Toward Achieving Rapid Estimation of Vitamin C in Citrus Peels by NIR Spectra Coupled with a Linear Algorithm
Abstract
1. Introduction
2. Results and Discussion
2.1. Statistical Values of Vitamin C
2.2. Spectral Profiles of Samples
2.3. Predicting Vitamin C Using Full Wavelength
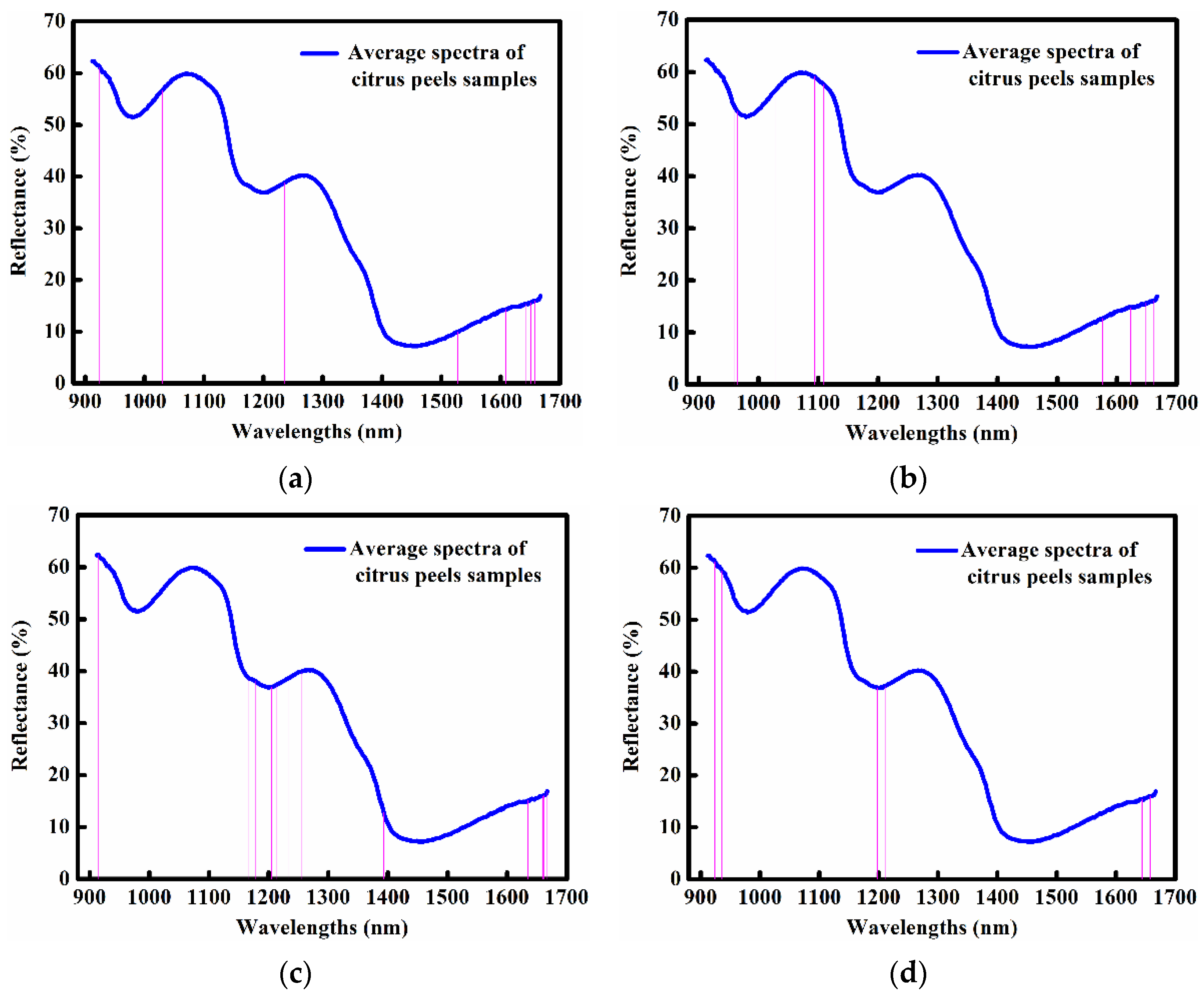
2.4. Optimal Wavelengths Selected by Four Different Methods
2.5. Predicting Vitamin C Using Optimal Wavelengths
2.6. F-Test and T-Test Analysis
2.7. Independent External Validation of Best Optimized Model
3. Materials and Methods
3.1. Preparation of Citrus Peel Samples
3.2. Spectral Collection and Preprocessing
3.3. Measurement of Vitamin C
3.4. Quantitative Relationship Establishment between Spectra and Vitamin C
3.5. Optimal Wavelength Selection and Model Optimization
3.6. Statistical Two-Sample Analysis
3.7. External Validation of Model
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- El-OtmaniA, M.; Ait-Oubahou, A.; Zacarías, L. Citrus spp.: Orange, mandarin, tangerine, clementine, grapefruit, pomelo, lemon and lime. In Postharvest Biology and Technology of Tropical and Subtropical Fruits; Yahia, E.M., Ed.; Woodhead Publishing Limited: Cambridge, UK, 2011; pp. 437–516e. [Google Scholar]
- Li, B.B.; Smith, B.; Hossain, M.M. Extraction of phenolics from citrus peels I. Solvent extraction method. Sep. Purif. Technol. 2006, 48, 182–188. [Google Scholar] [CrossRef]
- Rafiq, S.; Kaul, R.; Sofi, S.A.; Bashir, N.; Nazir, F.; Nayik, G.A. Citrus peel as a source of functional ingredient: A review. J. Saudi Soc. Agr. Sci. 2018, 17, 351–358. [Google Scholar] [CrossRef]
- Mamma, D.; Christakopoulos, P. Citrus peels: An excellent raw material for the bioconversion into value-added products. Tree Forest. Sci. Biotechnol. 2008, 2, 83–93. [Google Scholar]
- Nair, S.A.; SR, R.K.; Nair, A.S.; Baby, S. Citrus peels prevent cancer. Phytomedicine 2018, 50, 231–237. [Google Scholar] [CrossRef]
- Rehman, Z.U. Citrus peel extract—A natural source of antioxidant. Food Chem. 2006, 99, 450–454. [Google Scholar] [CrossRef]
- Güzel, M.; Akpınar, Ö. Production and characterization of bacterial cellulose from citrus peels. Waste Biomass. Valor. 2019, 10, 2165–2175. [Google Scholar] [CrossRef]
