Towards DNA-Based Methods Analysis for Honey: An Update
Abstract
1. Introduction
2. Methodology
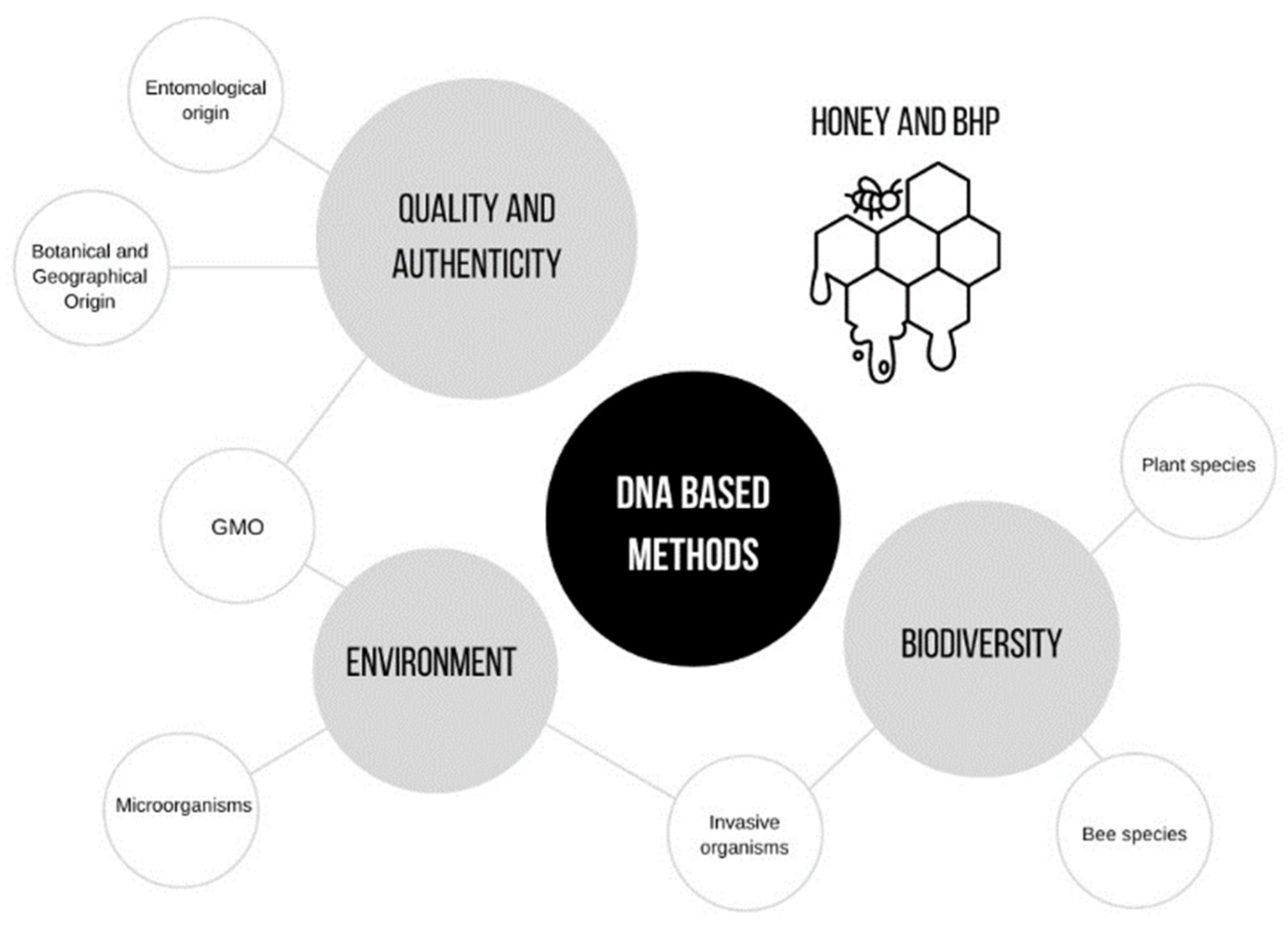
3. DNA-Based Methods
| Application | DNA Extraction | DNA Identification | DNA Yields | A260/A280 Ratio Range | Target | Reference |
|---|---|---|---|---|---|---|
| Botanical Origin | DNeasy® Blood and Tissue Kit (Qiagen) | Qualitative PCR and Real-time PCR | adh1, actin, LFY1, hmg, nr1, PAL, DXR, Profilin, ypr10, trnL | [43] | ||
| NucleoSpin® Plant (Macherey-Nagel), DNeasy® Plant Mini Kit (Qiagen), CTAB-based and Wizard® DNA-based | Qualitative PCR | nd–592.6 ng/uL | 1.0–2.1 | 18S rRNA and adh1 | [44] | |
| DNeasy® Isolation and Purification Kit (Qiagen) | Qualitative PCR | 10.0–25.0 ng/μL | ∼1.80 | rbcL and trnH-psbA plastid region | [45] | |
| DNeasy® Plant Mini Kit (Qiagen) | DNA Metabarcoding | rbcL | [46] | |||
| DNeasy® Plant Mini Kit (Qiagen) | DNA Metabarcoding | rbcL | [47] | |||
| CTAB-based Method | Next-generation Sequencing (NGS) | rbcL, matK and ITS2 | [37] | |||
| NucleoSpin® Plant II (Macherey-Nagel) | Qualitative PCR, Real-time PCR with HRM Analysis | 4.4–275.9 ng/μL | 1.9–2.3 | 18S rRNA and matK | [31] | |
| CTAB-based Method | Ion Torrent Sequencing (NGS) | trnL-UAA | [11] | |||
| DNeasy® Blood and Tissue kit (Qiagen), QIAcube Instrument (Qiagen) | DNA Metabarcoding | 0.1–29.7 ng/μL | 0.5–2.2 | trnL | [36] | |
| Pollen DNA | Automated CTAB Buffer-based Method, Maxwell® 16 FFS Nucleic Acid Extraction System, Custom-Kit (Promega GmbH), QIAQuick PCR Purification Kit (Qiagen) | Qualitative PCR, Real-time PCR | 4.1–10.7 ng/μL | 2.0 | actin | [48] |
| DNeasy® Power Plant Pro Kit (Qiagen) | Qualitative PCR and Sequencing | ITS2 | [49] | |||
| Botanical Origin and Entomological | In-house Method (Silica Membrane Spin Column) | Qualitative PCR and Ion Torrent Sequencing | ITS2, rbcLa, and COI | [39] | ||
| CTAB-based Method | Qualitative PCR, Real-time PCR and NGS Sequencing | trnL-UAA, Cox1, and COI | [13] | |||
| DNeasy® mericonTM Food Kit (Qiagen) | Qualitative PCR and Sanger Sequencing | 16S rRNA and COI | [42] | |||
| Entomological Origin | CTAB-based Method, Wizard® DNA-based and the Commercial Kits DNeasy® mericonTM Food Kit (Qiagen) and NucleoSpin® Isolation Food Kit (Macherey-Nagel) | Qualitative PCR | 0.1–1210.6 ng/μL | 0.6–2.6 | 16S rRNA | [25] |
| CTAB-based Method | Qualitative PCR and Sanger Sequencing | COI-COII intergenic spacer | [12] | |||
| NucleoSpin® Plant II Kit (Macherey-Nagel) | Qualitative PCR, Real-time PCR with HRM Analysis, Sanger Sequencing | 2.3–303.9 ng/µL | 1.1–2.6 | 18S rRNA, tRNAleu -cox2 intergenic region, and 16S rRNA | [50] | |
| NucleoSpin® Plant II Kit (Macherey-Nagel) | Real-time PCR with HRM Analysis and Sanger Sequencing | COI | [35] | |||
