Amyloid-like RIP1/RIP3 RHIM Fragments’ Characterization and Application as a Drug Depot
Abstract
1. Introduction
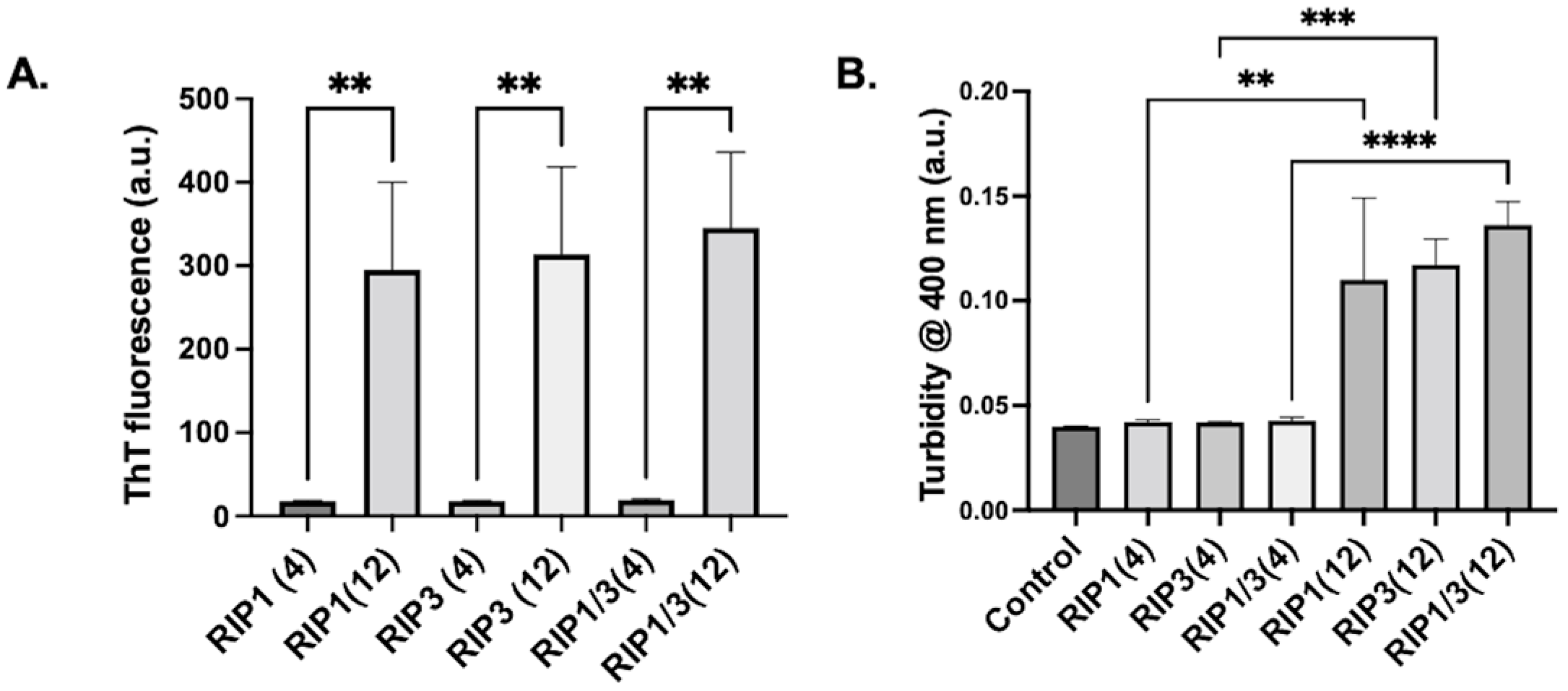
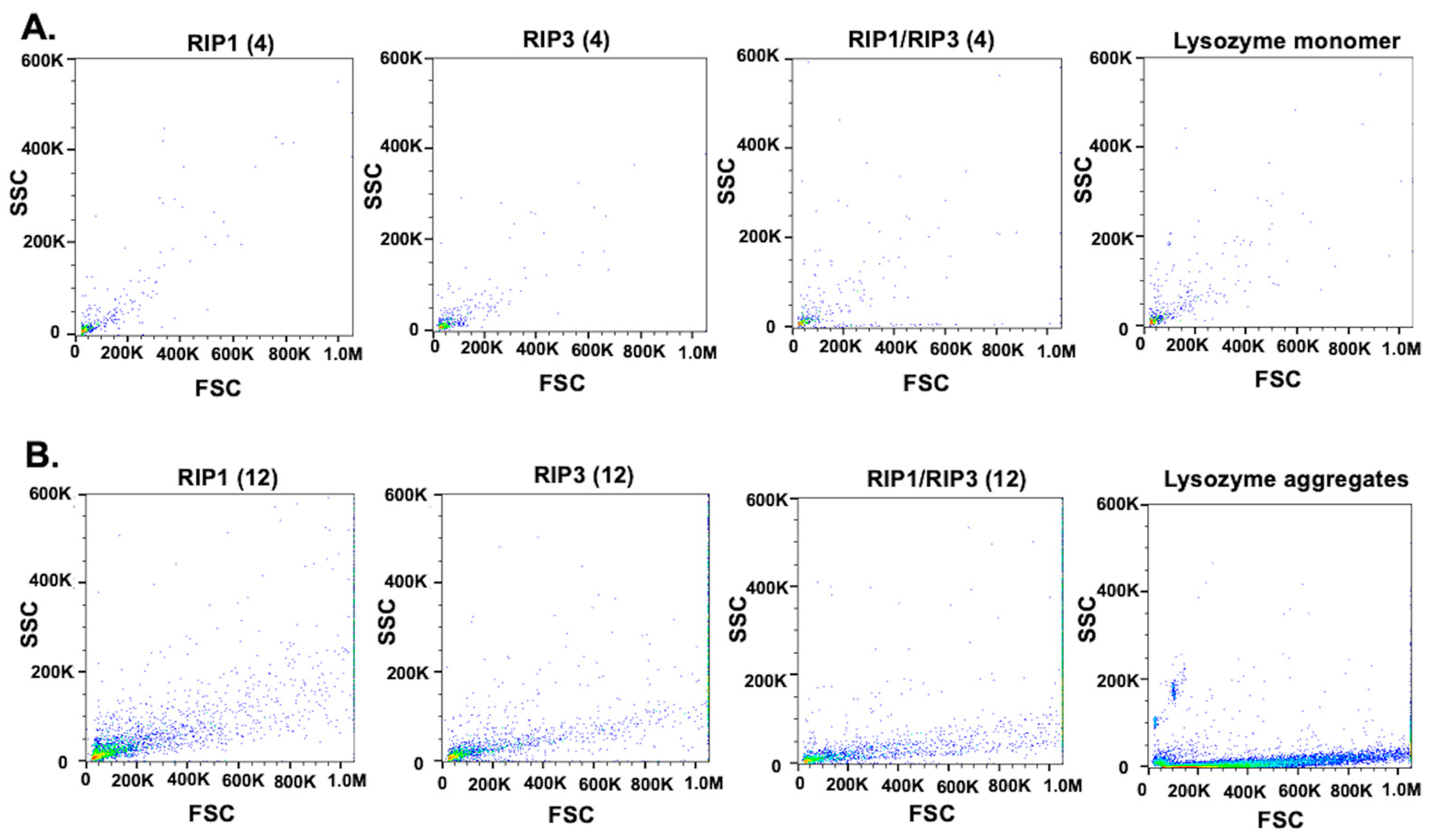