- Dhillon, S.S.; Gill, R.K.; Gill, S.S.; Singh, M. Studies on the utilization of citrus peel for pectinase production using fungus Aspergillus niger. Int. J. Environ. Stud. 2004, 61, 199–210. [Google Scholar] [CrossRef]
- Indulekha, J.; Gokul Siddarth, M.S.; Kalaichelvi, P.; Arunagiri, A. Characterization of citrus peels for bioethanol production. In Materials, Energy and Environment Engineering; Mohan, B.R., Srinikethan, G., Meikap, B., Eds.; Springer: Singapore, 2017; pp. 3–12. [Google Scholar]
- Sir Elkhatim, K.A.; Elagib, R.A.A.; Hassan, A.B. Content of phenolic compounds and vitamin C and antioxidant activity in wasted parts of Sudanese citrus fruits. Food Sci. Nutr. 2018, 6, 1214–1219. [Google Scholar] [CrossRef]
- Mditshwa, A.; Magwaza, L.S.; Tesfay, S.Z.; Opara, U.L. Postharvest factors affecting vitamin C content of citrus fruits: A review. Sci. Hortic. 2017, 218, 95–104. [Google Scholar] [CrossRef]
- Podmore, I.D.; Griffiths, H.R.; Herbert, K.E.; Mistry, N.; Mistry, P.; Lunec, J. Vitamin C exhibits pro-oxidant properties. Nature 1998, 392, 559. [Google Scholar] [CrossRef]
- Miller, P.; Ebert, B. Vitamin C regulates stem cells and cancer. Nature 2017, 549, 462–464. [Google Scholar] [CrossRef]
- Bozkir, H.; Tekgül, Y.; Erten, E.S. Effects of tray drying, vacuum infrared drying, and vacuum microwave drying techniques on quality characteristics and aroma profile of orange peels. J. Food Process Eng. 2021, 44, e13611. [Google Scholar] [CrossRef]
- Suri, S.; Singh, A.; Nema, P.K. Recent advances in valorization of citrus fruits processing waste: A way forward towards environmental sustainability. Food Sci. Biotechnol. 2021, 30, 1601–1626. [Google Scholar] [CrossRef]
- Nielsen, S.S. Vitamin C determination by indophenol method. In Food Analysis Laboratory Manual; Nielsen, S.S., Ed.; Springer: Boston, MA, USA, 2010; pp. 55–60. [Google Scholar]
- Card, D.J. Methods for assessment of vitamin C. In Laboratory Assessment of Vitamin Status; Harrington, D., Ed.; Elsevier: London, UK, 2019; pp. 301–316. [Google Scholar]