| Geographical Origin | Wizard® DNA-based | Machine Learning (Sequencing) | [51] | |||
| Arthropods, Plants, Fungi, Bacteria, and Viruses | CTAB-based Method | Qualitative PCR and Ion Torrent Sequencing | [15] | |||
| Plants, Bacteria, and Fungi | DNeasy® Plant Mini Kit (Qiagen) | DNA Metabarcoding | ITS2, rbcLa, trnL, 16S rRNA, and ITS | [38] | ||
| Dialysis and Phenol⁄chloroform⁄isoamyl Alcohol-based Method | Qualitative PCR and Sequencing | 0.07 ng ⁄μL | 1.35 | 16S rDNA and 18S rDNA | [52] | |
| Pathogens and Parasites | CTAB-based Method | Qualitative PCR and Sequencing | >1.6 | COI-COII, 16S rRNA, NapA, SSU rRNA, cytb, 18S rRNA, and Cox1 | [10] | |
| Viruses, Bacteria, Plants, Fungi, Protozoans, Arthropods, and Mammals | CTAB-based Method | Shotgun Sequencing | [16] | |||
| Parasite Lotmaria passim | CTAB-based Method | Qualitative PCR | cytb, 18S fragment and GAPDH fragment | [53] | ||
| Genetically Modified Organism | CTAB-based Method | DNA Concentration, DNA Integrity, PCR Amplification | 0.1–0.2 ng/μL | CaMV 35S promoter, 35S and Bt junction gene and Sad1 gene | [54] |
3.1. DNA Extraction Methods
3.2. DNA Identification Approaches
3.2.1. Target Genes
3.2.2. Honey Environmental DNA
3.2.3. Honey MicroRNAs
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scepankova, H.; Saraiva, J.A.; Estevinho, L.M. Honey Health Benefits and Uses in Medicine. In Bee Products—Chemical and Biological Properties, 1st ed.; Alvarez Suarez, J.M., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 83–96. [Google Scholar]
- Soares, S.; Amaral, J.S.; Oliveira, M.B.P.P.; Mafra, I. A Comprehensive Review on the Main Honey Authentication Issues: Production and Origin. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1072–1100. [Google Scholar] [CrossRef]
- Mandal, M.D.; Mandal, S. Honey: Its medicinal property and antibacterial activity. Asian Pac. J. Trop. Biomed. 2011, 1, 154–160. [Google Scholar] [CrossRef]
- Haoan, Z.; Ni, C.; Liangliang, H.; Guoxia, P.; Xiaofeng, X.; Liming, W.; Wei, C. Antioxidant and hepatoprotective effects of A. cerana honey against acute alcohol-induced liver damage in mice. Food Res. Int. 2017, 101, 35–44. [Google Scholar]
- Erejuwa, O.O.; Gurtu, S.; Sulaiman, S.A.; Ab Wahab, M.S.; Sirajudeen, K.N.S.; Salzihan, M.; Salleh, M. Hypoglycemic and Antioxidant Effects of Honey Supplementation in Streptozotocin-induced Diabetic Rats. Int. J. Vitam. Nutr. Res. 2010, 80, 74–82. [Google Scholar]
- Erejuwa, O.O.; Sulaiman, S.A.; Ab Wahab, M.S.; Sirajudeen, K.N.S.; Salleh, S.; Gurtu, S. Honey supplementation in spontaneously hypertensive rats elicits antihypertensive effect via amelioration of renal oxidative stress. Oxidative Med. Cell. Longev. 2012, 2012, 374037. [Google Scholar] [CrossRef]
