Probing the Mechanisms of Inhibitors Binding to Presenilin Homologue Using Molecular Dynamics Simulations
Abstract
1. Introduction
2. Results and Discussion
2.1. The Binding Mode of Inhibitors to PSH
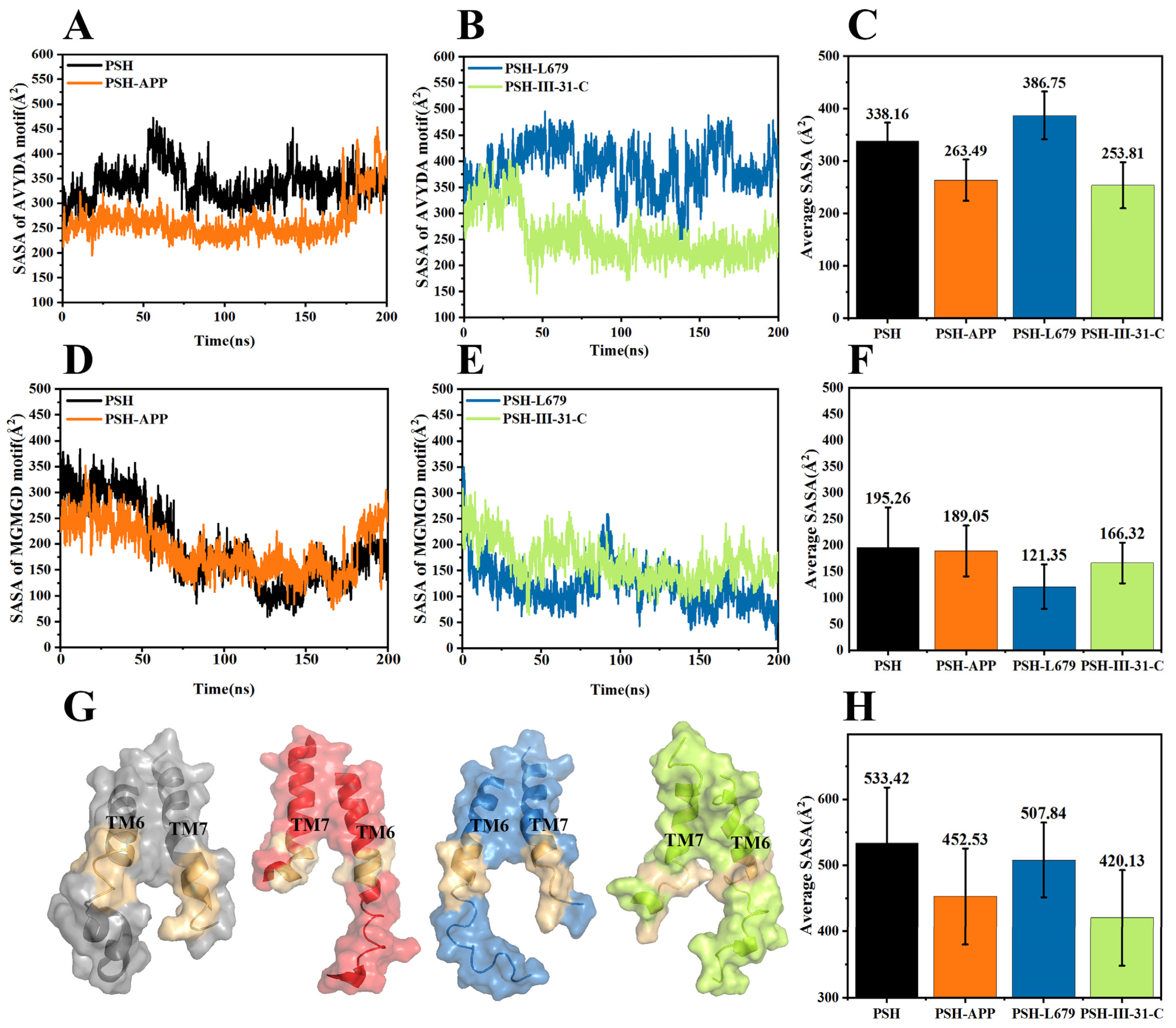
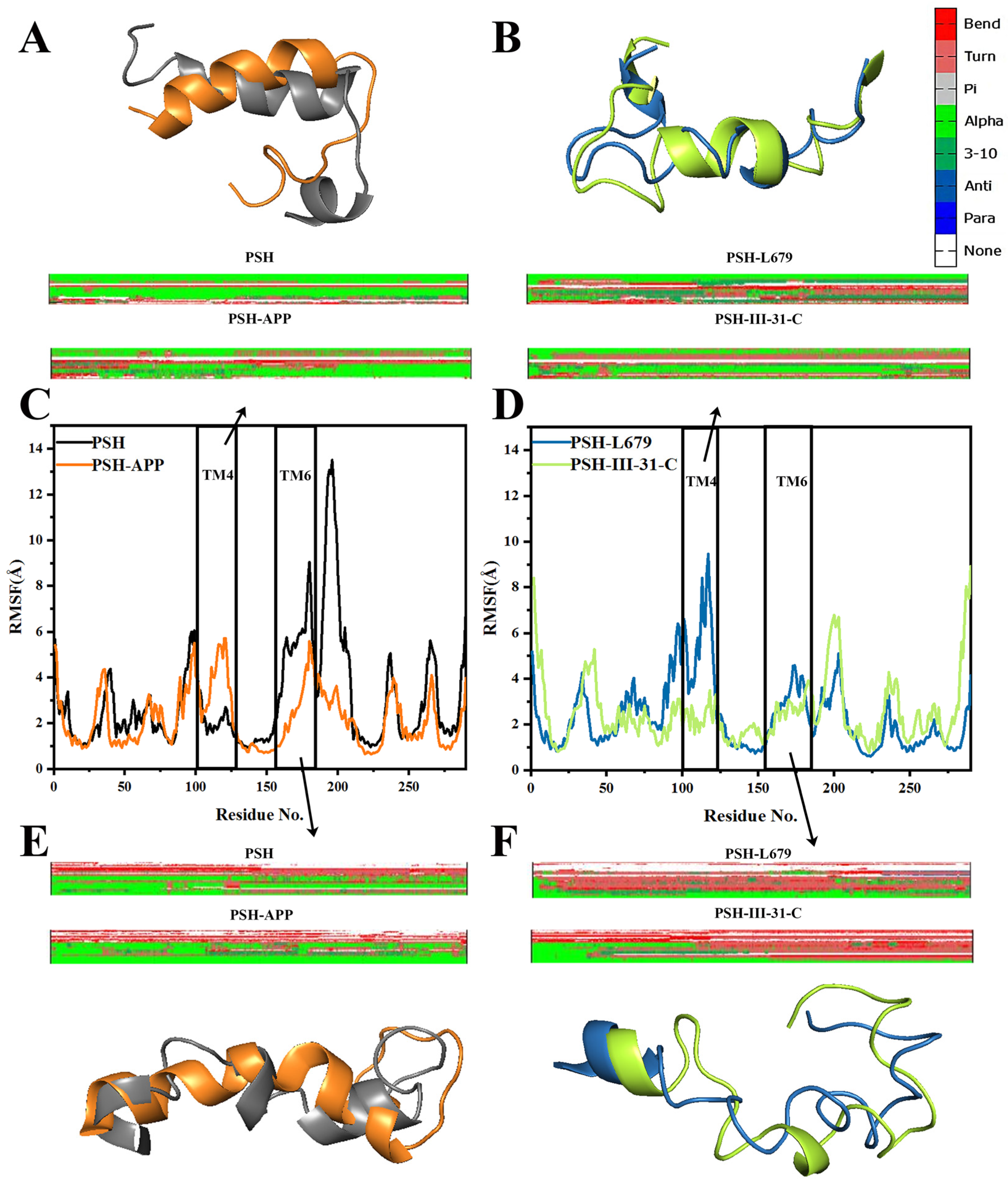
2.2. Structural Stability and Dynamics Properties of the Four Systems
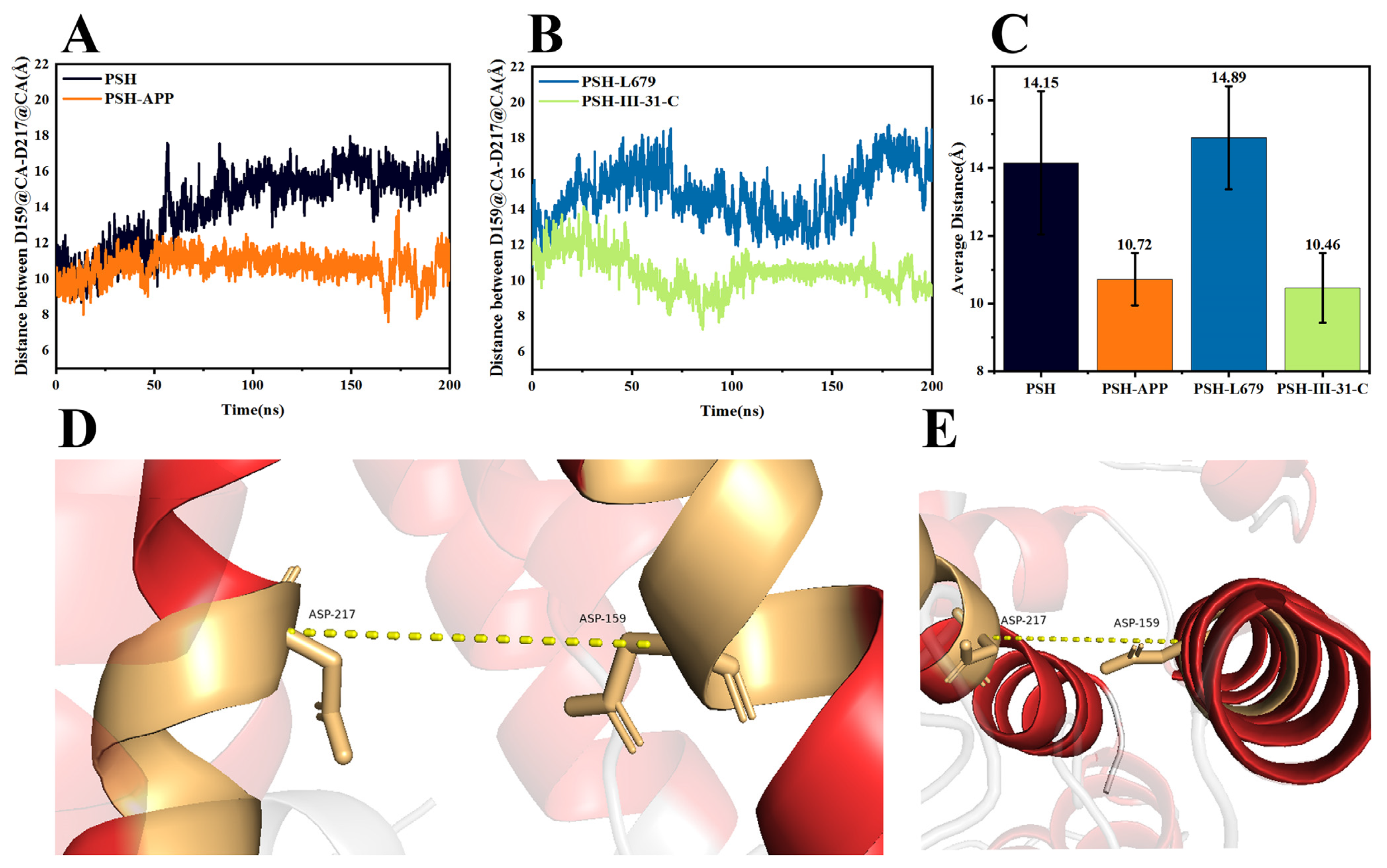
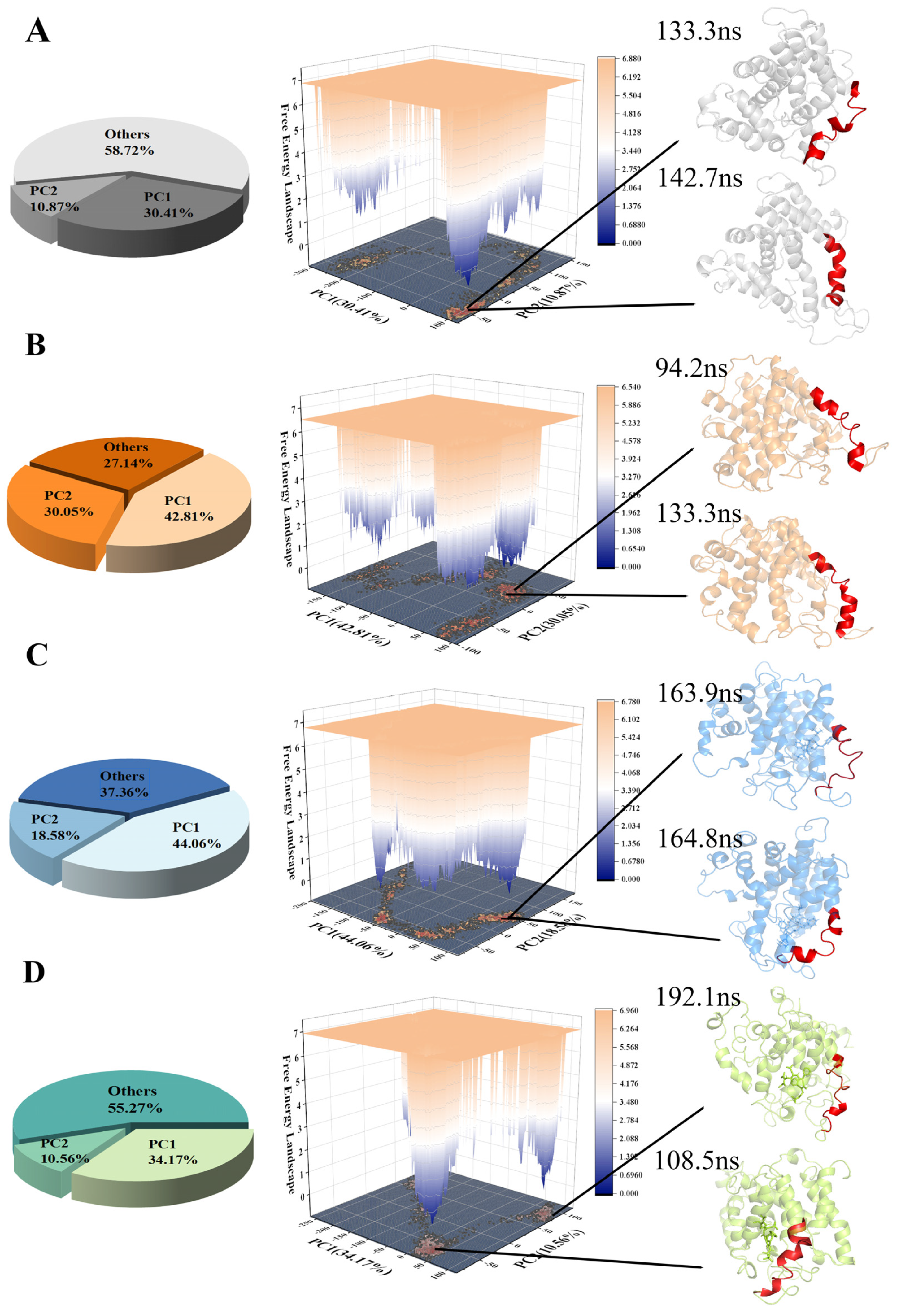
2.3. Comparison of the Conformational Changes of the Four Systems
2.4. Protein Network Analysis
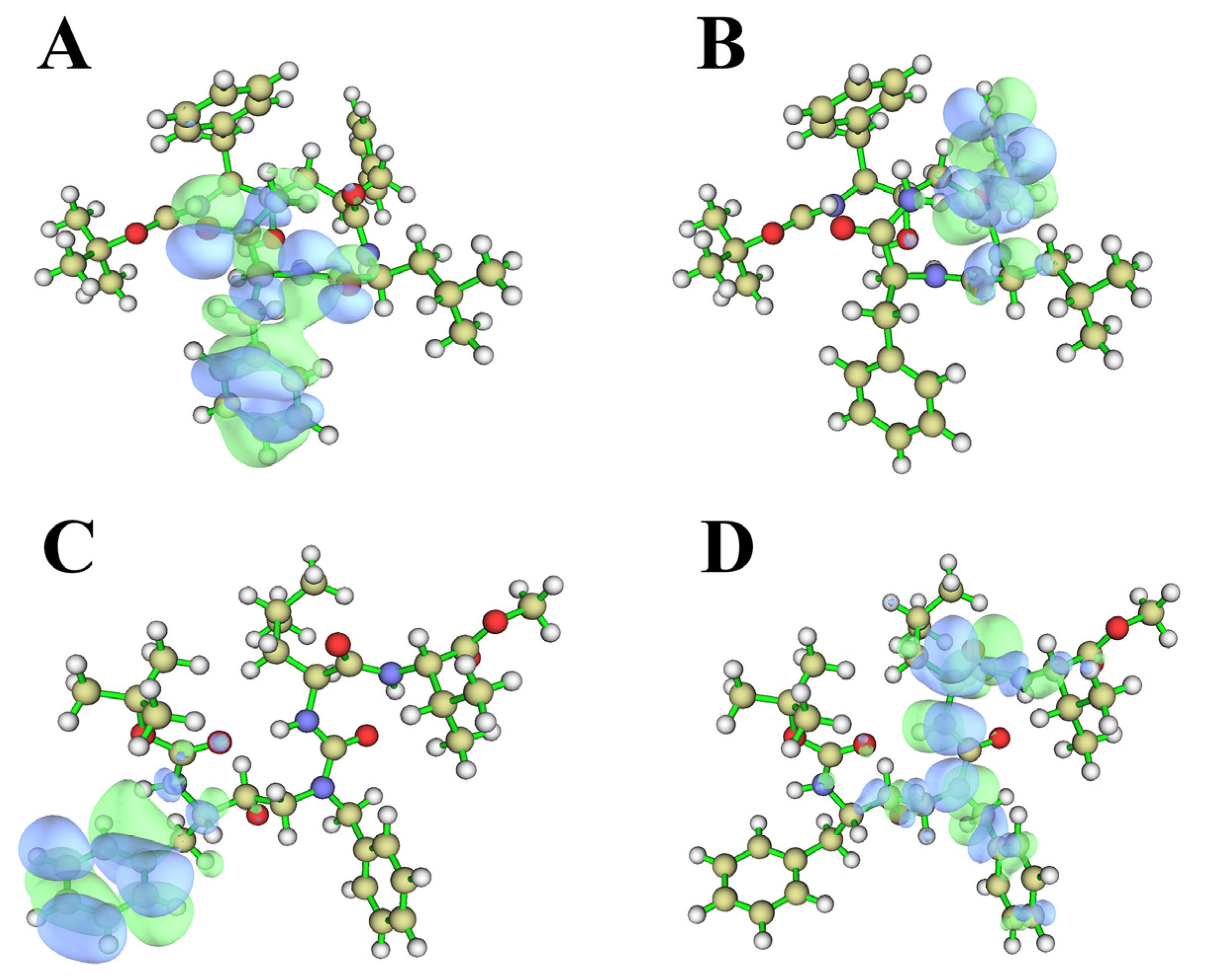
2.5. Quantum Chemical Calculation
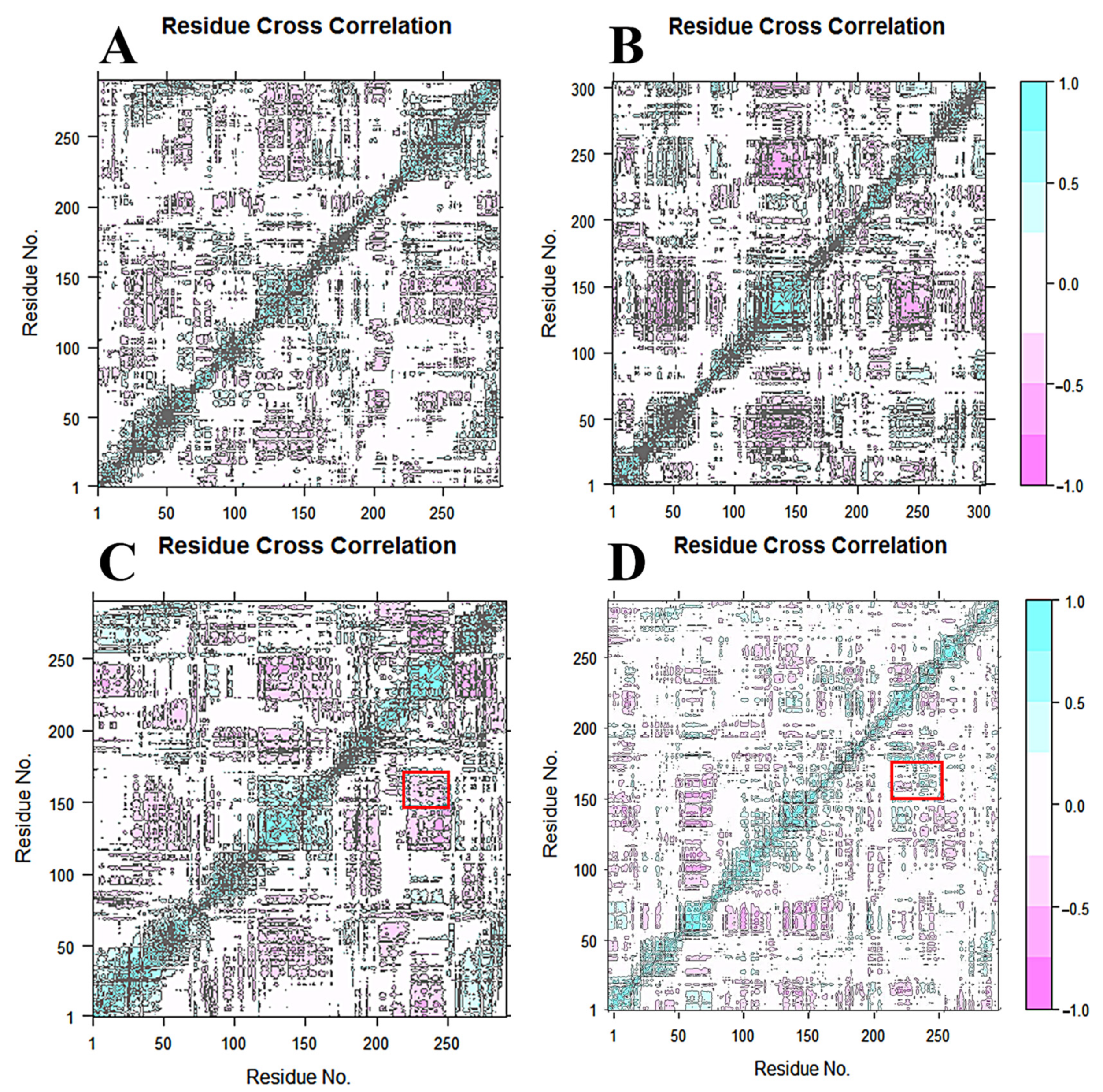
2.6. Dynamical Cross-Correlation Matrix and Principle Component Analysis
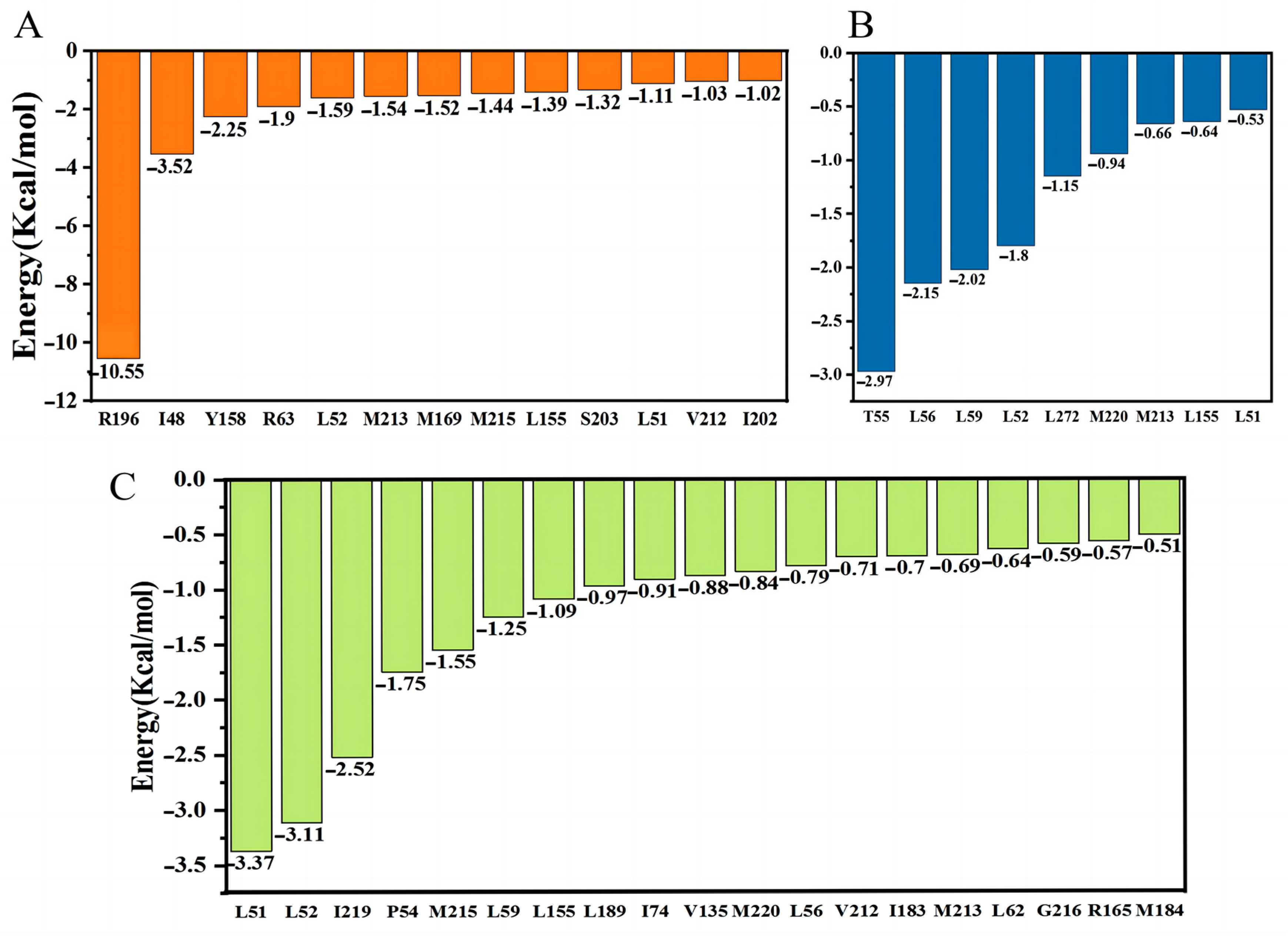
2.7. MM-PBSA Study
3. Materials and Methods
3.1. System Preparation
3.2. Molecular Dynamics Simulations
3.3. Trajectory Analysis
3.4. Protein Structure Network Analysis
3.5. Quantum Chemical Calculation
3.6. MM-PBSA Calculations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Bai, X.C.; Yan, C.; Yang, G.; Lu, P.; Ma, D.; Sun, L.; Zhou, R.; Scheres, S.H.; Shi, Y. An atomic structure of human γ-secretase. Nature 2015, 525, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhou, R.; Guo, X.; Yan, C.; Lei, J.; Shi, Y. Structural basis of γ-secretase inhibition and modulation by small molecule drugs. Cell 2021, 184, 521–533.e14. [Google Scholar] [CrossRef] [PubMed]
