Identification and Characterization of a Novel Cathelicidin from Hydrophis cyanocinctus with Antimicrobial and Anti-Inflammatory Activity
Abstract
1. Introduction
2. Results
2.1. Identification of Hydrostatin-AMP2
2.2. Molecular Structure of Hydrostatin-AMP2
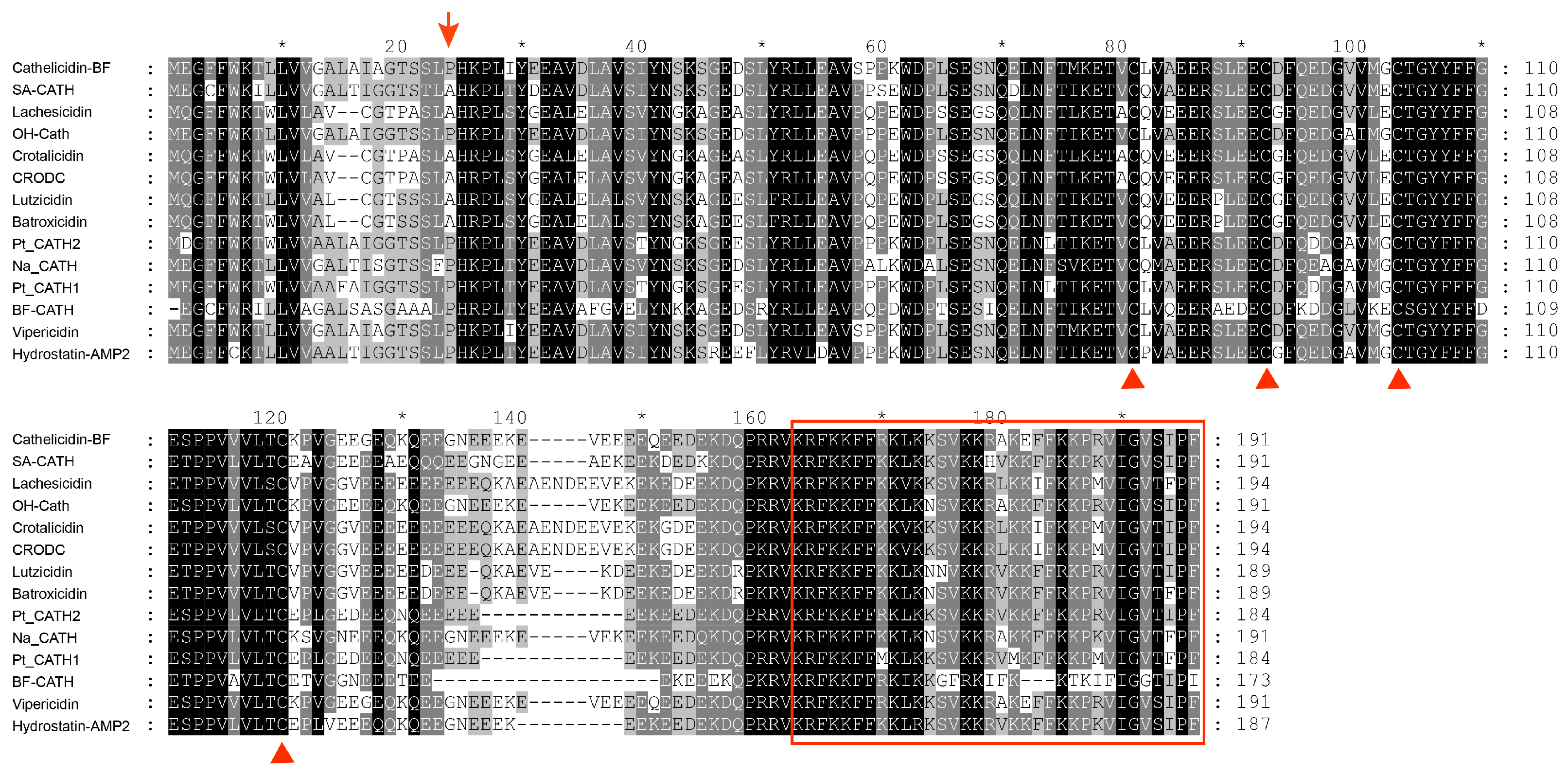
2.3. Sequence Comparison and Phylogenetic Analysis
2.4. Antimicrobial Activities of Hydrostatin-AMP2
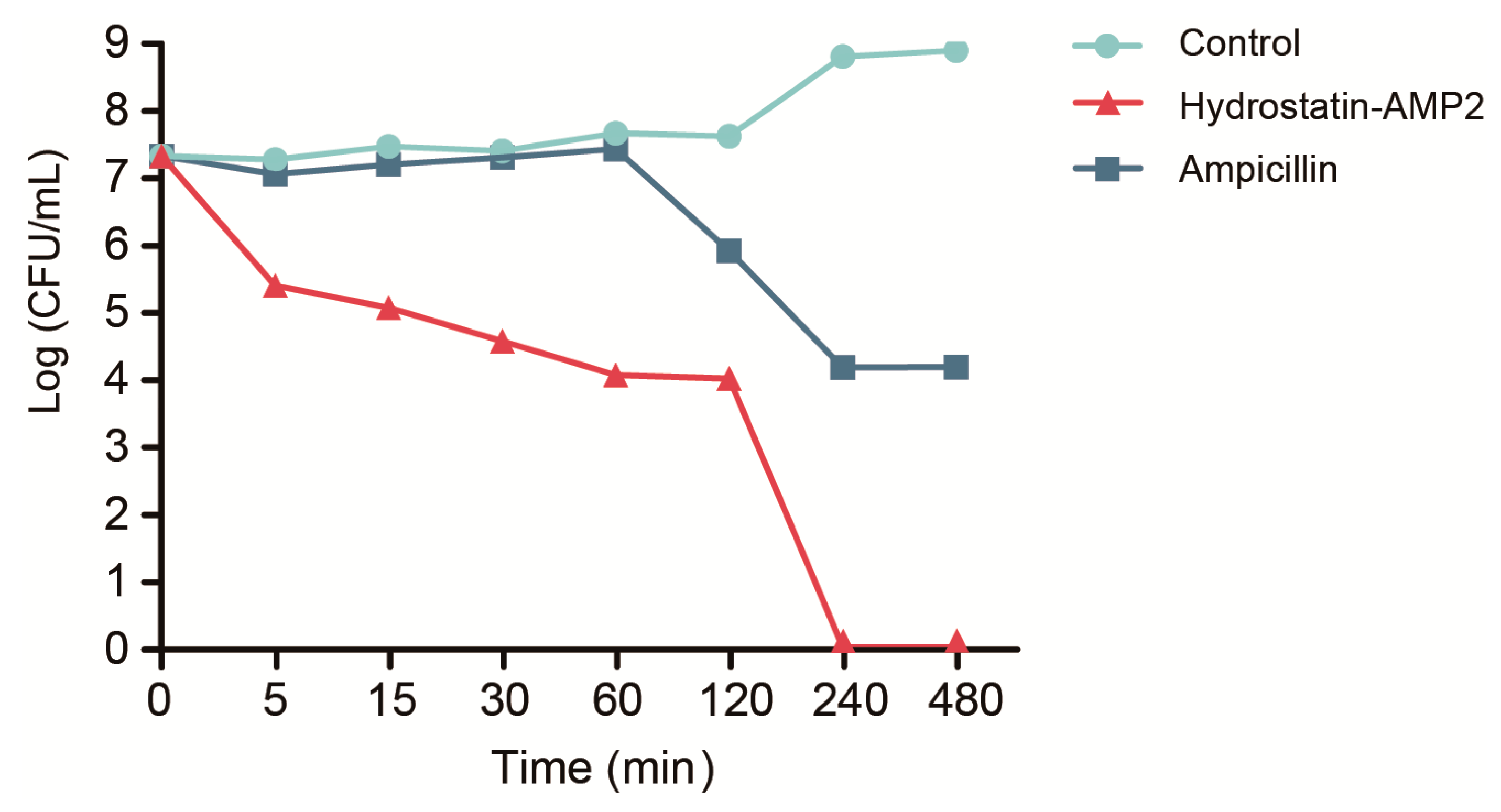
2.5. Time-Kill Kinetics of Hydrostatin-AMP2
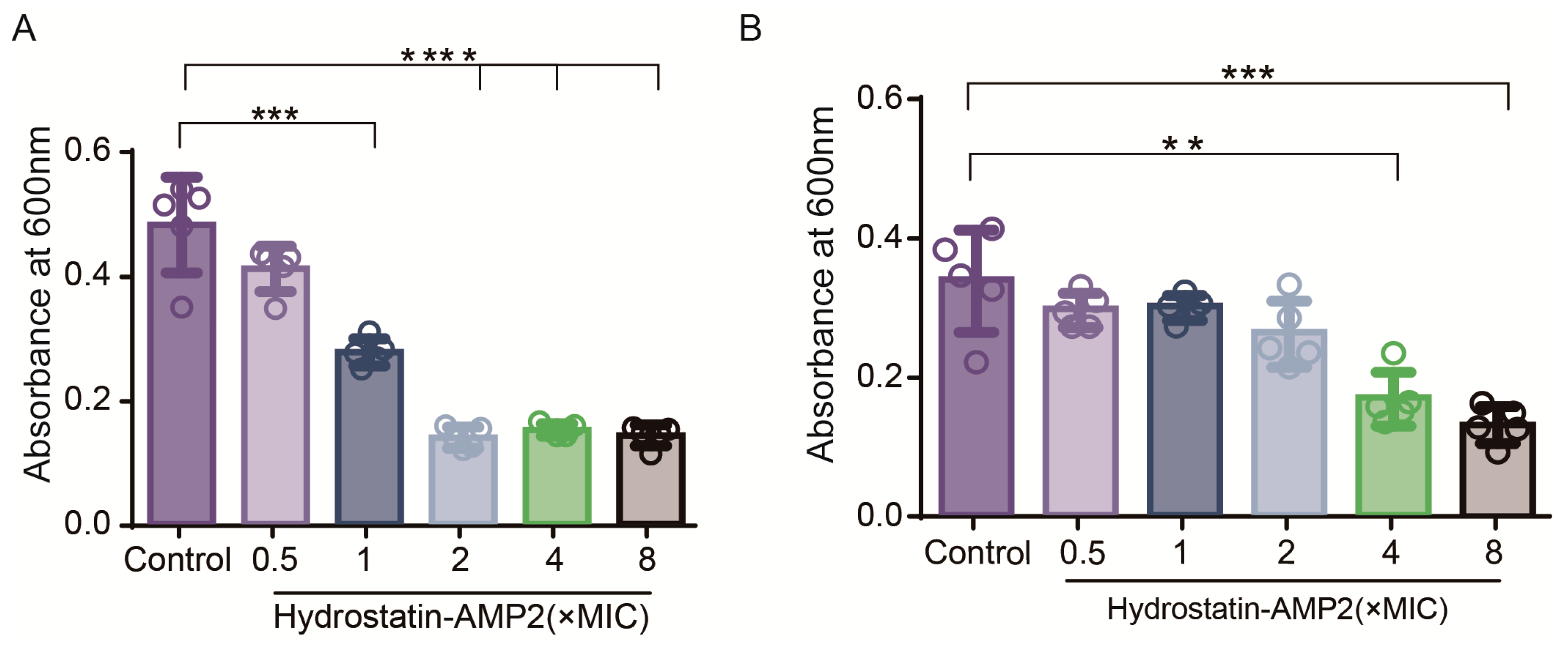
2.6. Inhibition and Eradication of Biofilms by Hydrostatin-AMP2
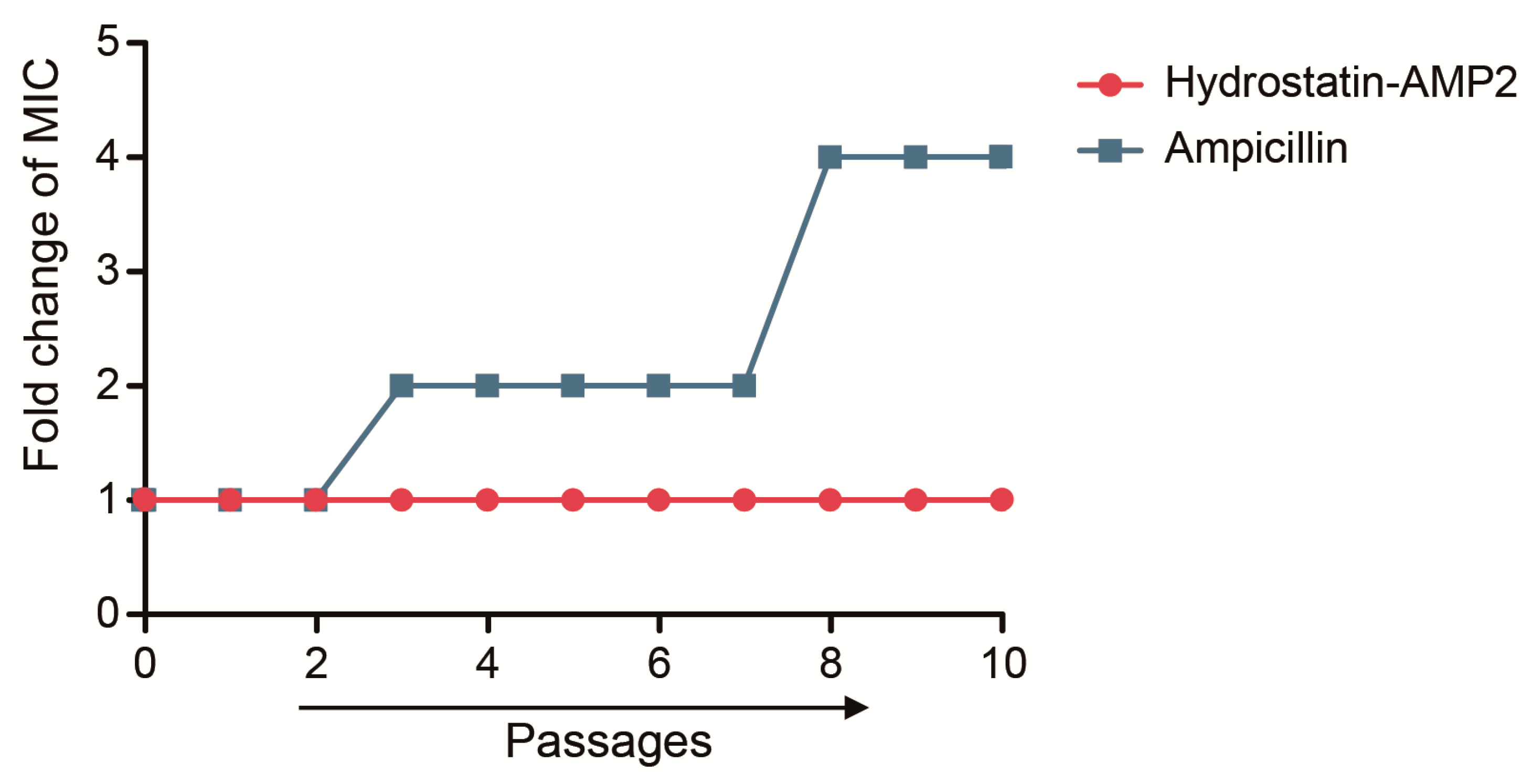
2.7. Induction of Drug-Resistance
2.8. Salt Tolerance, Thermal Tolerance, pH Stability and Serum Stability of Hydrostatin-AMP2
2.9. Hemolytic Activity and Cytotoxicity of Hydrostatin-AMP2
2.10. Hydrostatin-AMP2 Inhibited the Pro-Inflammatory Factor Expression Induced by LPS
3. Discussion
4. Materials and Methods
4.1. Identification and Activity Prediction of Hydrostatin-AMP2
4.2. Multi-Sequence Alignment and Phylogenetic Analysis
4.3. Biological Materials and Reagents
4.4. Circular Dichroism Spectroscopy
4.5. Antimicrobial Assays
4.6. Time-Killing Kinetics
4.7. Biofilm Inhibition Assay
4.8. Biofilm Eradication Assay
4.9. Induction of Resistance
4.10. Salt Stability Analysis
4.11. Thermal Stability Analysis
4.12. Serum Stability Analysis
4.13. pH Stability Analysis
4.14. Cytotoxicity Assays
4.15. Hemolysis Assays
4.16. RNA Extract and Real-Time PCR
4.17. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bush, K. The coming of age of antibiotics: Discovery and therapeutic value. Ann. N. Y. Acad. Sci. 2010, 1213, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Mwangi, J.; Hao, X.; Lai, R.; Zhang, Z.Y. Antimicrobial peptides: New hope in the war against multidrug resistance. Zool. Res. 2019, 40, 488–505. [Google Scholar] [CrossRef]