- Yusuf, E.; Tkacz, K.; Turkiewicz, I.P.; Wojdyło, A.; Nowicka, P. Analysis of chemical compounds’ content in different varieties of carrots, including qualification and quantification of sugars, organic acids, minerals, and bioactive compounds by UPLC. Eur. Food Res. Technol. 2021, 247, 3053–3062. [Google Scholar] [CrossRef]
- El-Hawiet, A.; Elessawy, F.M.; El Demellawy, M.A.; El-Yazbi, A.F. Green fast and simple UPLC-ESI-MRM/MS method for determination of trace water-soluble vitamins in honey: Greenness assessment using GAPI and analytical eco-scale. Microchem. J. 2022, 181, 107625. [Google Scholar] [CrossRef]
- Pisoschi, A.M.; Pop, A.; Serban, A.I.; Fafaneata, C. Electrochemical methods for ascorbic acid determination. Electrochim. Acta. 2014, 121, 443–460. [Google Scholar] [CrossRef]
- Antonelli, M.L.; D’Ascenzo, G.; Laganà, A.; Pusceddu, P. Food analyses: A new calorimetric method for ascorbic acid (vitamin C) determination. Talanta 2002, 58, 961–967. [Google Scholar] [CrossRef]
- Pisoschi, A.M.; Danet, A.F.; Kalinowski, S. Ascorbic acid determination in commercial fruit juice samples by cyclic voltammetry. J. Autom. Methods Manag. Chem. 2008, 2008, 937651. [Google Scholar] [CrossRef]
- Shekhovtsova, T.N.; Muginova, S.V.; Luchinina, J.A.; Galimova, A.Z. Enzymatic methods in food analysis: Determination of ascorbic acid. Anal. Chim. Acta 2006, 573–574, 125–132. [Google Scholar] [CrossRef]
- Herbig, A.L.; Renard, C.M.G.C. Factors that impact the stability of vitamin C at intermediate temperatures in a food matrix. Food Chem. 2017, 220, 444–451. [Google Scholar] [CrossRef]
- He, H.J.; Wang, Y.; Zhang, M.; Wang, Y.; Ou, X.; Guo, J. Rapid determination of reducing sugar content in sweet potatoes using NIR spectra. J. Food Compos. Anal. 2022, 111, 104641. [Google Scholar] [CrossRef]
- Kutsanedzie, F.Y.H.; Guo, Z.; Chen, Q. Advances in nondestructive methods for meat quality and safety monitoring. Food Rev. Int. 2019, 35, 536–562. [Google Scholar] [CrossRef]
- Chen, Q.; Lin, H.; Zhao, J. Near-Infrared Spectroscopy Technology in Food. In Advanced Nondestructive Detection Technologies in Food; Chen, Q., Lin, H., Zhao, J., Eds.; Springer: Singapore, 2021; pp. 23–58. [Google Scholar]