- Bukhari, M.H.; Khalil, J.; Qamar, S.; Qamar, Z.; Zahid, M.; Ansari, N.; Bakhshi, I.M. Comparative gastroprotective effects of natural honey, Nigella sativa and cimetidine against acetylsalicylic acid induced gastric ulcer in albino rats. J. Coll. Physicians Surg. Pak. 2011, 21, 151–156. [Google Scholar]
- Moussa, A.; Noureddine, D.; Saad, A.; Abdelmelek, M.; Abdelkader, B. Antifungal activity of four honeys of different types from Algeria against pathogenic yeast: Candida albicans and Rhodotorula sp. Asian Pac. J. Trop. Biomed. 2012, 2, 554–557. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Y.; Hu, Y.; Zhou, J.; Chen, L.; Lu, X. Systematic Review of the Characteristic Markers in Honey of Various Botanical, Geographic, and Entomological Origins. ACS Food Sci. Technol. 2022, 2, 206–220. [Google Scholar] [CrossRef]
- Ribani, A.; Utzeri, V.J.; Taurisano, V.; Fontanesi, L. Honey as a Source of Environmental DNA for the Detection and Monitoring of Honey Bee Pathogens and Parasites. Vet. Sci. 2020, 7, 113. [Google Scholar] [CrossRef]
- Utzeri, V.J.; Ribani, A.; Schiavo, G.; Bertolini, F.; Bovo, S.; Fontanesi, L. Application of next generation semiconductor-based sequencing to detect the botanical composition of monofloral, polyfloral and honeydew honey. Food Control 2018, 86, 342–349. [Google Scholar] [CrossRef]
- Utzeri, V.J.; Ribani, A.; Fontanesi, L. Authentication of honey based on a DNA method to differentiate Apis mellifera subspecies: Application to Sicilian honey bee (A. m. siciliana) and Iberian honey bee (A. m. iberiensis) honeys. Food Control 2018, 91, 294–301. [Google Scholar] [CrossRef]
- Utzeri, V.J.; Schiavo, G.; Ribani, A.; Tinarelli, S.; Bertolini, F.; Bovo, S.; Fontanesi, L. Entomological signatures in honey: An environmental DNA metabarcoding approach can disclose information on plant-sucking insects in agricultural and forest landscapes. Sci Rep. 2018, 8, 9996. [Google Scholar] [CrossRef] [PubMed]
- Utzeri, V.J.; Schiavo, G.; Ribani, A.; Bertolini, F.; Bovo, S.; Fontanesi, L. A next generation sequencing approach for targeted Varroa destructor (Acari: Varroidae) mitochondrial DNA analysis based on honey derived environmental DNA. J. Invertebr. Pathol. 2019, 161, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Bovo, S.; Ribani, A.; Utzeri, V.J.; Schiavo, G.; Bertolini, F.; Fontanesi, L. Shotgun metagenomics of honey DNA: Evaluation of a methodological approach to describe a multi-kingdom honey bee derived environmental DNA signature. PLoS ONE 2018, 13, e0205575. [Google Scholar] [CrossRef]