- Boman, H.G. Innate immunity and the normal microflora. Immunol. Rev. 2000, 173, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Steiner, H.; Hultmark, D.; Engström, A.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA 1987, 84, 5449–5453. [Google Scholar] [CrossRef]
- Wang, S.; Fan, L.; Pan, H.; Li, Y.; Qiu, Y.; Lu, Y. Antimicrobial peptides from marine animals: Sources, structures, mechanisms and the potential for drug development. Front. Mar. Sci. 2023, 9, 2880. [Google Scholar] [CrossRef]
- Sørensen, O.; Arnljots, K.; Cowland, J.B.; Bainton, D.F.; Borregaard, N. The human antibacterial cathelicidin, hCAP-18, is synthesized in myelocytes and metamyelocytes and localized to specific granules in neutrophils. Blood 1997, 90, 2796–2803. [Google Scholar] [CrossRef]
- Shinnar, A.E.; Butler, K.L.; Park, H.J. Cathelicidin family of antimicrobial peptides: Proteolytic processing and protease resistance. Bioorg. Chem. 2003, 31, 425–436. [Google Scholar] [CrossRef]
- Coorens, M.; van Dijk, A.; Bikker, F.; Veldhuizen, E.J.A.; Haagsman, H.P. Importance of Endosomal Cathelicidin Degradation To Enhance DNA-Induced Chicken Macrophage Activation. J. Immunol. 2015, 195, 3970–3977. [Google Scholar] [CrossRef]
- Du, H.; Samuel, R.L.; Massiah, M.A.; Gillmor, S.D. The structure and behavior of the NA-CATH antimicrobial peptide with liposomes. Biochim. Biophys. Acta 2015, 1848, 2394–2405. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Sun, X.; Dong, X.; Lin, H.; Tang, L.; Xue, M.; Zhong, G. Chlamydial plasmid-encoded virulence factor Pgp3 interacts with human cathelicidin peptide LL-37 to modulate immune response. Microbes Infect. 2019, 21, 50–55. [Google Scholar] [CrossRef]