- Miseo, E.V.; Meyer, F.; Ryan, J. Portable Near-Infrared Spectroscopy in Food Analysis. In Portable Spectroscopy and Spectrometry; Crocombe, R., Leary, P., Kammrath, B., Eds.; Wiley: Hoboken, NJ, USA, 2021; pp. 325–345. [Google Scholar]
- He, H.J.; Wang, Y.; Ou, X.; Ma, H.; Liu, H.; Yan, J. Rapid determination of chemical compositions in chicken flesh by mining hyperspectral data. J. Food Compos. Anal. 2023, 116, 105069. [Google Scholar] [CrossRef]
- Tirado-Kulieva, V.A.; Hernández-Martínez, E.; Suomela, J.-P. Non-destructive assessment of vitamin C in foods: A review of the main fndings and limitations of vibrational spectroscopic techniques. Eur. Food Res. Technol. 2022, 248, 2185–2195. [Google Scholar] [CrossRef]
- Valentini, M.; dos Santos, G.B.; Vieira, B.M. Multiple linear regression analysis (MLR) applied for modeling a new WQI equation for monitoring the water quality of Mirim Lagoon, in the state of Rio Grande do Sul-Brazil. SN Appl. Sci. 2021, 3, 70. [Google Scholar] [CrossRef]
- Albergaria, J.T.; Martins, F.G.; Alvim-Ferraz, M.C.M.; Delerue-Matos, C. Multiple linear regression and artificial neural networks to predict time and efficiency of soil vapor extraction. Water Air Soil Poll. 2014, 225, 2058. [Google Scholar] [CrossRef]
- Mohammadi-Moghaddam, T.; Razavi, S.; Sazgarnia, A.; Taghizadeh, M. Predicting the moisture content and textural characteristics of roasted pistachio kernels using Vis/NIR reflectance spectroscopy and PLSR analysis. J. Food Meas. Charact. 2018, 12, 346–355. [Google Scholar] [CrossRef]
- Kokalj, M.; Rihtarič, M.; Kreft, S. Commonly applied smoothing of IR spectra showed unappropriate for the identification of plant leaf samples. Chemom. Intell. Lab. Syst. 2011, 108, 154–161. [Google Scholar] [CrossRef]
- Rinnan, Å.; van den Berg, F.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC-Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Peng, J.; Peng, S.; Jiang, A.; Wei, J.; Li, C.; Tan, J. Asymmetric least squares for multiple spectra baseline correction. Anal. Chim. Acta 2010, 683, 63–68. [Google Scholar] [CrossRef]
- Roy, I.G. On computing first and second order derivative spectra. J. Comput. Phys. 2015, 295, 307–321. [Google Scholar] [CrossRef]