- Bovo, S.; Utzeri, V.J.; Ribani, A.; Cabbri, R.; Fontanesi, L. Shotgun sequencing of honey DNA can describe honey bee derived environmental signatures and the honey bee hologenome complexity. Sci. Rep. 2020, 10, 9279. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Willerslev, E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Lambert, O.; Piroux, M.; Puyo, S.; Thorin, C.; Larhantec, M.; Delbac, F.; Pouliquen, H. Bees, honey and pollen as sentinels for lead environmental contamination. Environ. Pollut. 2012, 170, 254–259. [Google Scholar] [CrossRef]
- Cunningham, M.M.; Tran, L.; McKee, C.G.; Ortega Polo, R.; Newman, T.; Lansing, L.; Griffiths, J.S.; Bilodeau, G.J.; Rott, M.; Marta Guarna, M. Honey bees as biomonitors of environmental contaminants, pathogens, and climate change. Ecol. Indic. 2022, 134, 108457. [Google Scholar] [CrossRef]
- Gates, D.M. Climate Change and Its Biological Consequences; Sinauer Associates: Sunderland, MA, USA, 1993. [Google Scholar]
- Villalba, A.; Maggi, M.; Ondarza, P.M.; Szawarski, N.; Miglioranza, K.S.B. Influence of land use on chlorpyrifos and persistent organic pollutant levels in honey bees, bee bread and honey: Beehive exposure assessment. Sci. Total Environ. 2020, 713, 136554. [Google Scholar] [CrossRef]
- Goretti, E.; Pallottini, M.; Rossi, R.; La Porta, G.; Gardi, T.; Cenci Goga, B.T.; Elia, A.C.; Galletti, M.; Moroni, B.; Petroselli, C.; et al. Heavy metal bioaccumulation in honey bee matrix, an indicator to assess the contamination level in terrestrial environments. Environ. Pollut. 2020, 256, 113388. [Google Scholar] [CrossRef]
- Cochard, P.; Laurie, M.; Veyrand, B.; Le Bizec, B.; Poirot, B.; Marchand, P. PAH7 concentration reflects anthropization: A study using environmental biomonitoring with honeybees. Sci. Total Environ. 2021, 751, 141831. [Google Scholar] [CrossRef] [PubMed]
- Lambert, O.; Veyrand, B.; Durand, S.; Marchand, P.; Bizec, B.L.; Piroux, M.; Puyo, S.; Thorin, C.; Delbac, F.; Pouliquen, H. Polycyclic aromatic hydrocarbons: Bees, honey and pollen as sentinels for environmental chemical contaminants. Chemosphere 2012, 86, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Kek, S.P.; Chin, N.L.; Tan, S.W.; Yusof, Y.A.; Chua, L.S. Comparison of DNA extraction methods for entomological origin identification of honey using simple additive weighting method. Int. J. Food Sci. Technol. 2018, 53, 2490–2499. [Google Scholar] [CrossRef]
- Eftekhari, A.; Alipour, M.; Chodari, L.; Maleki Dizaj, S.; Ardalan, M.; Samiei, M.; Sharifi, S.; Zununi Vahed, S.; Huseynova, I.; Khalilov, R. A comprehensive review of detection methods for SARS-CoV-2. Microorganisms 2021, 9, 232. [Google Scholar] [CrossRef]
- Sadighbayan, D.; Sadighbayan, K.; Khosroushahi, A.Y.; Hasanzadeh, M. Recent advances on the DNA-based electrochemical biosensing of cancer biomarkers: Analytical approach. TrAC Trends Anal. Chem. 2019, 119, 115609. [Google Scholar] [CrossRef]