- Miranda, A.; Montiel, E.; Ulrich, H.; Paz, C. Selective Secretase Targeting for Alzheimer’s Disease Therapy. J. Alzheimer’s Dis. 2021, 81, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhou, R.; Yang, G.; Shi, Y. Analysis of 138 pathogenic mutations in presenilin-1 on the in vitro production of Aβ42 and Aβ40 peptides by γ-secretase. Proc. Natl. Acad. Sci. USA 2017, 114, E476–E485. [Google Scholar] [CrossRef] [PubMed]
- Güner, G.; Lichtenthaler, S.F. The substrate repertoire of γ-secretase/presenilin. Semin. Cell Dev. Biol. 2020, 105, 27–42. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, R.J.; Wong, P.C. Amyloid precursor protein processing and Alzheimer’s disease. Annu. Rev. Neurosci. 2011, 34, 185–204. [Google Scholar] [CrossRef] [PubMed]
- Reiss, A.B.; Arain, H.A.; Stecker, M.M.; Siegart, N.M.; Kasselman, L.J. Amyloid toxicity in Alzheimer’s disease. Rev. Neurosci. 2018, 29, 613–627. [Google Scholar] [CrossRef]
- Wang, H.; Zang, C.; Liu, X.S.; Aster, J.C. The role of Notch receptors in transcriptional regulation. J. Cell. Physiol. 2015, 230, 982–988. [Google Scholar] [CrossRef]