- Gautam, R.; Vanga, S.; Ariese, F.; Umapathy, S. Review of multidimensional data processing approaches for Raman and infrared spectroscopy. EPJ Techn. Instrum. 2015, 2, 8. [Google Scholar] [CrossRef]
- Jiao, Y.; Li, Z.; Chen, X.; Fei, S. Preprocessing methods for near-infrared spectrum calibration. J. Chemom. 2020, 34, e3306. [Google Scholar] [CrossRef]
- Oliveri, P.; Malegori, C.; Simonetti, R.; Casale, M. The Impact of Signal Pre-Processing on the Final Interpretation of Analytical Outcomes—A tutorial. Anal. Chim. Acta 2019, 1058, 9–17. [Google Scholar] [CrossRef]
- AOAC. Method 967.21: Vitamin C (total) in food. In Official Methods of Analysis of the Association of Official Analytical Chemists; AOAC International: Washington, DC, USA, 2007; pp. 24–25. [Google Scholar]
- Gatius, F.; Miralbés, C.; David, C.; Puy, J. Comparison of CCA and PLS to explore and model NIR data. Chemom. Intell. Lab. Syst. 2017, 164, 76–82. [Google Scholar] [CrossRef]
- Li, B.; Martin, E.; Morris, J. Latent variable selection in partial least squares modelling. IFAC Proc. Vol. 2001, 34, 463–468. [Google Scholar] [CrossRef]
- Gowen, A.A.; Downey, G.; Esquerre, C.; O’Donnell, C.P. Preventing over-fitting in PLS calibration models of near-infrared (NIR) spectroscopy data using regression coefficients. J. Chemometr. 2011, 25, 375–381. [Google Scholar] [CrossRef]
- Wang, Y.Y.; He, H.J.; Jiang, S.Q.; Ma, H.J. Nondestructive determination of IMP content in chilled chicken based on hyperspectral data combined with chemometrics. Int. J. Agr. Biol. Eng. 2022, 15, 23–27. [Google Scholar] [CrossRef]
- Kasemsumran, S.; Du, Y.P.; Li, B.Y.; Maruo, K.; Ozaki, Y. Moving window cross validation: A new cross validation method for the selection of a rational number of components in a partial least squares calibration model. Analyst 2006, 131, 529–537. [Google Scholar] [CrossRef]
- Jiang, S.Q.; He, H.J.; Ma, H.J.; Chen, F.S.; Xu, B.C.; Liu, H.; Zhu, M.M.; Kang, Z.L.; Zhao, S.M. Quick assessment of chicken spoilage based on hyperspectral NIR spectra combined with partial least squares regression. Int. J. Agr. Biol. Eng. 2021, 14, 243–250. [Google Scholar] [CrossRef]
