Potential of siRNA-Bearing Subtilosomes in the Treatment of Diethylnitrosamine-Induced Hepatocellular Carcinoma
Abstract
1. Introduction
2. Results
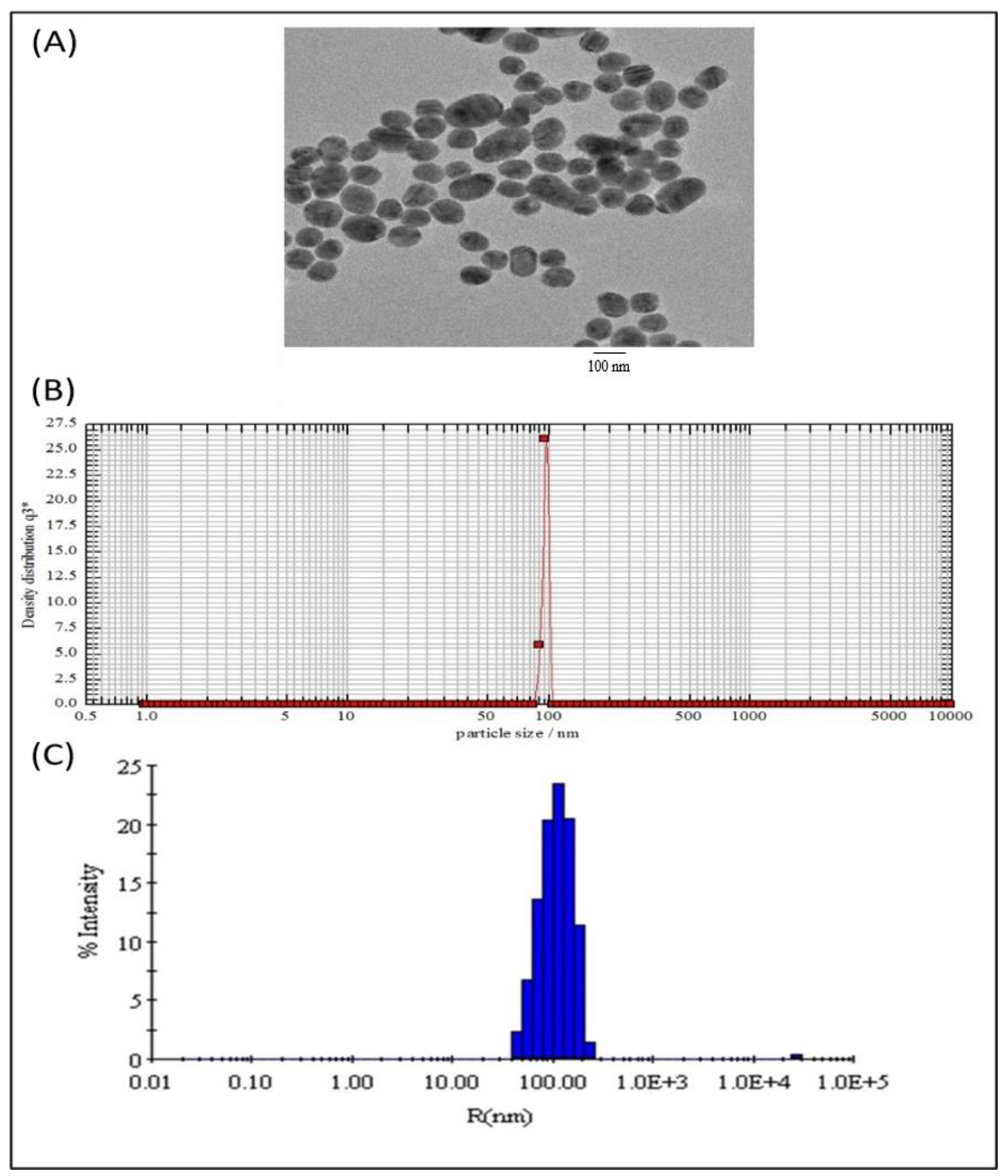
2.1. Characterization of Subtilosomes-siRNA Based Nanoparticles
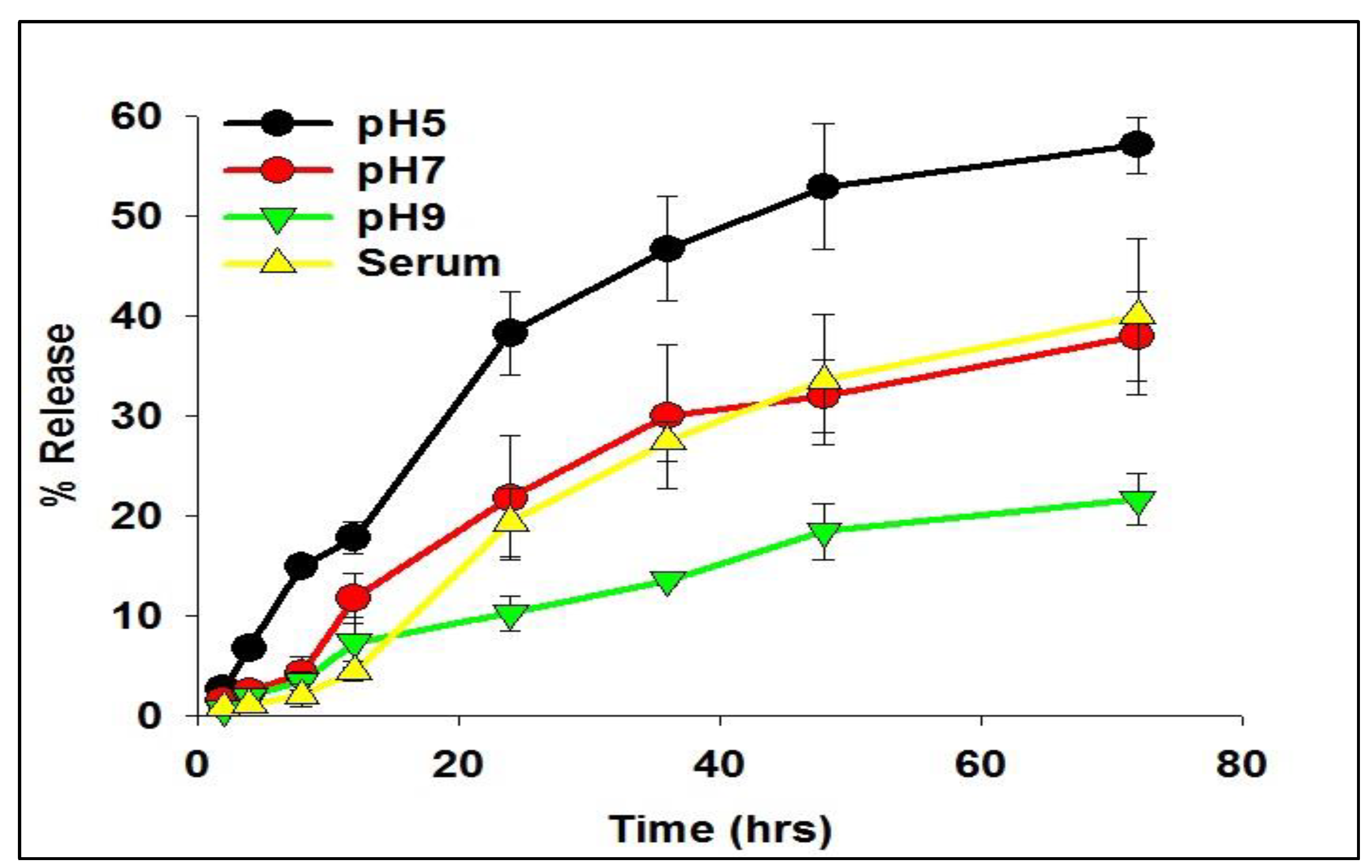
2.2. Release Kinetics of siRNA from As-Synthesized Nanoparticles
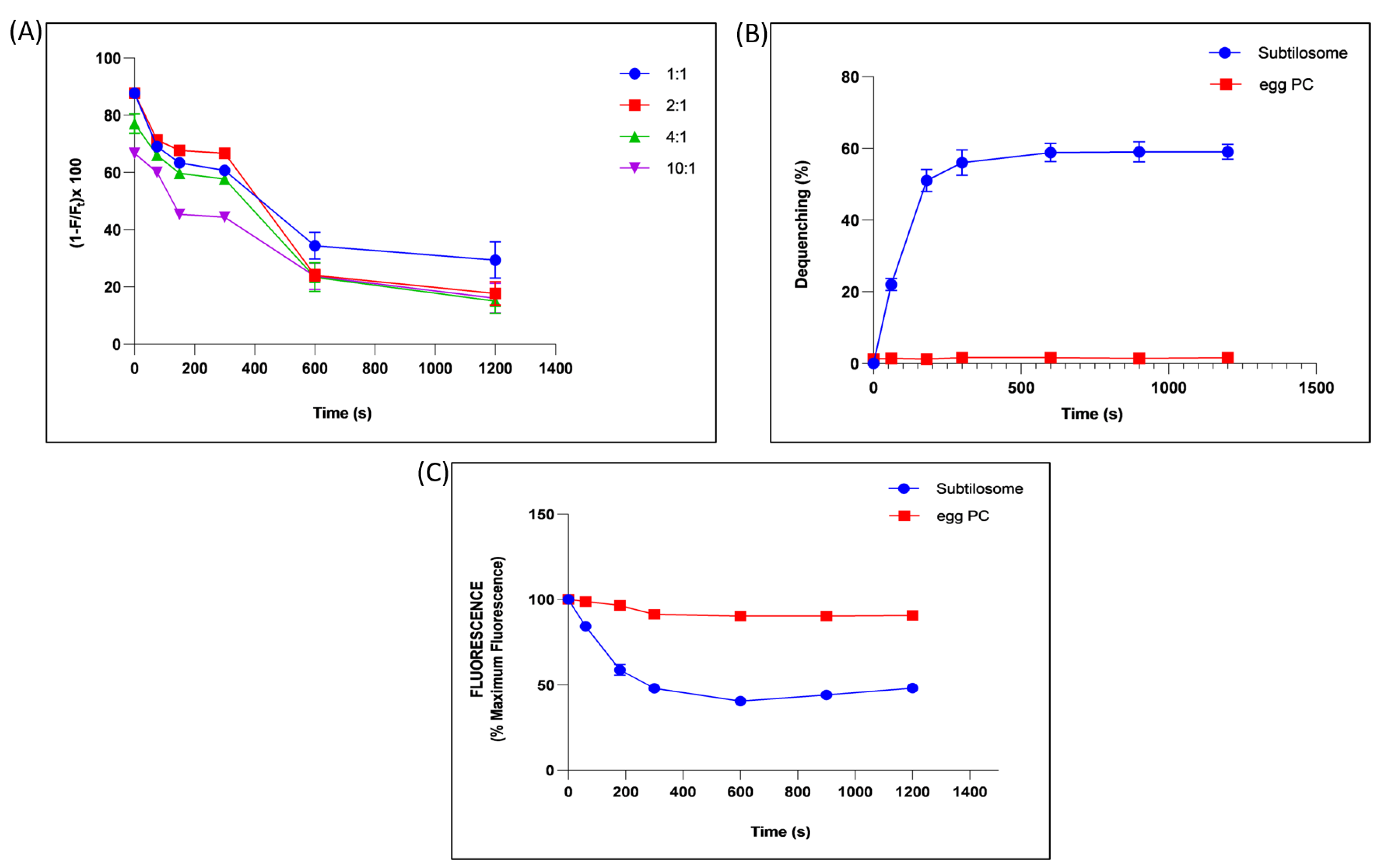
2.3. Analysis of Fusogenic Property of Subtilosome
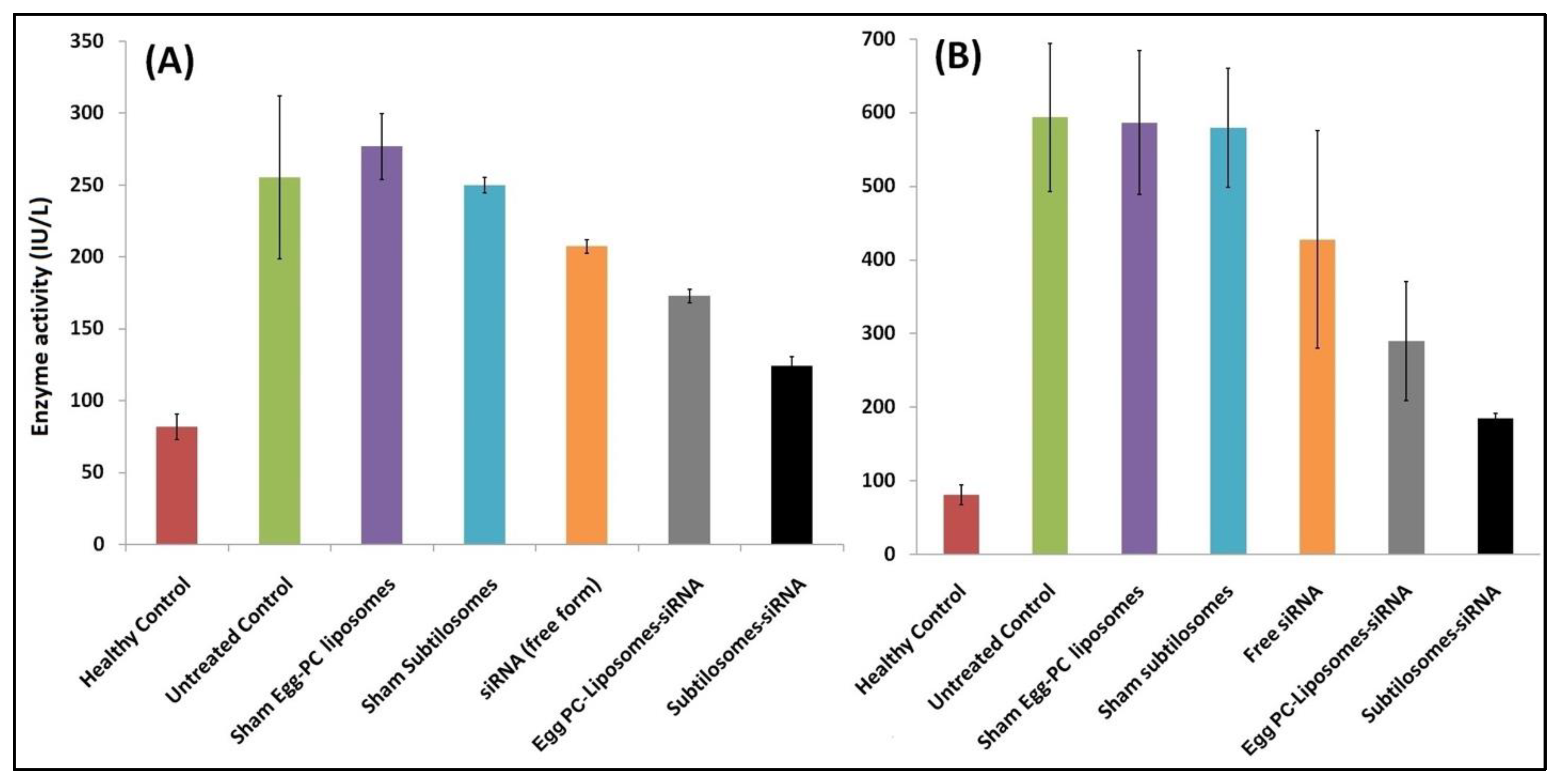
2.4. COX-2-siRNA-Bearing Subtilosomes Mediated Liver-Enzyme Depletion in the Experimental Animals
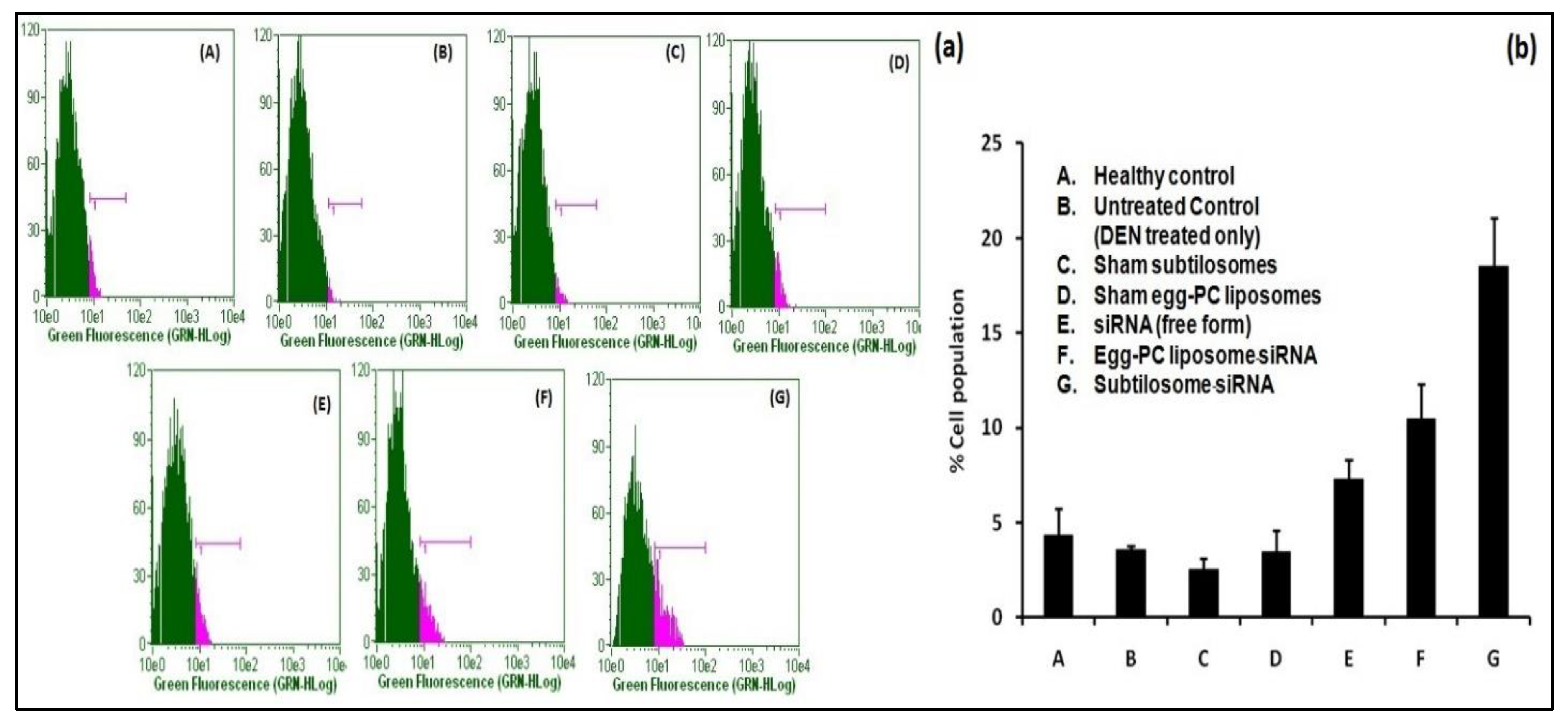
2.5. The siRNA-Bearing Subtilosomes Induce Apoptosis in Hepatocellular Carcinoma Cells
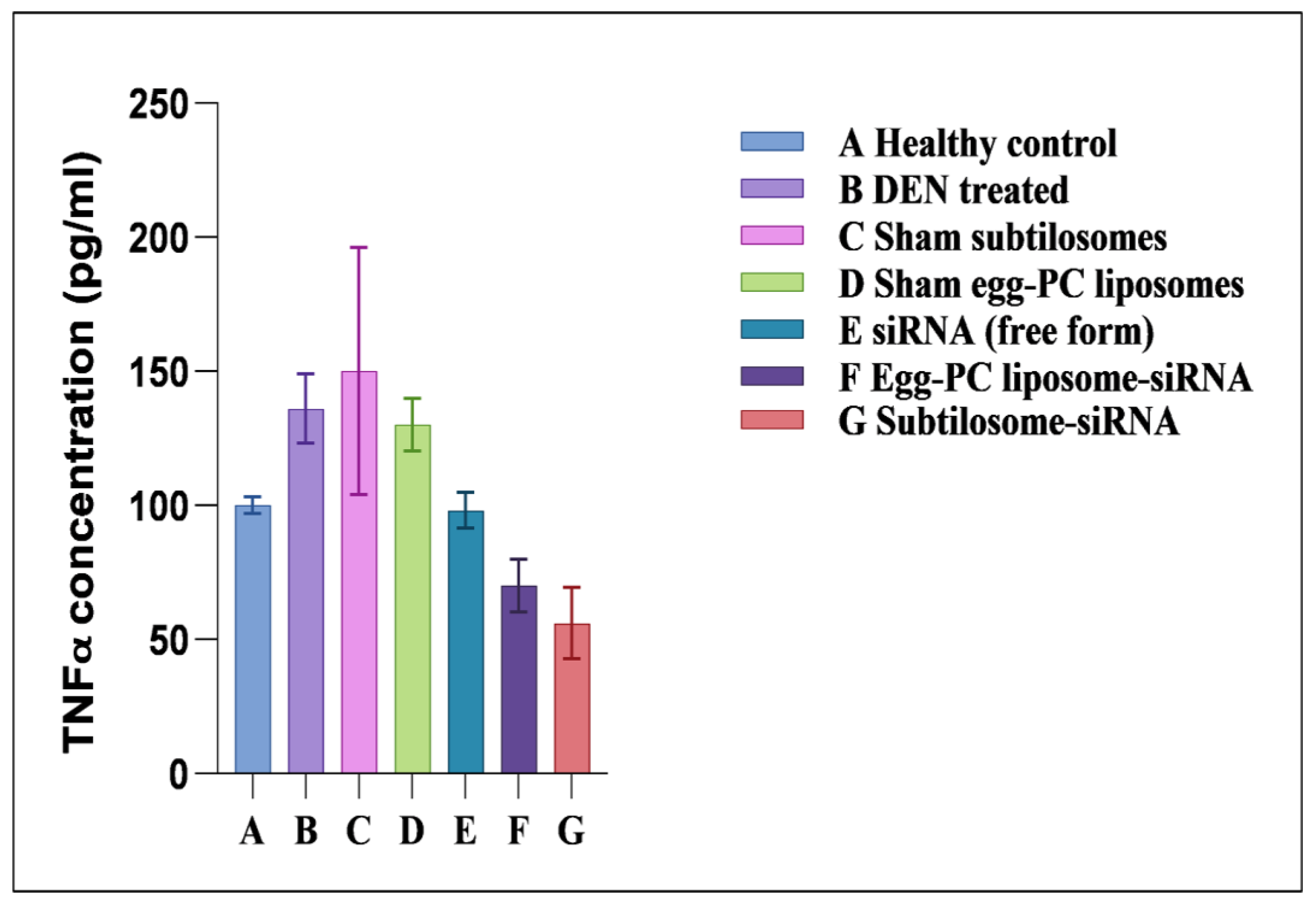
2.6. Effect of siRNA-Bearing Subtilosomes on the Expression of TNF-α
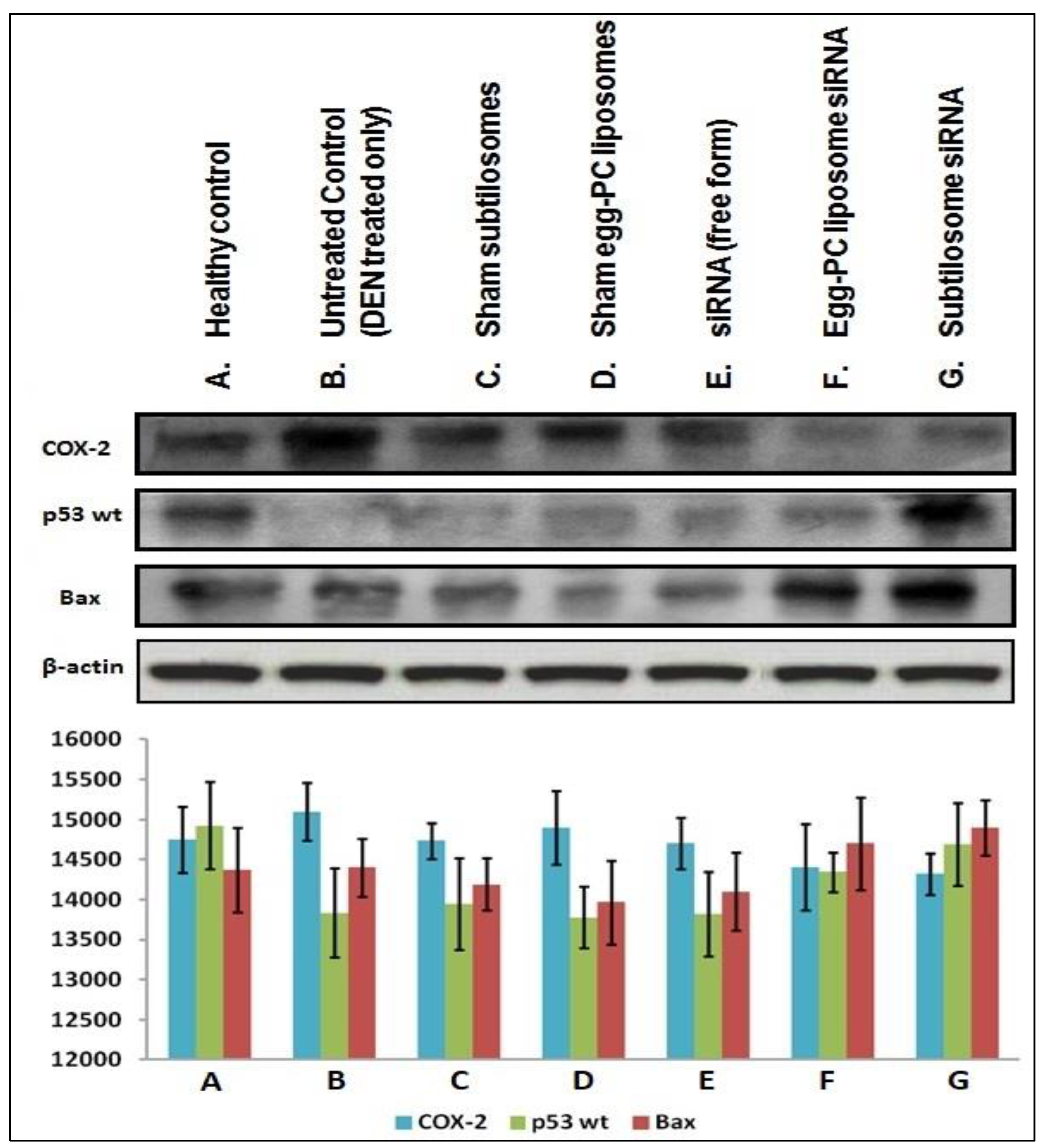
2.7. Expression of Pro/Anti-Apoptotic Factors
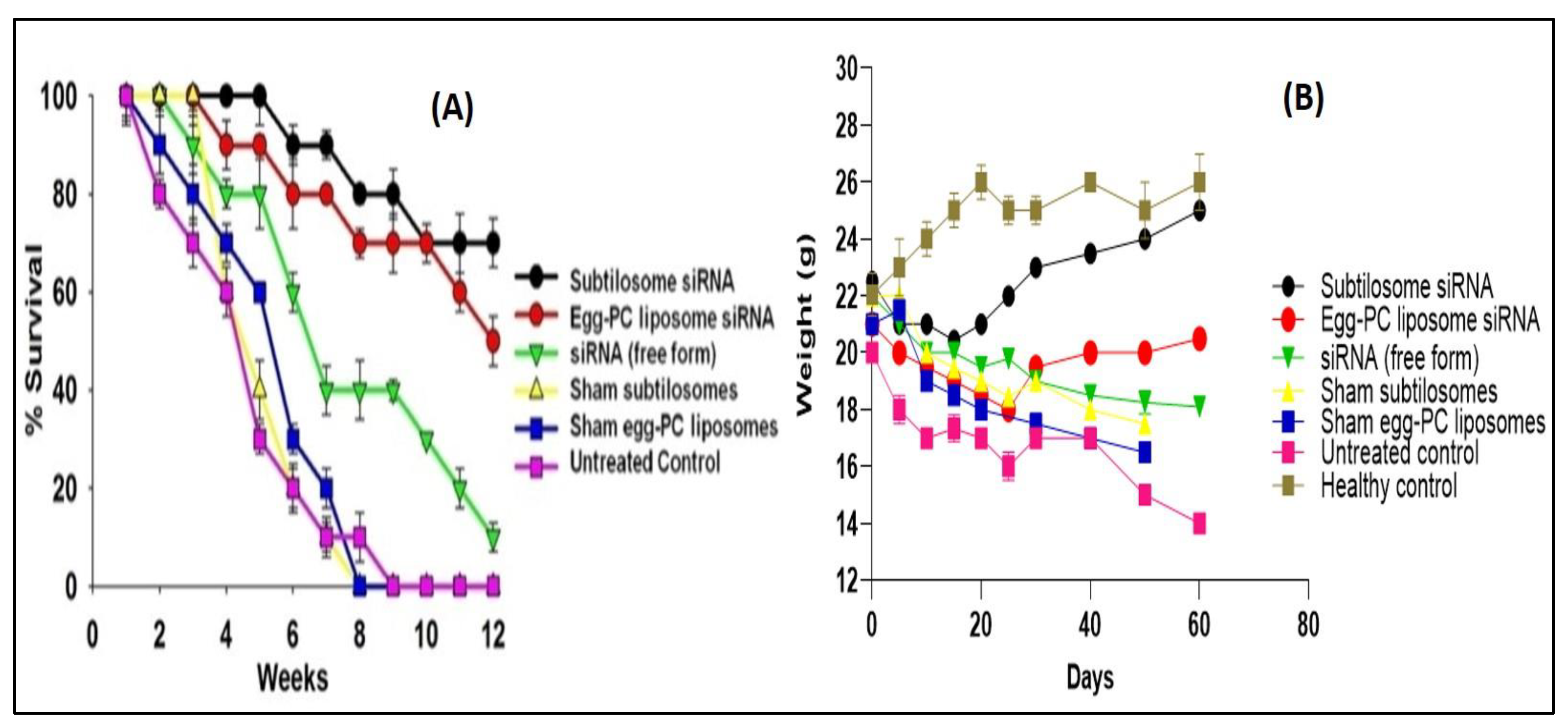
2.8. Survival Study
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Isolation of B. subtilis Membrane Lipids and Preparation of siRNA-Bearing Subtilosome Formulation
4.3. Preparation of Egg-PC-Liposome-Encapsulated siRNA
4.4. Characterization of the siRNA-Bearing Formulations
4.5. Entrapment Efficiency of siRNA
4.6. In Vitro Release Kinetics
4.7. High-Performance Liquid Chromatography (HPLC) Analysis
4.8. Determination of Fusogenic Property of Subtilosomes
4.8.1. Fluorescence-Resonance Energy Transfer (FRET) Assay
4.8.2. Dequenching Assay
4.8.3. Aqueous-Content-Mixing Assay
4.9. Induction of Liver Cancer with Diethylnitrosamine (DEN)
4.10. Assessment of Anticancer Efficacy
- Group I Healthy control;
- Group II Untreated control (DEN-treated only);
- Group III Sham liposomes;
- Group IV Sham subtilosomes;
- Group V siRNA (free form);
- Group VI PC-liposome siRNA; and
- Group VII Subtilosome-siRNA
4.11. Assessment of Liver Enzymes
4.12. Measurement of TNF-α Level
4.13. Preparation of Cell Homogenate
4.14. Western Blotting
4.15. Apoptosis Detection with APO-BRDUTM Labeling
4.16. Survival Analysis
4.17. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
References
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef]