- Bashir, A.; Lu, Q.; Carson, D.; Raphael, B.J.; Liu, Y.T.; Bafna, V. Optimizing PCR assays for DNA-based cancer diagnostics. J. Comput. Biol. 2010, 17, 369–381. [Google Scholar] [CrossRef]
- Soares, S.; Amaral, J.S.; Oliveira, M.B.; Mafra, I. A SYBR Green real-time PCR assay to detect and quantify pork meat in processed poultry meat products. Meat Sci. 2013, 94, 115–120. [Google Scholar] [CrossRef]
- Soares, S.; Amaral, J.S.; Oliveira, M.B.; Mafra, I. Quantitative detection of soybean in meat products by a TaqMan real-time PCR assay. Meat Sci. 2014, 98, 41–46. [Google Scholar] [CrossRef]
- Soares, S.; Grazina, L.; Costa, J.; Amaral, J.S.; Oliveira, M.B.P.P.; Mafra, I. Botanical authentication of lavender (Lavandula spp.) honey by a novel DNA-barcoding approach coupled to high resolution melting analysis. Food Control 2018, 86, 367–373. [Google Scholar] [CrossRef]
- Soares, S.; Grazina, L.; Mafra, I.; Costa, J.; Pinto, M.A.; Duc, H.P.; Oliveira, M.B.P.P.; Amaral, J.S. Novel diagnostic tools for Asian (Apis cerana) and European (Apis mellifera) honey authentication. Food Res. Int. 2018, 105, 686–693. [Google Scholar] [CrossRef]
- Villa, C.; Costa, J.; Gondar, C.; Oliveira, M.B.P.P.; Mafra, I. Effect of food matrix and thermal processing on the performance of a normalised quantitative real-time PCR approach for lupine (Lupinus albus) detection as a potential allergenic food. Food Chem. 2018, 262, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F.; Amorim, A.; van Asch, B. Chapter 8—Genetic and DNA-Based Techniques. In Comprehensive Analytical Chemistry; de la Guardia, M., Gonzálvez, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 195–220. [Google Scholar]
- Honrado, M.; Lopes, A.R.; Pinto, M.A.; Amaral, J.S. A novel real-time PCR coupled with high resolution melting analysis as a simple and fast tool for the entomological authentication of honey by targeting Apis mellifera mitochondrial DNA. Food Res. Int. 2022, 161, 111761. [Google Scholar] [CrossRef]
- Chiara, B.; Francesco, C.; Fulvio, B.; Paola, M.; Annalisa, G.; Stefania, S.; Luigi, A.P.; Simone, P. Exploring the botanical composition of polyfloral and monofloral honeys through DNA metabarcoding. Food Control 2021, 128, 108175. [Google Scholar] [CrossRef]
- Laha, R.C.; De Mandal, S.; Ralte, L.; Ralte, L.; Kumar, N.S.; Gurusubramanian, G.; Satishkumar, R.; Mugasimangalam, R.; Kuravadi, N.A. Meta-barcoding in combination with palynological inference is a potent diagnostic marker for honey floral composition. AMB Express 2017, 7, 132. [Google Scholar] [CrossRef] [PubMed]
- Wirta, H.; Abrego, N.; Miller, K.; Roslin, T.; Vesterinen, E. DNA traces the origin of honey by identifying plants, bacteria and fungi. Sci. Rep. 2021, 11, 4798. [Google Scholar] [CrossRef] [PubMed]