- He, H.J.; Wu, D.; Sun, D.W. Nondestructive spectroscopic and imaging techniques for quality evaluation and assessment of fish and fish products. Crit. Rev. Food Sci. 2015, 55, 864–886. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Zhao, J.; Malcolm, J.W.P.; Mel, H.; Mao, H. Variables selection methods in near-infrared spectroscopy. Anal. Chim. Acta 2010, 667, 14–32. [Google Scholar]
- Zhu, Y.D.; He, H.J.; Jiang, S.Q.; Ma, H.J.; Chen, F.S.; Xu, B.C.; Liu, H.; Zhu, M.M.; Zhao, S.M.; Kang, Z.L. Mining hyperspectral data for non-destructive and rapid prediction of nitrite content in ham sausages. Int. J. Agr. Biol. Eng. 2021, 14, 182–187. [Google Scholar] [CrossRef]
- Mehmood, T.; Liland, K.H.; Snipen, L.; Sæbø, S. A review of variable selection methods in Partial Least Squares Regression. Chemom. Intell. Lab. Syst. 2012, 118, 62–69. [Google Scholar] [CrossRef]
- Soares, S.F.C.; Gomes, A.A.; Araujo, M.C.U.; Filho, A.R.G.; Galvão, R.K.H. The successive projections algorithm. TrAC-Trends Anal. Chem. 2013, 42, 84–98. [Google Scholar] [CrossRef]
- Zheng, K.; Li, Q.; Wang, J.; Geng, J.; Cao, P.; Sui, T.; Wang, X.; Du, Y. Stability competitive adaptive reweighted sampling (SCARS) and its applications to multivariate calibration of NIR spectra. Chemom. Intell. Lab. Syst. 2012, 112, 48–54. [Google Scholar] [CrossRef]
- Glatting, G.; Kletting, P.; Reske, S.N.; Hohl, K.; Ring, C. Choosing the informative fit function: Comparison of the Akaike information criterion and the F-test. Med. Phys. 2007, 34, 4285–4292. [Google Scholar] [CrossRef]
- Adusah, A.K.; Brooks, G.P. Type I error inflation of the separate-variances welch t test with very small sample sizes when assumptions are met. J. Mod. Appl. Stat. Meth. 2011, 10, 362–372. [Google Scholar] [CrossRef]
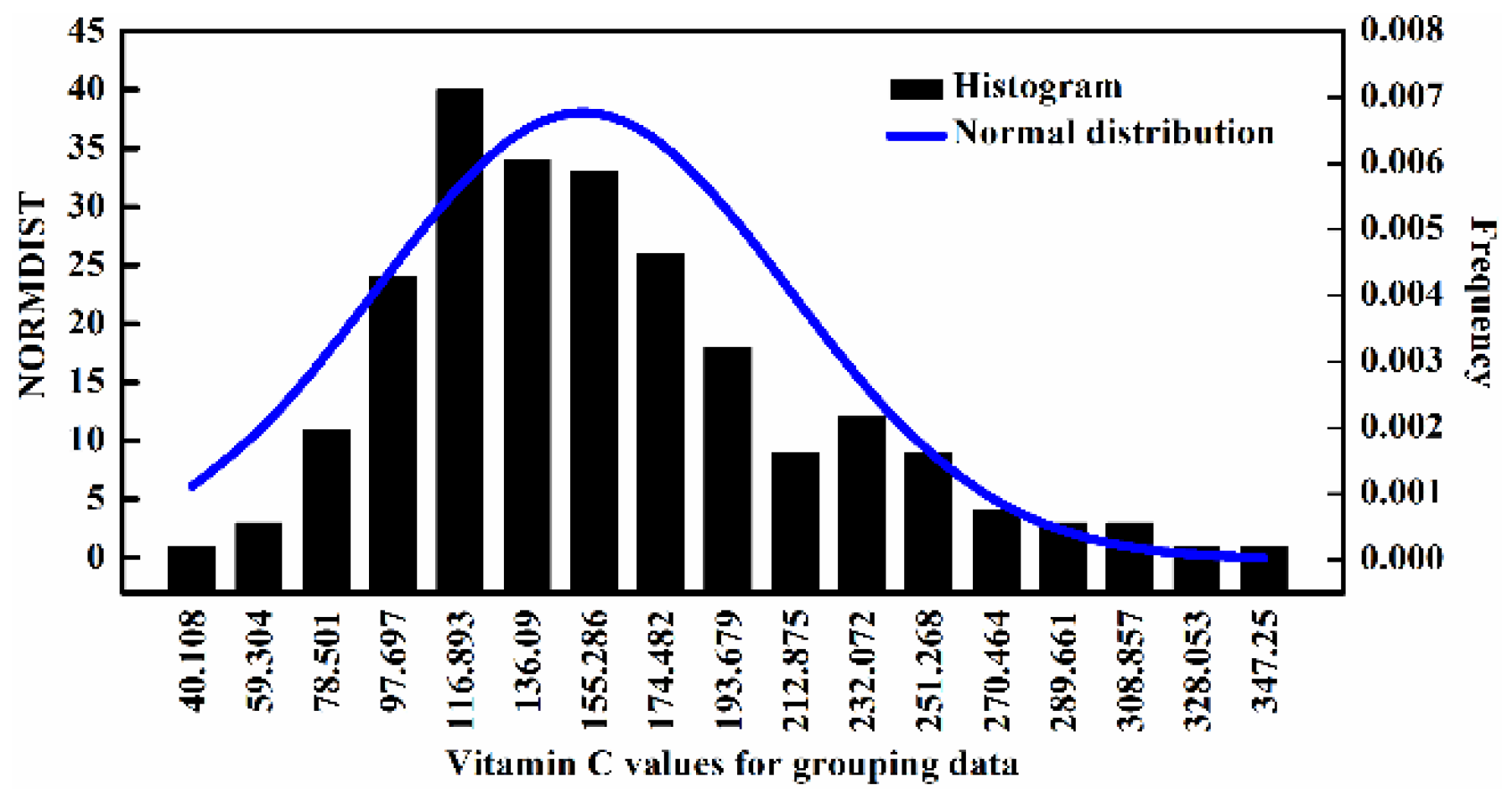
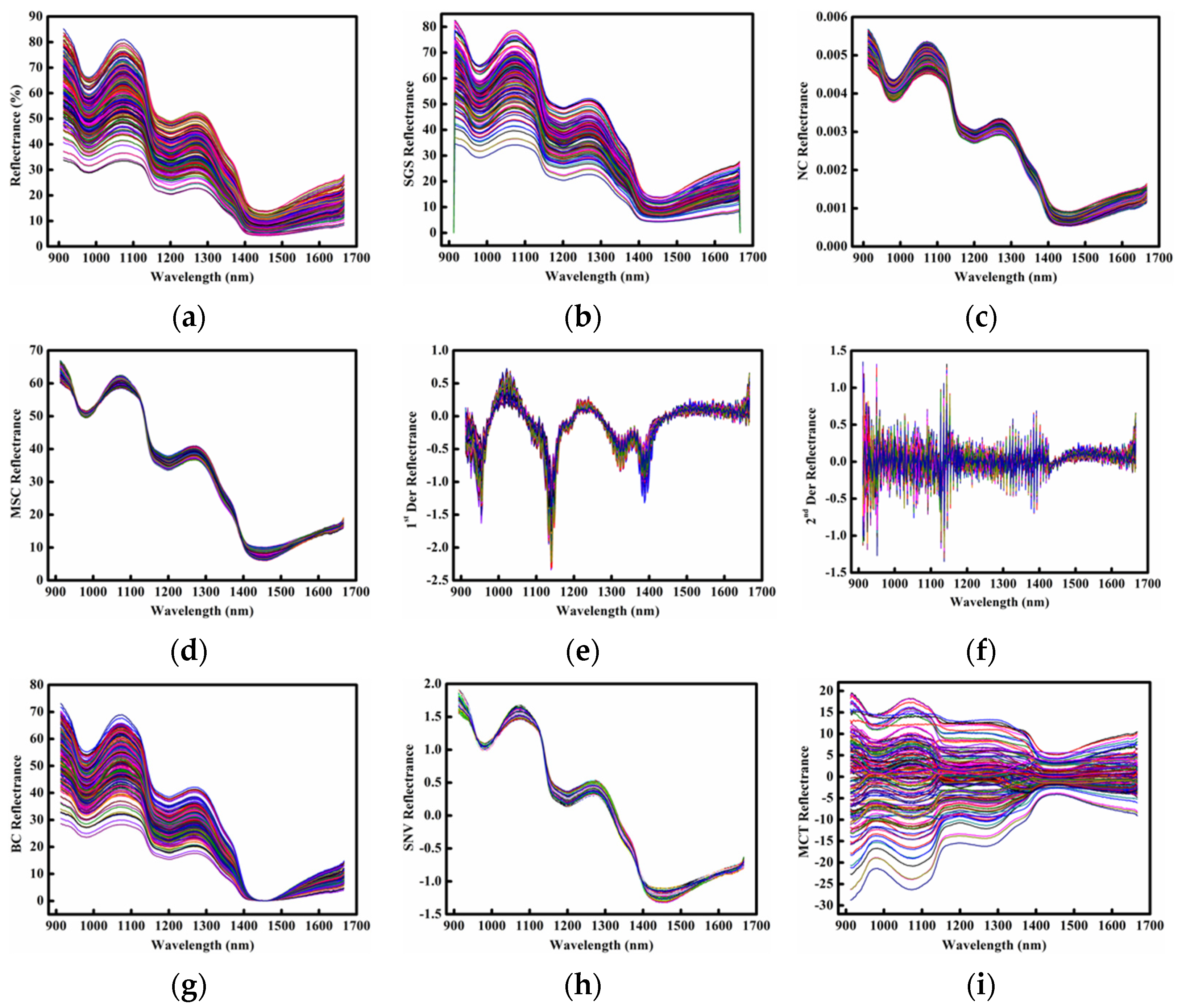

| Sample Set | Number of Sample | Minimum | Maximum | Mean | Standard Deviation |
|---|---|---|---|---|---|
| Calibration set | 166 | 40.108 | 342.413 | 152.379 | 59.085 |
| Prediction set | 83 | 43.304 | 333.110 | 151.582 | 58.106 |