- Mak, L.-Y.; Cruz-Ramón, V.; Chinchilla-López, P.; Torres, H.A.; LoConte, N.K.; Rice, J.P.; Foxhall, L.E.; Sturgis, E.M.; Merrill, J.K.; Bailey, H.H.; et al. Global Epidemiology, Prevention, and Management of Hepatocellular Carcinoma. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 262–279. [Google Scholar] [CrossRef]
- Venook, A.P.; Papandreou, C.; Furuse, J.; de Guevara, L.L. The incidence and epidemiology of hepatocellular carcinoma: A global and regional perspective. The Oncologist 2010, 15 (Suppl. 4), 5–13. [Google Scholar] [CrossRef]
- Hoshida, Y.; Fuchs, B.C.; Tanabe, K.K. Prevention of hepatocellular carcinoma: Potential targets, experimental models, and clinical challenges. Curr. Cancer Drug Targets 2012, 12, 1129–1159. [Google Scholar]
- Alhosaini, K.; Azhar, A.; Alonazi, A.; Al-Zoghaibi, F. GPCRs: The most promiscuous druggable receptor of the mankind. Saudi Pharm. J. 2021, 29, 539–551. [Google Scholar] [CrossRef]
- Nieto Gutierrez, A.; McDonald, P.H. GPCRs: Emerging anti-cancer drug targets. Cell. Signal. 2018, 41, 65–74. [Google Scholar] [CrossRef]
- Xu, X.; Zhu, G.-Q.; Zhang, K.; Zhou, Y.-C.; Li, X.-L.; Xu, W.; Zhang, H.; Shao, Y.; Zhang, Z.-Y.; Sun, W.-H. Cyclooxygenase-2 mediated synergistic effect of ursolic acid in combination with paclitaxel against human gastric carcinoma. Oncotarget 2017, 8, 92770–92777. [Google Scholar] [CrossRef]
- Tołoczko-Iwaniuk, N.; Dziemiańczyk-Pakieła, D.; Nowaszewska, B.K.; Celińska-Janowicz, K.; Miltyk, W. Celecoxib in Cancer Therapy and Prevention—Review. Curr. Drug Targets 2019, 20, 302–315. [Google Scholar] [CrossRef]
- Jendrossek, V. Targeting apoptosis pathways by Celecoxib in cancer. Cancer Lett. 2013, 332, 313–324. [Google Scholar] [CrossRef]
- Manu, K.A.; Shanmugam, M.K.; Li, F.; Chen, L.; Siveen, K.S.; Ahn, K.S.; Kumar, A.P.; Sethi, G. Simvastatin sensitizes human gastric cancer xenograft in nude mice to capecitabine by suppressing nuclear factor-kappa B-regulated gene products. J. Mol. Med. 2014, 92, 267–276. [Google Scholar] [CrossRef]
- Martín-Sanz, P.; Casado, M.; Boscá, L. Cyclooxygenase 2 in liver dysfunction and carcinogenesis: Facts and perspectives. World J. Gastroenterol. 2017, 23, 3572–3580. [Google Scholar] [CrossRef]