- Bustos, V.; Pulina, M.V.; Ledo, J. Amyloidogenic and anti-amyloidogenic properties of presenilin 1. Adv. Pharmacol. 2021, 90, 239–251. [Google Scholar] [CrossRef]
- Ledo, J.H.; Liebmann, T.; Zhang, R.; Chang, J.C.; Azevedo, E.P.; Wong, E.; Silva, H.M.; Troyanskaya, O.G.; Bustos, V.; Greengard, P. Presenilin 1 phosphorylation regulates amyloid-β degradation by microglia. Mol. Psychiatry 2021, 26, 5620–5635. [Google Scholar] [CrossRef]
- Fan, C.; Huang, X.; Song, H.; Jia, J. Presenilin 1 intronic polymorphism in sporadic Alzheimer’s disease in a Northern Chinese population. Neuroreport 2020, 31, 37–40. [Google Scholar] [CrossRef]
- Li, Y.; Bohm, C.; Dodd, R.; Chen, F.; Qamar, S.; Schmitt-Ulms, G.; Fraser, P.E.; St George-Hyslop, P.H. Structural biology of presenilin 1 complexes. Mol. Neurodegener. 2014, 9, 59. [Google Scholar] [CrossRef]
- Stanga, S.; Vrancx, C.; Tasiaux, B.; Marinangeli, C.; Karlström, H.; Kienlen-Campard, P. Specificity of presenilin-1- and presenilin-2-dependent γ-secretases towards substrate processing. J. Cell. Mol. Med. 2018, 22, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Yurrita, M.M.; Fernández-Muñoz, B.; Del Castillo, G.; Martín-Villar, E.; Renart, J.; Quintanilla, M. Podoplanin is a substrate of presenilin-1/γ-secretase. Int. J. Biochem. Cell Biol. 2014, 46, 68–75. [Google Scholar] [CrossRef] [PubMed]