| Spectra | Model | Number of LV | Calibration | Cross-Validation | Prediction | ΔE | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| rC | RMSEC | rCV | RMSECV | rP | RMSEP | RPD | ||||
| RAW | RAW-PLS 1 | 13 | 0.957 | 13.077 | 0.930 | 15.703 | 0.928 | 16.343 | 3.537 | 3.266 |
| SGS | SGS-PLS 2 | 14 | 0.971 | 11.314 | 0.934 | 15.235 | 0.926 | 16.319 | 3.542 | 5.005 |
| NC | NC-PLS 3 | 14 | 0.974 | 10.674 | 0.953 | 13.174 | 0.956 | 13.798 | 4.189 | 3.124 |
| MSC | MSC-PLS 4 | 14 | 0.974 | 10.671 | 0.932 | 15.499 | 0.918 | 16.428 | 3.518 | 5.757 |
| 1st Der | 1st Der-PLS 5 | 8 | 0.944 | 13.170 | 0.889 | 17.907 | 0.877 | 18.114 | 3.191 | 4.944 |
| 2nd Der | 2nd Der-PLS 6 | 5 | 0.915 | 18.949 | 0.892 | 21.566 | 0.881 | 22.335 | 2.588 | 3.386 |
| BC | BC-PLS 7 | 12 | 0.950 | 14.237 | 0.906 | 20.135 | 0.913 | 20.253 | 2.854 | 6.016 |
| SNV | SNV-PLS 8 | 11 | 0.947 | 14.611 | 0.894 | 21.421 | 0.887 | 23.916 | 2.417 | 9.305 |
| MCT | MCT-PLS 9 | 13 | 0.957 | 13.077 | 0.910 | 20.703 | 0.893 | 21.744 | 2.658 | 8.667 |
| Method | Number of Optimal Wavelengths | Specific Wavelengths | Wavelength Reduction |
|---|---|---|---|
| RC | 22 | 915, 924, 938, 950, 997, 1030, 1100, 1102, 1161, 1176, 1236, 1385, 1528, 1596, 1609, 1616, 1623, 1642, 1651, 1655, 1657, 1658 nm | 94% |
| SR | 15 | 915, 927, 960, 965, 1016, 1028, 1094, 1109, 1397, 1576, 1623, 1642, 1648, 1662, 1664 nm | 96% |
| SPA | 20 | 912, 915, 986, 1162, 1167, 1178, 1205, 1209, 1214, 1234, 1255, 1352, 1393, 1635, 1642, 1658,1660, 1662, 1664, 1667 nm | 95% |
| CARS | 11 | 924, 927, 936, 954, 1198, 1211, 1635, 1637, 1642, 1644, 1658 nm | 97% |
| Model | Number of Wavelength | Number of LV | Calibration | Cross-Validation | Prediction | ΔE | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| rC | RMSEC | rCV | RMSECV | rP | RMSEP | RPD | ||||
| RC-NC-PLS 1 | 22 | 9 | 0.936 | 16.316 | 0.908 | 19.887 | 0.901 | 20.354 | 2.855 | 4.038 |
| SR-NC-PLS 2 | 15 | 10 | 0.942 | 15.432 | 0.937 | 16.418 | 0.936 | 16.689 | 3.482 | 1.257 |
| SPA-NC-PLS 3 | 20 | 11 | 0.901 | 20.499 | 0.849 | 25.655 | 0.835 | 26.768 | 2.245 | 6.269 |
| CARS-NC-PLS 4 | 11 | 7 | 0.814 | 24.494 | 0.785 | 25.767 | 0.787 | 25.916 | 2.358 | 1.422 |