- Ekor, M.; Odewabi, A.O.; Kale, O.E.; Adesanoye, O.A.; Bamidele, T.O. Celecoxib, a selective cyclooxygenase-2 inhibitor, lowers plasma cholesterol and attenuates hepatic lipid peroxidation during carbon-tetrachloride-associated hepatotoxicity in rats. Drug Chem. Toxicol. 2013, 36, 1–8. [Google Scholar] [CrossRef]
- Swaminathan, G.; Shigna, A.; Kumar, A.; Byroju, V.V.; Durgempudi, V.R.; Dinesh Kumar, L. RNA Interference and Nanotechnology: A Promising Alliance for Next Generation Cancer Therapeutics. Front. Nanotechnol. 2021, 3, 694838. [Google Scholar] [CrossRef]
- Devanapally, S.; Ravikumar, S.; Jose, A.M. Double-stranded RNA made in C. elegans neurons can enter the germline and cause transgenerational gene silencing. Proc. Natl. Acad. Sci. USA 2015, 112, 2133–2138. [Google Scholar] [CrossRef]
- Davis, M.E.; Zuckerman, J.E.; Choi, C.H.J.; Seligson, D.; Tolcher, A.; Alabi, C.A.; Yen, Y.; Heidel, J.D.; Ribas, A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 2010, 464, 1067–1070. [Google Scholar] [CrossRef]
- Fu, Z.; Zhang, X.; Zhou, X.; Ur-Rehman, U.; Yu, M.; Liang, H.; Guo, H.; Guo, X.; Kong, Y.; Su, Y.; et al. In vivo self-assembled small RNAs as a new generation of RNAi therapeutics. Cell Res. 2021, 31, 631–648. [Google Scholar] [CrossRef]
- Whitehead, K.A.; Langer, R.; Anderson, D.G. Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 2009, 8, 129–138. [Google Scholar] [CrossRef]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.-J. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef]
- Roscigno, G.; Scognamiglio, I.; Ingenito, F.; Chianese, R.V.; Palma, F.; Chan, A.; Condorelli, G. Modulating the Crosstalk between the Tumor and the Microenvironment Using SiRNA: A Flexible Strategy for Breast Cancer Treatment. Cancers 2020, 12, 3744. [Google Scholar] [CrossRef]
- Meng, Z.; Lu, M. RNA Interference-Induced Innate Immunity, Off-Target Effect, or Immune Adjuvant? Front. Immunol. 2017, 8, 331. [Google Scholar] [CrossRef]
- Singh, A.P.; Biswas, A.; Shukla, A.; Maiti, P. Targeted therapy in chronic diseases using nanomaterial-based drug delivery vehicles. Signal Transduct. Target. Ther. 2019, 4, 33. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, S.; Koide, H.; Asai, T. Recent advances in siRNA delivery mediated by lipid-based nanoparticles. Adv. Drug Deliv. Rev. 2020, 154–155, 64–78. [Google Scholar] [CrossRef] [PubMed]