- Cheung, G.Y.; Otto, M. Do antimicrobial peptides and antimicrobial-peptide resistance play important roles during bacterial infection? Future Microbiol. 2018, 13, 1073–1075. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Cerri, F.; Saliu, F.; Maggioni, D.; Montano, S.; Seveso, D.; Lavorano, S.; Zoia, L.; Gosetti, F.; Lasagni, M.; Orlandi, M.; et al. Cytotoxic Compounds from Alcyoniidae: An Overview of the Last 30 Years. Mar. Drugs 2022, 20, 134. [Google Scholar] [CrossRef]
- Minutti-Zanella, C.; Gil-Leyva, E.J.; Vergara, I. Immunomodulatory properties of molecules from animal venoms. Toxicon 2021, 191, 54–68. [Google Scholar] [CrossRef]
- Li, A.; Wang, J.; Sun, K.; Wang, S.; Zhao, X.; Wang, T.; Xiong, L.; Xu, W.; Qiu, L.; Shang, Y.; et al. Two Reference-Quality Sea Snake Genomes Reveal Their Divergent Evolution of Adaptive Traits and Venom Systems. Mol. Biol. Evol. 2021, 38, 4867–4883. [Google Scholar] [CrossRef]
- Torrent, M.; Di Tommaso, P.; Pulido, D.; Nogués, M.V.; Notredame, C.; Boix, E.; Andreu, D. AMPA: An automated web server for prediction of protein antimicrobial regions. Bioinformatics 2012, 28, 130–131. [Google Scholar] [CrossRef]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016, 44, 1094–1097. [Google Scholar] [CrossRef] [PubMed]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 2021, 49, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef] [PubMed]
- Mortuza, S.M.; Zheng, W.; Zhang, C.; Li, Y.; Pearce, R.; Zhang, Y. Improving fragment-based ab initio protein structure assembly using low-accuracy contact-map predictions. Nat. Commun. 2021, 12, 5011. [Google Scholar] [CrossRef] [PubMed]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific alpha-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef] [PubMed]