- Prosser, S.W.J.; Hebert, P.D.N. Rapid identification of the botanical and entomological sources of honey using DNA metabarcoding. Food Chem. 2017, 214, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Richardson, R.T.; Lin, C.H.; Sponsler, D.B.; Quijia, J.O.; Goodell, K.; Johnson, R.M. Application of ITS2 metabarcoding to determine the provenance of pollen collected by honey bees in an agroecosystem. Appl. Plant Sci. 2015, 3, 1400066. [Google Scholar] [CrossRef]
- CODEX STAN 12-1981; Codex Alimentarius, Revised Standard for Honey. FAO: Rome, Italy, 2001; pp. 19–26.
- Kek, S.P.; Chin, N.L.; Tan, S.W.; Yusof, Y.A.; Chua, L.S. Molecular identification of honey entomological origin based on bee mitochondrial 16S rRNA and COI gene sequences. Food Control 2017, 78, 150–159. [Google Scholar] [CrossRef]
- Laube, I.; Hird, H.; Brodmann, P.; Ullmann, S.; Schöne-Michling, M.; Chisholm, J.; Broll, H. Development of primer and probe sets for the detection of plant species in honey. Food Chem. 2010, 118, 979–986. [Google Scholar] [CrossRef]
- Soares, S.; Amaral, J.S.; Oliveira, M.B.; Mafra, I. Improving DNA isolation from honey for the botanical origin identification. Food Control 2015, 48, 130–136. [Google Scholar] [CrossRef]
- Bruni, I.; Galimberti, A.; Caridi, L.; Scaccabarozzi, D.; De Mattia, F.; Casiraghi, M.; Labra, M. A DNA barcoding approach to identify plant species in multiflower honey. Food Chem. 2015, 170, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Danner, N.; Grimmer, G.; Ankenbrand, M.; von der Ohe, K.; von der Ohe, W.; Rost, S.; Härtel, S.; Steffan-Dewenter, I. Evaluating multiplexed next-generation sequencing as a method in palynology for mixed pollen samples. Plant Biol. 2015, 17, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lang, D.; Meng, G.; Hu, J.; Tang, M.; Zhou, X. Tracing the origin of honey products based on metagenomics and machine learning. Food Chem. 2022, 371, 131066. [Google Scholar] [CrossRef] [PubMed]
- Guertler, P.; Eicheldinger, A.; Muschler, P.; Goerlich, O.; Busch, U. Automated DNA extraction from pollen in honey. Food Chem. 2014, 149, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.E.; Newbold, L.K.; Gweon, H.S.; Read, D.S.; Woodcock, B.A.; Pywell, R.F. Integration of DNA extraction, metabarcoding and an informatics pipeline to underpin a national citizen science honey monitoring scheme. MethodsX 2021, 8, 101303. [Google Scholar] [CrossRef] [PubMed]
- Lauro, F.M.; Favaretto, M.; Covolo, L.; Rassu, M.; Bertoloni, G. Rapid detection of Paenibacillus larvae from honey and hive samples with a novel nested PCR protocol. Int. J. Food Microbiol. 2003, 81, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, J.; de Vere, N.; Griffith, A.; Ford, C.R.; Allainguillaume, J.; Hegarty, M.J.; Baillie, L.; Adams-Groom, B. Using DNA Metabarcoding to Identify the Floral Composition of Honey: A New Tool for Investigating Honey Bee Foraging Preferences. PLoS ONE 2015, 10, e0134735. [Google Scholar] [CrossRef]