- Deeba, F.; Tahseen, H.N.; Sharad, K.S.; Ahmad, N.; Akhtar, S.; Saleemuddin, M.; Mohammad, O. Phospholipid diversity: Correlation with membrane-membrane fusion events. Biochim. Biophys. Acta 2005, 1669, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Shukla, Y.; Kalra, N.; Alam, M.; Ahmad, M.G.; Hakim, S.R.; Owais, M. Potential of diallyl sulfide bearing pH-sensitive liposomes in chemoprevention against DMBA-induced skin papilloma. Mol. Med. Camb. Mass 2007, 13, 443–451. [Google Scholar] [CrossRef]
- Maroof, A.; Farazuddin, M.; Owais, M. Potential use of liposomal diallyl sulfide in the treatment of experimental murine candidiasis. Biosci. Rep. 2010, 30, 223–231. [Google Scholar] [CrossRef]
- Ansari, M.A.; Zubair, S.; Mahmood, A.; Gupta, P.; Khan, A.A.; Gupta, U.D.; Arora, A.; Owais, M. RD antigen based nanovaccine imparts long term protection by inducing memory response against experimental murine tuberculosis. PLoS ONE 2011, 6, e22889. [Google Scholar] [CrossRef]
- Ansari, M.A.; Zubair, S.; Tufail, S.; Ahmad, E.; Khan, M.R.; Quadri, Z.; Owais, M. Ether lipid vesicle-based antigens impart protection against experimental listeriosis. Int. J. Nanomed. 2012, 7, 2433–2447. [Google Scholar] [CrossRef]
- Chauhan, A.; Zubair, S.; Nadeem, A.; Ansari, S.A.; Ansari, M.Y.; Mohammad, O. Escheriosome-mediated cytosolic delivery of PLK1-specific siRNA: Potential in treatment of liver cancer in BALB/c mice. Nanomedicine 2014, 9, 407–420. [Google Scholar] [CrossRef]
- Rouzer, C.A.; Marnett, L.J. Cyclooxygenases: Structural and functional insights. J. Lipid Res. 2009, 50, S29–S34. [Google Scholar] [CrossRef]
- Shiotani, H.; Denda, A.; Yamamoto, K.; Kitayama, W.; Endoh, T.; Sasaki, Y.; Tsutsumi, N.; Sugimura, M.; Konishi, Y. Increased expression of cyclooxygenase-2 protein in 4-nitroquinoline-1-oxide-induced rat tongue carcinomas and chemopreventive efficacy of a specific inhibitor, nimesulide. Cancer Res. 2001, 61, 1451–1456. [Google Scholar]