- Klemperer, R.M.; Gilbert, P.; Meier, A.M.; Cozens, R.M.; Brown, M.R. Influence of suspending media upon the susceptibility of Pseudomonas aeruginosa NCTC 6750 and its spheroplasts to polymyxin B. Antimicrob. Agents Chemother. 1979, 15, 147–151. [Google Scholar] [CrossRef]
- Liang, X.; Liu, K.; Zhao, P.; Zhou, J.; Zhang, F.; He, Y.; Zhang, H.; Fareed, M.S.; Lu, Y.; Xu, Y.; et al. The effects of incorporation of the counterparts and mimics of L-lysine on the antimicrobial activity, hemolytic activity, cytotoxicity and tryptic stability of antimicrobial peptide polybia-MPII. Amino Acids 2022, 54, 123–135. [Google Scholar] [CrossRef]
- Pirkhezranian, Z.; Tahmoorespur, M.; Daura, X.; Monhemi, H.; Sekhavati, M.H. Interaction of camel Lactoferrin derived peptides with DNA: A molecular dynamics study. BMC Genom. 2020, 21, 60. [Google Scholar] [CrossRef]
- Pirkhezranian, Z.; Tahmoorespur, M.; Monhemi, H.; Sekhavati, M.H. Computational Peptide Engineering Approach for Selection the Best Engendered Camel Lactoferrin-Derive Peptide with Potency to Interact with DNA. Int. J. Pept. Res. Ther. 2020, 26, 2203–2212. [Google Scholar] [CrossRef]
- Sani, M.A.; Separovic, F. How Membrane-Active Peptides Get into Lipid Membranes. Acc. Chem. Res. 2016, 49, 1130–1138. [Google Scholar] [CrossRef]
- Falanga, A.; Lombardi, L.; Franci, G.; Vitiello, M.; Iovene, M.R.; Morelli, G.; Galdiero, M.; Galdiero, S. Marine Antimicrobial Peptides: Nature Provides Templates for the Design of Novel Compounds against Pathogenic Bacteria. Int. J. Mol. Sci. 2016, 17, 785. [Google Scholar] [CrossRef]
- Mwangi, J.; Yin, Y.; Wang, G.; Yang, M.; Li, Y.; Zhang, Z.; Lai, R. The antimicrobial peptide ZY4 combats multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii infection. Proc. Natl. Acad. Sci. USA 2019, 116, 26516–26522. [Google Scholar] [CrossRef]