- Olivieri, C.; Marota, I.; Rollo, F.; Luciani, S. Tracking plant, fungal, and bacterial DNA in honey specimens. J. Forensic Sci. 2012, 57, 222–227. [Google Scholar] [CrossRef]
- Kast, C.; Roetschi, A. Evaluation of baker’s yeast in honey using a real-time PCR assay. Food Microbiol. 2017, 62, 282–288. [Google Scholar] [CrossRef]
- Cheng, H.; Jin, W.; Wu, H.; Wang, F.; You, C.; Peng, Y.; Jia, S. Isolation and PCR Detection of Foreign DNA Sequences in Bee Honey Raised on Genetically Modified Bt (Cry 1 Ac) Cotton. Food Bioprod. Process. 2007, 85, 141–145. [Google Scholar] [CrossRef]
- Soares, S.; Grazina, L.; Mafra, I.; Costa, J.; Pinto, M.A.; Oliveira, M.B.P.P.; Amaral, J.S. Towards honey authentication: Differentiation of Apis mellifera subspecies in European honeys based on mitochondrial DNA markers. Food Chem. 2019, 283, 294–301. [Google Scholar] [CrossRef]
- Raime, K.; Krjutškov, K.; Remm, M. Method for the Identification of Plant DNA in Food Using Alignment-Free Analysis of Sequencing Reads: A Case Study on Lupin. Front. Plant Sci. 2020, 11, 646. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Latif, A.; Osman, G. Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods 2017, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Baksay, S.; Pornon, A.; Burrus, M.; Mariette, J.; Andalo, C.; Escaravage, N. Experimental quantification of pollen with DNA metabarcoding using ITS1 and trnL. Sci. Rep. 2020, 10, 4202. [Google Scholar] [CrossRef]
- Demeke, T.; Jenkins, G.R. Influence of DNA extraction methods, PCR inhibitors and quantification methods on real-time PCR assay of biotechnology-derived traits. Anal. Bioanal. Chem. 2010, 396, 1977–1990. [Google Scholar] [CrossRef]
- Jain, S.; Jesus, F.; Marchioro, G.; Araújo, E. Extraction of DNA from honey and its amplification by PCR for botanical identification. Food Sci. Technol. 2013, 33, 753–756. [Google Scholar] [CrossRef]
- Sajali, N.; Wong, S.C.; Hanapi, U.K.; Abu Bakar Jamaluddin, S.; Tasrip, N.A.; Mohd Desa, M.N. The Challenges of DNA Extraction in Different Assorted Food Matrices: A Review. J. Food Sci. 2018, 83, 2409–2414. [Google Scholar] [CrossRef]
- Soares, S.; Pinto, D.; Rodrigues, F.; Alves, R.C.; Oliveira, M.B.P.P. Portuguese Honeys from Different Geographical and Botanical Origins: A 4-Year Stability Study Regarding Quality Parameters and Antioxidant Activity. Molecules 2017, 22, 1338. [Google Scholar] [CrossRef]
- Valentini, A.; Miquel, C.; Taberlet, P. DNA Barcoding for Honey Biodiversity. Diversity 2010, 2, 610–617. [Google Scholar] [CrossRef]