- Stolina, M.; Sharma, S.; Lin, Y.; Dohadwala, M.; Gardner, B.; Luo, J.; Zhu, L.; Kronenberg, M.; Miller, P.W.; Portanova, J.; et al. Specific inhibition of cyclooxygenase 2 restores antitumor reactivity by altering the balance of IL-10 and IL-12 synthesis. J. Immunol. 2000, 164, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Mohr, S.; Bakal, C.; Perrimon, N. Genomic screening with RNAi: Results and challenges. Annu. Rev. Biochem. 2010, 79, 37–64. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.R.; Kim, I.K.; Bae, K.H.; Lee, S.H.; Lee, Y.; Park, T.G. Cationic solid lipid nanoparticles reconstituted from low density lipoprotein components for delivery of siRNA. Mol. Pharm. 2008, 5, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Sherwani, M.A.; Tufail, S.; Khan, A.A.; Owais, M. Dendrosome mediated topical gene silencing by PLK-1 specific siRNA: Implication in treatment of skin cancer in mouse model. RSC Adv. 2016, 6, 6843–6857. [Google Scholar] [CrossRef]
- Ahmad, I.; Owais, M.; Shahid, M.; Aqil, F. (Eds.) Combating Fungal Infections; Springer: Berlin/Heidelberg, Germany, 2010; ISBN 978-3-642-12172-2. [Google Scholar]
- Malamas, A.S.; Gujrati, M.; Kummitha, C.M.; Xu, R.; Lu, Z.-R. Design and evaluation of new pH-sensitive amphiphilic cationic lipids for siRNA delivery. J. Control. Release 2013, 171, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Ozawa, S.; Miyamoto, C.; Maehata, Y.; Suzuki, A.; Maeda, T.; Baba, Y. Acidic extracellular microenvironment and cancer. Cancer Cell Int. 2013, 13, 89. [Google Scholar] [CrossRef]
- Aggarwal, B.B. Signalling pathways of the TNF superfamily: A double-edged sword. Nat. Rev. Immunol. 2003, 3, 745–756. [Google Scholar] [CrossRef]
- Budhu, A.; Wang, X.W. The role of cytokines in hepatocellular carcinoma. J. Leukoc. Biol. 2006, 80, 1197–1213. [Google Scholar] [CrossRef]
- Shiraki, K.; Yamanaka, T.; Inoue, H.; Kawakita, T.; Enokimura, N.; Okano, H.; Sugimoto, K.; Murata, K.; Nakano, T. Expression of TNF-related apoptosis-inducing ligand in human hepatocellular carcinoma. Int. J. Oncol. 2005, 26, 1273–1281. [Google Scholar] [CrossRef]
- Pietsch, E.C.; Sykes, S.M.; McMahon, S.B.; Murphy, M.E. The p53 family and programmed cell death. Oncogene 2008, 27, 6507–6521. [Google Scholar] [CrossRef]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011, 2, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Menendez, D.; Inga, A.; Resnick, M.A. The expanding universe of p53 targets. Nat. Rev. Cancer 2009, 9, 724–737. [Google Scholar] [CrossRef] [PubMed]