- Hochdörfer, T.; Tiedje, C.; Stumpo, D.J.; Blackshear, P.J.; Gaestel, M.; Huber, M. LPS-induced production of TNF-α and IL-6 in mast cells is dependent on p38 but independent of TTP. Cell Signal. 2013, 25, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Bilgic, H.; Ytterberg, S.R.; Amin, S.; McNallan, K.T.; Wilson, J.C.; Koeuth, T.; Ellingson, S.; Newman, B.; Bauer, J.W.; Peterson, E.J.; et al. Interleukin-6 and type I interferon-regulated genes and chemokines mark disease activity in dermatomyositis. Arthritis Rheum. 2009, 60, 3436–3446. [Google Scholar] [CrossRef] [PubMed]
- Vallabhapurapu, S.; Karin, M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 2009, 27, 693–733. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, C.; Meng, J.; Li, N.; Xu, Z.; Liu, X.; Hou, S. Galectin-3 regulates microglial activation and promotes inflammation through TLR4/MyD88/NF-kB in experimental autoimmune uveitis. Clin. Immunol. 2022, 236, 108939. [Google Scholar] [CrossRef]
- Wang, Y.C.; Zhou, Y.; Fang, H.; Lin, S.; Wang, P.F.; Xiong, R.P.; Chen, J.; Xiong, X.Y.; Lv, F.L.; Liang, Q.L.; et al. Toll-like receptor 2/4 heterodimer mediates inflammatory injury in intracerebral hemorrhage. Ann. Neurol. 2014, 75, 876–889. [Google Scholar] [CrossRef]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, 276–279. [Google Scholar] [CrossRef]
- Lanave, C.; Licciulli, F.; De Robertis, M.; Marolla, A.; Attimonelli, M. Update of AMmtDB: A database of multi-aligned Metazoa mitochondrial DNA sequences. Nucleic Acids Res. 2002, 30, 174–175. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- CLSI. M07: Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015; pp. 3–110. [Google Scholar]