| RC-NC-MLR 5 | 22 | - | 0.913 | 19.148 | 0.852 | 25.112 | 0.848 | 26.776 | 2.319 | 7.628 |
| SR-NC-MLR 6 | 15 | - | 0.955 | 13.430 | 0.948 | 14.651 | 0.949 | 14.814 | 4.260 | 1.384 |
| SPA-NC-MLR 7 | 20 | - | 0.910 | 19.447 | 0.855 | 24.865 | 0.829 | 27.326 | 2.346 | 7.879 |
| CARS-NC-MLR 8 | 11 | - | 0.842 | 23.578 | 0.836 | 26.663 | 0.818 | 29.409 | 2.214 | 5.831 |
| Model | Double Sample Analysis | Index | Predicted Value | Measured Value |
|---|---|---|---|---|
| SR-NC-MLR | F-test | average | 149.861 | 150.334 |
| variance | 3478.621 | 3464.791 | ||
| observed value | 83 | 83 | ||
| df | 82 | 82 | ||
| F | 1.004 | |||
| P (F <= f) one-tailed | 0.493 | |||
| F (one-tailed critical value) | 1.441 | |||
| t-test | average | 149.861 | 150.334 | |
| variance | 3478.621 | 3464.791 | ||
| observed value | 83 | 83 | ||
| pooled variance | 3471.706 | |||
| assumed mean difference | 0 | |||
| df | 164 | |||
| t Stat | 0.0517 | |||
| P (T <= t) one-tailed | 0.479 | |||
| t (one-tailed critical value) | 1.654 | |||
| P (T <= t) two-tailed | 0.959 | |||
| t (two-tailed critical value) | 1.975 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Lin, M.; He, H.; Wang, Y.; Wang, J.; Liu, H. Toward Achieving Rapid Estimation of Vitamin C in Citrus Peels by NIR Spectra Coupled with a Linear Algorithm. Molecules 2023, 28, 1681. https://doi.org/10.3390/molecules28041681
Zhang W, Lin M, He H, Wang Y, Wang J, Liu H. Toward Achieving Rapid Estimation of Vitamin C in Citrus Peels by NIR Spectra Coupled with a Linear Algorithm. Molecules. 2023; 28(4):1681. https://doi.org/10.3390/molecules28041681
Chicago/Turabian StyleZhang, Weiqing, Mei Lin, Hongju He, Yuling Wang, Jingru Wang, and Hongjie Liu. 2023. "Toward Achieving Rapid Estimation of Vitamin C in Citrus Peels by NIR Spectra Coupled with a Linear Algorithm" Molecules 28, no. 4: 1681. https://doi.org/10.3390/molecules28041681
APA StyleZhang, W., Lin, M., He, H., Wang, Y., Wang, J., & Liu, H. (2023). Toward Achieving Rapid Estimation of Vitamin C in Citrus Peels by NIR Spectra Coupled with a Linear Algorithm. Molecules, 28(4), 1681. https://doi.org/10.3390/molecules28041681