- Yip, K.W.; Reed, J.C. Bcl-2 family proteins and cancer. Oncogene 2008, 27, 6398–6406. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.C. Bcl-2-family proteins and hematologic malignancies: History and future prospects. Blood 2008, 111, 3322–3330. [Google Scholar] [CrossRef] [PubMed]
- Saelens, X.; Festjens, N.; Vande Walle, L.; van Gurp, M.; van Loo, G.; Vandenabeele, P. Toxic proteins released from mitochondria in cell death. Oncogene 2004, 23, 2861–2874. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.-M.; Heo, J.-I.; Oh, J.-Y.; Kim, Y.-M.; Ha, K.-S.; Kim, J.-I.; Han, J.A. COX-2 regulates p53 activity and inhibits DNA damage-induced apoptosis. Biochem. Biophys. Res. Commun. 2005, 328, 1107–1112. [Google Scholar] [CrossRef]
- Emoto, K.; Umeda, M. An essential role for a membrane lipid in cytokinesis. Regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J. Cell Biol. 2000, 149, 1215–1224. [Google Scholar] [CrossRef]
- Haque, M.E.; Chakraborty, H.; Koklic, T.; Komatsu, H.; Axelsen, P.H.; Lentz, B.R. Hemagglutinin fusion peptide mutants in model membranes: Structural properties, membrane physical properties, and PEG-mediated fusion. Biophys. J. 2011, 101, 1095–1104. [Google Scholar] [CrossRef]
- Otte, A.; Báez-Santos, Y.M.; Mun, E.A.; Soh, B.-K.; Lee, Y.-N.; Park, K. The in vivo transformation and pharmacokinetic properties of a liquid crystalline drug delivery system. Int. J. Pharm. 2017, 532, 345–351. [Google Scholar] [CrossRef]
- Kozlovsky, Y.; Chernomordik, L.V.; Kozlov, M.M. Lipid intermediates in membrane fusion: Formation, structure, and decay of hemifusion diaphragm. Biophys. J. 2002, 83, 2634–2651. [Google Scholar] [CrossRef]
- Han, J.; Pluhackova, K.; Böckmann, R.A. The Multifaceted Role of SNARE Proteins in Membrane Fusion. Front. Physiol. 2017, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Salzberg, L.I.; Helmann, J.D. Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition. J. Bacteriol. 2008, 190, 7797–7807. [Google Scholar] [CrossRef] [PubMed]