- Zhong, L.; Liu, J.; Teng, S.; Xie, Z. Identification of a Novel Cathelicidin from the Deinagkistrodon acutus Genome with Antibacterial Activity by Multiple Mechanisms. Toxins 2020, 12, 771. [Google Scholar] [CrossRef]
- Furtado, A.A.; Daniele-Silva, A.; de Oliveira, I.R.R.; Mendes, R.F.V.; Santos, E.; Carvalho, E.; Damasceno, I.Z.; Parente, A.; Sena, K.; Silva-Júnior, A.A.D.; et al. In silico and in vitro structure-stability-function relationship of analog peptides of Stigmurin and its antibacterial and antibiofilm activities. Pharmacol. Res. 2022, 181, 106245. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Bu, S.; Wang, Z.; Zhou, H.; Li, X.; Wei, J.; He, X.; Wan, J. Click Chemistry Actuated Exponential Amplification Reaction Assisted CRISPR-Cas12a for the Electrochemical Detection of MicroRNAs. ACS Omega 2022, 7, 35515–35522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, S.; Li, J.; Bu, X.; Dong, X.; Chen, N.; Li, F.; Zhu, J.; Sang, L.; Zeng, Y.; et al. Dual-sensitive antibacterial peptide nanoparticles prevent dental caries. Theranostics 2022, 12, 4818–4833. [Google Scholar] [CrossRef]
- Chen, J.; Hao, D.; Mei, K.; Li, X.; Li, T.; Ma, C.; Xi, X.; Li, L.; Wang, L.; Zhou, M.; et al. In Vitro and In Vivo Studies on the Antibacterial Activity and Safety of a New Antimicrobial Peptide Dermaseptin-AC. Microbiol. Spectr. 2021, 9, e0131821. [Google Scholar] [CrossRef]
- Wang, Y.; Hong, J.; Liu, X.; Yang, H.; Liu, R.; Wu, J.; Wang, A.; Lin, D.; Lai, R. Snake cathelicidin from Bungarus fasciatus is a potent peptide antibiotics. PLoS ONE 2008, 3, e3217. [Google Scholar] [CrossRef] [PubMed]