- De Strooper, B.; Saftig, P.; Craessaerts, K.; Vanderstichele, H.; Guhde, G.; Annaert, W.; Von Figura, K.; Van Leuven, F. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature 1998, 391, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.S.; Xia, W.; Ostaszewski, B.L.; Diehl, T.S.; Kimberly, W.T.; Selkoe, D.J. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature 1999, 398, 513–517. [Google Scholar] [CrossRef] [PubMed]
- De Strooper, B. Nicastrin: Gatekeeper of the gamma-secretase complex. Cell 2005, 122, 318–320. [Google Scholar] [CrossRef]
- Urban, S. Nicastrin guards Alzheimer’s gate. Proc. Natl. Acad. Sci. USA 2016, 113, 1112–1114. [Google Scholar] [CrossRef]
- De Strooper, B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active gamma-Secretase complex. Neuron 2003, 38, 9–12. [Google Scholar] [CrossRef]
- Kimberly, W.T.; Wolfe, M.S. Identity and function of gamma-secretase. J. Neurosci. Res. 2003, 74, 353–360. [Google Scholar] [CrossRef]
- Dehury, B.; Kepp, K.P. Membrane dynamics of γ-secretase with the anterior pharynx-defective 1B subunit. J. Cell. Biochem. 2021, 122, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Zeng, L.; Li, T.; Meyer, M.A.; Cui, M.Z.; Xu, X. Nicastrin is required for amyloid precursor protein (APP) but not Notch processing, while anterior pharynx-defective 1 is dispensable for processing of both APP and Notch. J. Neurochem. 2016, 136, 1246–1258. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Jeong, Y.H.; Kim, H.M.; Park, H.Y.; Yoon, D.; Kim, D.H.; Saeki, S.; Moon, S.J.; Kang, M.J. Presenilin enhancer-2 (PSENEN), a component of the gamma-secretase complex, is involved in adipocyte differentiation. Domest. Anim. Endocrinol. 2009, 37, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Andreoli, V.; Trecroci, F.; La Russa, A.; Cittadella, R.; Liguori, M.; Spadafora, P.; Caracciolo, M.; Di Palma, G.; Colica, C.; Gambardella, A.; et al. Presenilin enhancer-2 gene: Identification of a novel promoter mutation in a patient with early-onset familial Alzheimer’s disease. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2011, 7, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Liu, Z.; Ilagan, M.X.; Kopan, R. Gamma-secretase composed of PS1/Pen2/Aph1a can cleave notch and amyloid precursor protein in the absence of nicastrin. J. Neurosci. Off. J. Soc. Neurosci. 2010, 30, 1648–1656. [Google Scholar] [CrossRef]