- Pueyo, M.T.; Bloch, C.; Carmona-Ribeiro, A.M.; di Mascio, P. Lipopeptides produced by a soil Bacillus megaterium strain. Microb. Ecol. 2009, 57, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Shukla, M.K.; Mishra, A.; Kumari, P.; Reddy, C.R.K.; Jha, B. Isolation and characterization of exopolysaccharides from seaweed associated bacteria Bacillus licheniformis. Carbohydr. Polym. 2011, 84, 1019–1026. [Google Scholar] [CrossRef]
- Bottone, E.J. Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 2010, 23, 382–398. [Google Scholar] [CrossRef]
- Mallick, A.I.; Singha, H.; Chaudhuri, P.; Nadeem, A.; Khan, S.A.; Dar, K.A.; Owais, M. Liposomised recombinant ribosomal L7/L12 protein protects BALB/c mice against Brucella abortus 544 infection. Vaccine 2007, 25, 3692–3704. [Google Scholar] [CrossRef]
- Ahmad, N.; Deeba, F.; Faisal, S.M.; Khan, A.; Agrewala, J.N.; Dwivedi, V.; Owais, M. Role of fusogenic non-PC liposomes in elicitation of protective immune response against experimental murine salmonellosis. Biochimie 2006, 88, 1391–1400. [Google Scholar] [CrossRef]
- Gidden, J.; Denson, J.; Liyanage, R.; Ivey, D.M.; Lay, J.O. Lipid Compositions in Escherichia coli and Bacillus subtilis During Growth as Determined by MALDI-TOF and TOF/TOF Mass Spectrometry. Int. J. Mass Spectrom. 2009, 283, 178–184. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Kumar, A.; Gupta, C.M. Red cell membrane abnormalities in chronic myeloid leukaemia. Nature 1983, 303, 632–633. [Google Scholar] [CrossRef]
- Khan, M.A.; Nasti, T.H.; Saima, K.; Mallick, A.I.; Firoz, A.; Wajahul, H.; Ahmad, N.; Mohammad, O. Co-administration of immunomodulator tuftsin and liposomised nystatin can combat less susceptible Candida albicans infection in temporarily neutropenic mice. FEMS Immunol. Med. Microbiol. 2004, 41, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, T.; Chatterjee, A.; Rana, A.; Srivastawa, S.; Damodaran, S.; Chatterjee, M. Cell proliferation and hepatocarcinogenesis in rat initiated by diethylnitrosamine and promoted by phenobarbital: Potential roles of early DNA damage and liver metallothionein expression. Life Sci. 2007, 81, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Carr, B.I.; Pancoska, P.; Branch, R.A. Significance of increased serum GGTP levels in HCC patients. Hepato-Gastroenterology 2010, 57, 869–874. [Google Scholar] [PubMed]