- Kraaijeveld, K.; de Weger, L.A.; Ventayol García, M.; Buermans, H.; Frank, J.; Hiemstra, P.S.; den Dunnen, J.T. Efficient and sensitive identification and quantification of airborne pollen using next-generation DNA sequencing. Mol. Ecol. Resour. 2015, 15, 8–16. [Google Scholar] [CrossRef]
- De Vere, N.; Jones, L.E.; Gilmore, T.; Moscrop, J.; Lowe, A.; Smith, D.; Hegarty, M.J.; Creer, S.; Ford, C.R. Using DNA metabarcoding to investigate honey bee foraging reveals limited flower use despite high floral availability. Sci. Rep. 2017, 7, 42838. [Google Scholar] [CrossRef] [PubMed]
- Danner, N.; Keller, A.; Härtel, S.; Steffan-Dewenter, I. Honey bee foraging ecology: Season but not landscape diversity shapes the amount and diversity of collected pollen. PLoS ONE 2017, 12, e0183716. [Google Scholar] [CrossRef] [PubMed]
- Porter, T.M.; Hajibabaei, M. Scaling up: A guide to high-throughput genomic approaches for biodiversity analysis. Mol. Ecol. 2018, 27, 313–338. [Google Scholar] [CrossRef] [PubMed]
- CBOL Plant Working Group. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797.
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and Using a Plant DNA Barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef]
- Ribani, A.; Utzeri, V.J.; Taurisano, V.; Galuppi, R.; Fontanesi, L. Analysis of honey environmental DNA indicates that the honey bee (Apis mellifera L.) trypanosome parasite Lotmaria passim is widespread in the apiaries of the North of Italy. J. Invertebr. Pathol. 2021, 184, 107628. [Google Scholar] [CrossRef]
- Giersch, T.; Berg, T.; Galea, F.; Hornitzky, M. Nosema ceranae infects honey bees (Apis mellifera) and contaminates honey in Australia. Apidologie 2009, 40, 117–123. [Google Scholar] [CrossRef]
- Gismondi, A.; Di Marco, G.; Canini, A. Detection of plant microRNAs in honey. PLoS ONE 2017, 12, e0172981. [Google Scholar] [CrossRef]
- Smith, C.; Cokcetin, N.; Truong, T.; Harry, E.; Hutvagner, G.; Bajan, S. Cataloguing the small RNA content of honey using next generation sequencing. Food Chem. 2021, 2, 100014. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soares, S.; Rodrigues, F.; Delerue-Matos, C. Towards DNA-Based Methods Analysis for Honey: An Update. Molecules 2023, 28, 2106. https://doi.org/10.3390/molecules28052106
Soares S, Rodrigues F, Delerue-Matos C. Towards DNA-Based Methods Analysis for Honey: An Update. Molecules. 2023; 28(5):2106. https://doi.org/10.3390/molecules28052106
Chicago/Turabian StyleSoares, Sónia, Francisca Rodrigues, and Cristina Delerue-Matos. 2023. "Towards DNA-Based Methods Analysis for Honey: An Update" Molecules 28, no. 5: 2106. https://doi.org/10.3390/molecules28052106
APA StyleSoares, S., Rodrigues, F., & Delerue-Matos, C. (2023). Towards DNA-Based Methods Analysis for Honey: An Update. Molecules, 28(5), 2106. https://doi.org/10.3390/molecules28052106