- Dang, S.; Wu, S.; Wang, J.; Li, H.; Huang, M.; He, W.; Li, Y.M.; Wong, C.C.; Shi, Y. Cleavage of amyloid precursor protein by an archaeal presenilin homologue PSH. Proc. Natl. Acad. Sci. USA 2015, 112, 3344–3349. [Google Scholar] [CrossRef]
- Esler, W.P.; Kimberly, W.T.; Ostaszewski, B.L.; Ye, W.; Diehl, T.S.; Selkoe, D.J.; Wolfe, M.S. Activity-dependent isolation of the presenilin- gamma -secretase complex reveals nicastrin and a gamma substrate. Proc. Natl. Acad. Sci. USA 2002, 99, 2720–2725. [Google Scholar] [CrossRef]
- Shearman, M.S.; Beher, D.; Clarke, E.E.; Lewis, H.D.; Harrison, T.; Hunt, P.; Nadin, A.; Smith, A.L.; Stevenson, G.; Castro, J.L. L-685,458, an aspartyl protease transition state mimic, is a potent inhibitor of amyloid beta-protein precursor gamma-secretase activity. Biochemistry 2000, 39, 8698–8704. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Xie, X.; Yan, X.; Wang, Z.; Zhou, H.; Diao, W.; Zhou, W.; Long, J.; Shen, Y. Open-closed motion of Mint2 regulates APP metabolism. J. Mol. Cell Biol. 2013, 5, 48–56. [Google Scholar] [CrossRef]
- Accelrys Software Inc. Discovery Studio Visualizer v21.1.0. Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 20 November 2020).