- Farazuddin, M.; Dua, B.; Zia, Q.; Khan, A.A.; Joshi, B.; Owais, M. Chemotherapeutic potential of curcumin-bearing microcells against hepatocellular carcinoma in model animals. Int. J. Nanomed. 2014, 9, 1139–1152. [Google Scholar] [CrossRef]
- Waterborg, J.H.; Matthews, H.R. The lowry method for protein quantitation. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 1984; Volume 1, pp. 1–3. [Google Scholar] [CrossRef]
- Goldman, A.; Ursitti, J.A.; Mozdzanowski, J.; Speicher, D.W. Electroblotting from Polyacrylamide Gels. Curr. Protoc. Protein Sci. 2015, 82, 10.7.1–10.7.16. [Google Scholar] [CrossRef]
- Li, W.-C.; Ralphs, K.L.; Tosh, D. Isolation and culture of adult mouse hepatocytes. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2010; Volume 633, pp. 185–196. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jamal, F.; Ahmed, G.; Farazuddin, M.; Altaf, I.; Farheen, S.; Zia, Q.; Azhar, A.; Ahmad, H.; Khan, A.A.; Somavarapu, S.; et al. Potential of siRNA-Bearing Subtilosomes in the Treatment of Diethylnitrosamine-Induced Hepatocellular Carcinoma. Molecules 2023, 28, 2191. https://doi.org/10.3390/molecules28052191
Jamal F, Ahmed G, Farazuddin M, Altaf I, Farheen S, Zia Q, Azhar A, Ahmad H, Khan AA, Somavarapu S, et al. Potential of siRNA-Bearing Subtilosomes in the Treatment of Diethylnitrosamine-Induced Hepatocellular Carcinoma. Molecules. 2023; 28(5):2191. https://doi.org/10.3390/molecules28052191
Chicago/Turabian StyleJamal, Fauzia, Ghufran Ahmed, Mohammad Farazuddin, Ishrat Altaf, Saba Farheen, Qamar Zia, Asim Azhar, Hira Ahmad, Aijaz Ahmed Khan, Satyanarayana Somavarapu, and et al. 2023. "Potential of siRNA-Bearing Subtilosomes in the Treatment of Diethylnitrosamine-Induced Hepatocellular Carcinoma" Molecules 28, no. 5: 2191. https://doi.org/10.3390/molecules28052191
APA StyleJamal, F., Ahmed, G., Farazuddin, M., Altaf, I., Farheen, S., Zia, Q., Azhar, A., Ahmad, H., Khan, A. A., Somavarapu, S., Agrawal, A., & Owais, M. (2023). Potential of siRNA-Bearing Subtilosomes in the Treatment of Diethylnitrosamine-Induced Hepatocellular Carcinoma. Molecules, 28(5), 2191. https://doi.org/10.3390/molecules28052191





