, C.; Bottoni, A.; Zerbetto, F. Blocking the Passage: C60 Geometrically Clogs K+ Channels. ACS Nano 2015, 9, 4827–4834. [Google Scholar] [CrossRef]
- Calvaresi, M.; Arnesano, F.; Bonacchi, S.; Bottoni, A.; Calò, V.; Conte, S.; Falini, G.; Fermani, S.; Losacco, M.; Montalti, M.; et al. C60@Lysozyme: Direct Observation by Nuclear Magnetic Resonance of a 1:1 Fullerene Protein Adduct. ACS Nano 2014, 8, 1871–1877. [Google Scholar] [CrossRef]
- di Giosia, M.; Valle, F.; Cantelli, A.; Bottoni, A.; Zerbetto, F.; Fasoli, E.; Calvaresi, M. High-Throughput Virtual Screening to Rationally Design Protein—Carbon Nanotube Interactions. Identification and Preparation of Stable Water Dispersions of Protein—Carbon Nanotube Hybrids and Efficient Design of New Functional Materials. Carbon 2019, 147, 70–82. [Google Scholar] [CrossRef]
- Berto, M.; di Giosia, M.; Giordani, M.; Sensi, M.; Valle, F.; Alessandrini, A.; Menozzi, C.; Cantelli, A.; Gazzadi, G.C.; Zerbetto, F.; et al. Green Fabrication of (6,5)Carbon Nanotube/Protein Transistor Endowed with Specific Recognition. Adv. Electron. Mater. 2021, 7, 2001114. [Google Scholar] [CrossRef]
- di Giosia, M.; Marforio, T.D.; Cantelli, A.; Valle, F.; Zerbetto, F.; Su, Q.; Wang, H.; Calvaresi, M. Inhibition of α-Chymotrypsin by Pristine Single-Wall Carbon Nanotubes: Clogging up the Active Site. J. Colloid Interface Sci. 2020, 571, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Calvaresi, M.; Hoefinger, S.; Zerbetto, F. Probing the Structure of Lysozyme-Carbon-Nanotube Hybrids with Molecular Dynamics. Chem. A Eur. J. 2012, 18, 4308–4313. [Google Scholar] [CrossRef] [PubMed]
- Calvaresi, M.; Zerbetto, F. In Silico Carborane Docking to Proteins and Potential Drug Targets. J. Chem. Inf. Model. 2011, 51, 1882–1896. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. Ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from Ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Case, D.; Betz, R.; Botello-Smith, W.; Cerutti, D.; Cheatham, T., III; Darden, T.; Duke, R.; Giese, T.; Gohlke, H.; Goetz, A.; et al. Amber 16; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.Py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Montalti, M.; Credi, A.; Prodi, L.; Gandolfi, M.T. Handbook of Photochemistry; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]



| Secondary Structures | HSA | Ce6@HSA (SI Site) | Ce6@HSA (Heme Site) |
|---|---|---|---|
| Coil | 13.1 | 13.0 | 12.9 |
| π Helix | 0.0 | 0.1 | 0.1 |
| 3–10 Helix | 2.9 | 3.6 | 3.3 |
| α Helix | 65.6 | 68.8 | 68.4 |
| Isolated β Bridge | 0.2 | 0.0 | 0.0 |
| β Sheet | 0.0 | 0.0 | 0.0 |
| Turn | 18.3 | 14.6 | 15.3 |
| Absorption | Emission | ||||
|---|---|---|---|---|---|
| λmax,Soret (nm) | λmax,Q-band (nm) | λmax (nm) | Φ | t (ns) | |
| Ce6 | 403 | 657 | 661 | 0.17 | 4.5 |
| Ce6@HSA | 409 | 665 | 667 | 0.17 | 5.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marconi, A.; Mattioli, E.J.; Ingargiola, F.; Giugliano, G.; Marforio, T.D.; Prodi, L.; Di Giosia, M.; Calvaresi, M. Dissecting the Interactions between Chlorin e6 and Human Serum Albumin. Molecules 2023, 28, 2348. https://doi.org/10.3390/molecules28052348
Marconi A, Mattioli EJ, Ingargiola F, Giugliano G, Marforio TD, Prodi L, Di Giosia M, Calvaresi M. Dissecting the Interactions between Chlorin e6 and Human Serum Albumin. Molecules. 2023; 28(5):2348. https://doi.org/10.3390/molecules28052348
Chicago/Turabian StyleMarconi, Alessia, Edoardo Jun Mattioli, Filippo Ingargiola, Giulia Giugliano, Tainah Dorina Marforio, Luca Prodi, Matteo Di Giosia, and Matteo Calvaresi. 2023. "Dissecting the Interactions between Chlorin e6 and Human Serum Albumin" Molecules 28, no. 5: 2348. https://doi.org/10.3390/molecules28052348
APA StyleMarconi, A., Mattioli, E. J., Ingargiola, F., Giugliano, G., Marforio, T. D., Prodi, L., Di Giosia, M., & Calvaresi, M. (2023). Dissecting the Interactions between Chlorin e6 and Human Serum Albumin. Molecules, 28(5), 2348. https://doi.org/10.3390/molecules28052348