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scaknabu, G.; Baronw, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Revision, A.02; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Case, D.A.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; Izadi, S.; Janowski, P.; et al. AMBER; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.C.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Vangunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Davidchack, R.L.; Handel, R.; Tretyakov, M.V. Langevin thermostat for rigid body dynamics. J. Chem. Phys. 2009, 130, 234101. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. 3rd PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Cheatham, T.E., 3rd; Srinivasan, J.; Case, D.A.; Kollman, P.A. Molecular dynamics and continuum solvent studies of the stability of polyG-polyC and polyA-polyT DNA duplexes in solution. J. Biomol. Struct. Dyn. 1998, 16, 265–280. [Google Scholar] [CrossRef] [PubMed]
- King, E.; Aitchison, E.; Li, H.; Luo, R. Recent Developments in Free Energy Calculations for Drug Discovery. Front. Mol. Biosci. 2021, 8, 712085. [Google Scholar] [CrossRef] [PubMed]



| System | PSH-APP | PSH-L679 | PSH-III-31-C |
|---|---|---|---|
| ΔEvdW | −92.80 ± 2.11 | −70.08 ± 0.42 | −90.25 ± 0.43 |
| ΔEele | −177.48 ± 4.53 | −30.80 ± 0.76 | −20.07 ± 0.40 |
| ΔGsolv | 221.95 ± 5.44 | 69.09 ± 0.78 | 45.31 ± 0.64 |
| ΔGgas | −270.27 ± 6.47 | −100.88 ± 0.90 | −110.33 ± 0.68 |
| ΔGtotal | −48.32 ± 1.35 | −31.78 ± 0.54 | −65.02 ± 0.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Liu, K.; Ma, Y.; Han, W. Probing the Mechanisms of Inhibitors Binding to Presenilin Homologue Using Molecular Dynamics Simulations. Molecules 2023, 28, 2076. https://doi.org/10.3390/molecules28052076
Wang M, Liu K, Ma Y, Han W. Probing the Mechanisms of Inhibitors Binding to Presenilin Homologue Using Molecular Dynamics Simulations. Molecules. 2023; 28(5):2076. https://doi.org/10.3390/molecules28052076
Chicago/Turabian StyleWang, Min, Kaifeng Liu, Yingying Ma, and Weiwei Han. 2023. "Probing the Mechanisms of Inhibitors Binding to Presenilin Homologue Using Molecular Dynamics Simulations" Molecules 28, no. 5: 2076. https://doi.org/10.3390/molecules28052076
APA StyleWang, M., Liu, K., Ma, Y., & Han, W. (2023). Probing the Mechanisms of Inhibitors Binding to Presenilin Homologue Using Molecular Dynamics Simulations. Molecules, 28(5), 2076. https://doi.org/10.3390/molecules28052076