- Higgins, D.L.; Chang, R.; Debabov, D.V.; Leung, J.; Wu, T.; Krause, K.M.; Sandvik, E.; Hubbard, J.M.; Kaniga, K.; Schmidt, D.E., Jr.; et al. Telavancin, a multifunctional lipoglycopeptide, disrupts both cell wall synthesis and cell membrane integrity in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2005, 49, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, Q.J.; Qiao, Y.; Chen, J.; Li, M.Y. The host defense peptide β-defensin confers protection against Vibrio anguillarum in ayu, Plecoglossus altivelis. Dev. Comp. Immunol. 2020, 103, 103511. [Google Scholar] [CrossRef]
- Qi, X.; Zhou, C.; Li, P.; Xu, W.; Cao, Y.; Ling, H.; Ning Chen, W.; Ming Li, C.; Xu, R.; Lamrani, M.; et al. Novel short antibacterial and antifungal peptides with low cytotoxicity: Efficacy and action mechanisms. Biochem. Biophys. Res. Commun. 2010, 398, 594–600. [Google Scholar] [CrossRef]



| Microorganisms | MIC/MBC (μg/mL) | ||
|---|---|---|---|
| Hydrostatin-AMP2 | Ampicillin | ||
| G− | Escherichia coli | 16/32 | 8/16 |
| Klebsiella pneumoniae 44 | 32 | >128 | |
| K. pneumoniae 48 | 16 | >128 | |
| K. pneumoniae 49 | 32 | >128 | |
| K. pneumoniae 50 | 32 | >128 | |
| K. pneumoniae 51 | 32 | >128 | |
| G+ | Propionibacterium acnes | 32 | 0.25 |
| Staphylococcus aureus | 32 | 0.25 | |
| Conditions | MIC (μg/mL) a | Conditions | MIC (μg/mL) | ||
|---|---|---|---|---|---|
| Control | 16 | ||||
| NaCl | 50 mM | 32 | Human serum | 1 h | 32 |
| 100 mM | 32 | 2 h | 32 | ||
| 150 mM | 32 | 3 h | 32 | ||
| 200 mM | 64 | 4 h | 32 | ||
| CaCl2 | 1 mM | 64 | Temperature | 4 °C | 32 |
| 2 mM | 64 | 37 °C | 32 | ||
| 4 mM | 128 | 80 °C | 32 | ||
| 100 °C | 64 | ||||
| pH | 2 | 64 | |||
| 12 | 64 | ||||
| Cells | Cell Death/Hemolysis (%) a |
|---|---|
| L929 | 8.08 |
| HaCaT | 5.89 |
| RAW264.7 | 10.81 |
| Erythrocytes | 4.6 |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| GAPDH | 5′-AGGTCGGTGTGAACGGATTTG-3′ | 5′-TGTAGACCATGTAGTTGAGGT-3′ |
| IL-6 | 5′-CCAATGCTCTCCTAACAGAT-3′ | 5′-TGTCCACAAACTGATATGCT-3′ |
| iNOS | 5′-GCCAGTCAGGTCTCAGCAAG-3′ | 5′-CGCATGCAATGTGTGCTTGT-3′ |
| IL-1β | 5′-CACTACAGGCTCCGAGATGAACAAC-3′ | 5′-TGTCGTTGCTTGGTTCTCCTTGTAC-3′ |
| TNF-α | 5′-CACCACGCTCTTCTGTCTACTGAAC-3′ | 5′-AGATGATCTGAGTGTGAGGGTCTGG-3′ |
| IFN-γ | 5′-CTGGAGGAACTGGCAAAAGGATGG-3′ | 5′-GACGCTTATGTTGTTGCTGATGGC-3′ |
| IL-10 | 5′-TGCCAAGCCTTATCGGAAATGATCC-3′ | 5′-AGCCGCATCCTGAGGGTCTTC-3′ |
| MCP-1 | 5′-CACTCACCTGCTGCTACTCATTCAC-3′ | 5′-CTTCTTTGGGACACCTGCTGCTG-3′ |
| IL-8 | 5′-CTTCTTTGGGACACCTGCTGCTG-3′ | 5′-CTTCTTTGGGACACCTGCTGCTG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Fan, L.; Pan, H.; Li, Y.; Zhao, X.; Qiu, Y.; Lu, Y. Identification and Characterization of a Novel Cathelicidin from Hydrophis cyanocinctus with Antimicrobial and Anti-Inflammatory Activity. Molecules 2023, 28, 2082. https://doi.org/10.3390/molecules28052082
Wang S, Fan L, Pan H, Li Y, Zhao X, Qiu Y, Lu Y. Identification and Characterization of a Novel Cathelicidin from Hydrophis cyanocinctus with Antimicrobial and Anti-Inflammatory Activity. Molecules. 2023; 28(5):2082. https://doi.org/10.3390/molecules28052082
Chicago/Turabian StyleWang, Shuocun, Liming Fan, Hanyu Pan, Yingying Li, Xin Zhao, Yan Qiu, and Yiming Lu. 2023. "Identification and Characterization of a Novel Cathelicidin from Hydrophis cyanocinctus with Antimicrobial and Anti-Inflammatory Activity" Molecules 28, no. 5: 2082. https://doi.org/10.3390/molecules28052082
APA StyleWang, S., Fan, L., Pan, H., Li, Y., Zhao, X., Qiu, Y., & Lu, Y. (2023). Identification and Characterization of a Novel Cathelicidin from Hydrophis cyanocinctus with Antimicrobial and Anti-Inflammatory Activity. Molecules, 28(5), 2082. https://doi.org/10.3390/molecules28052082