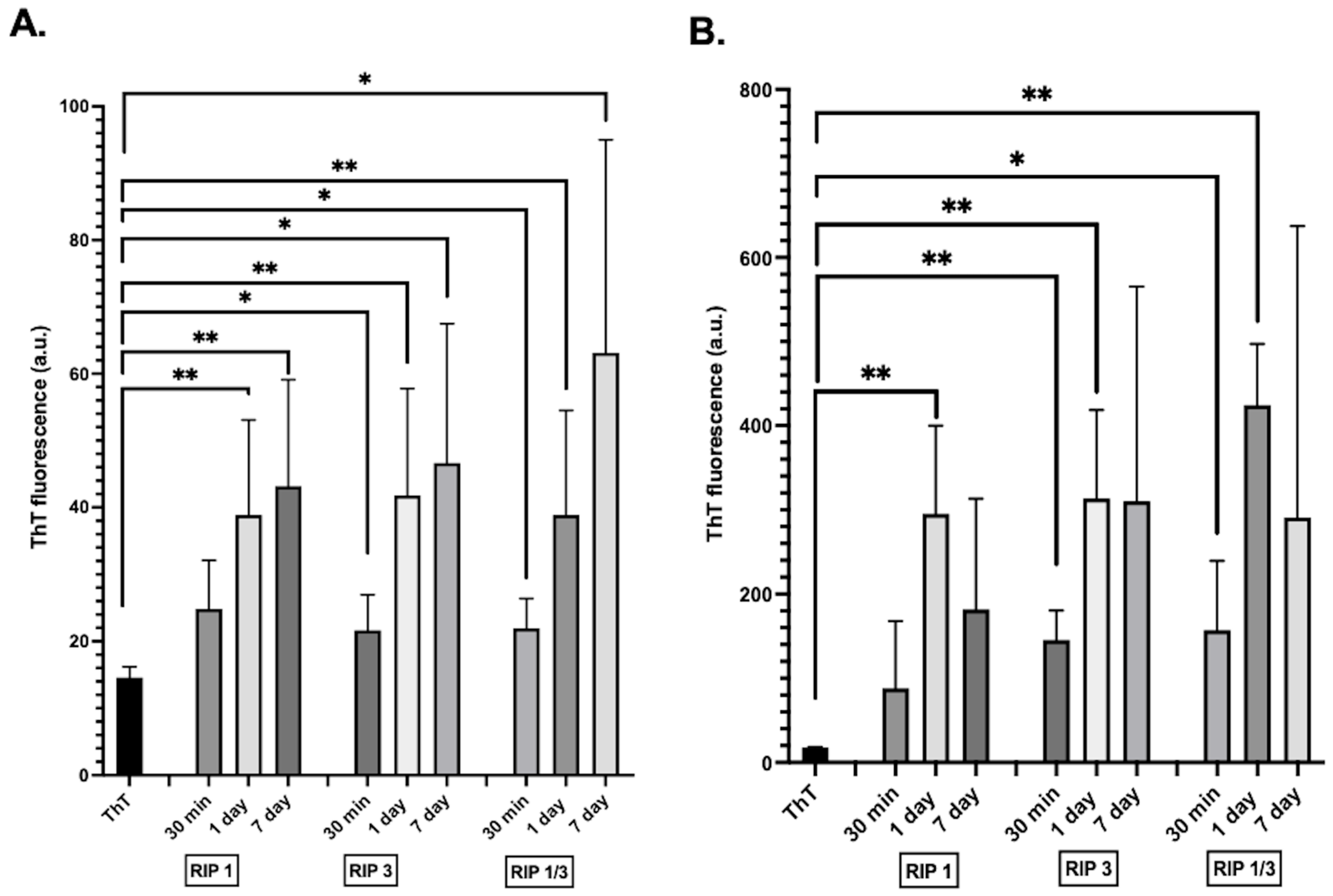
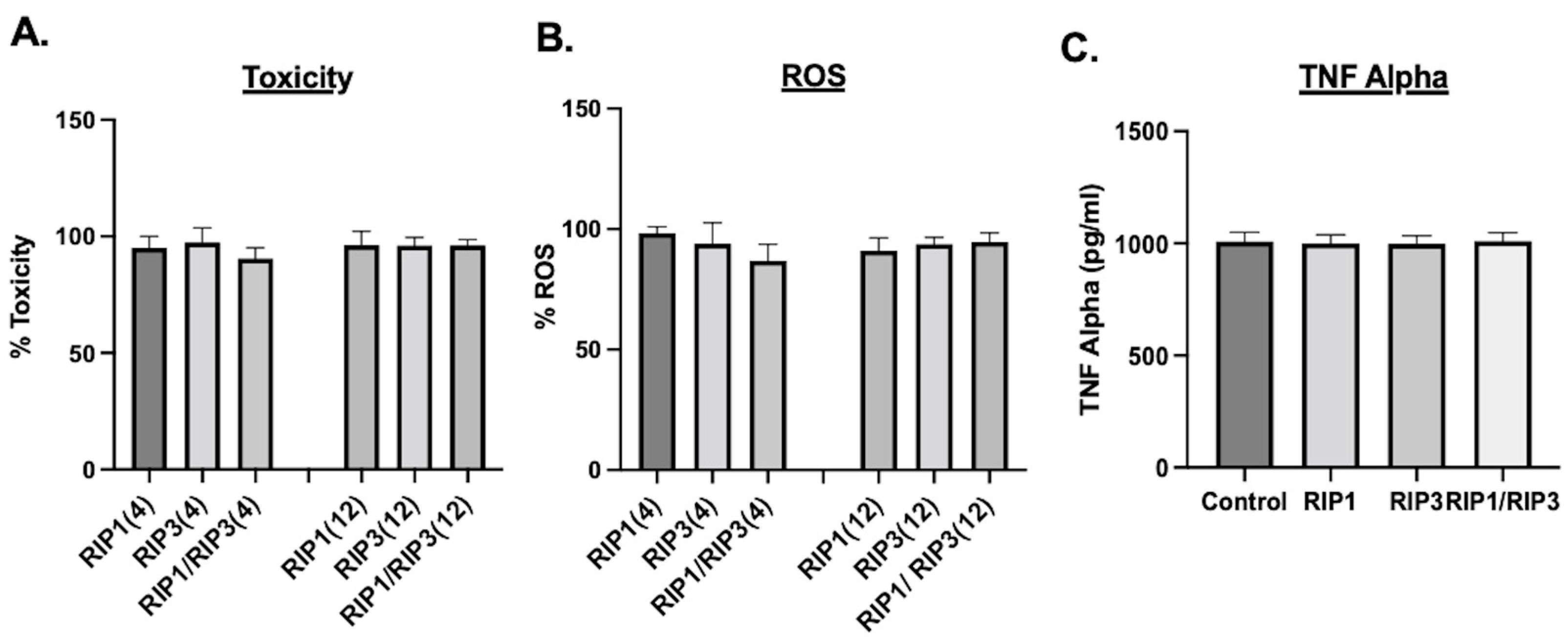
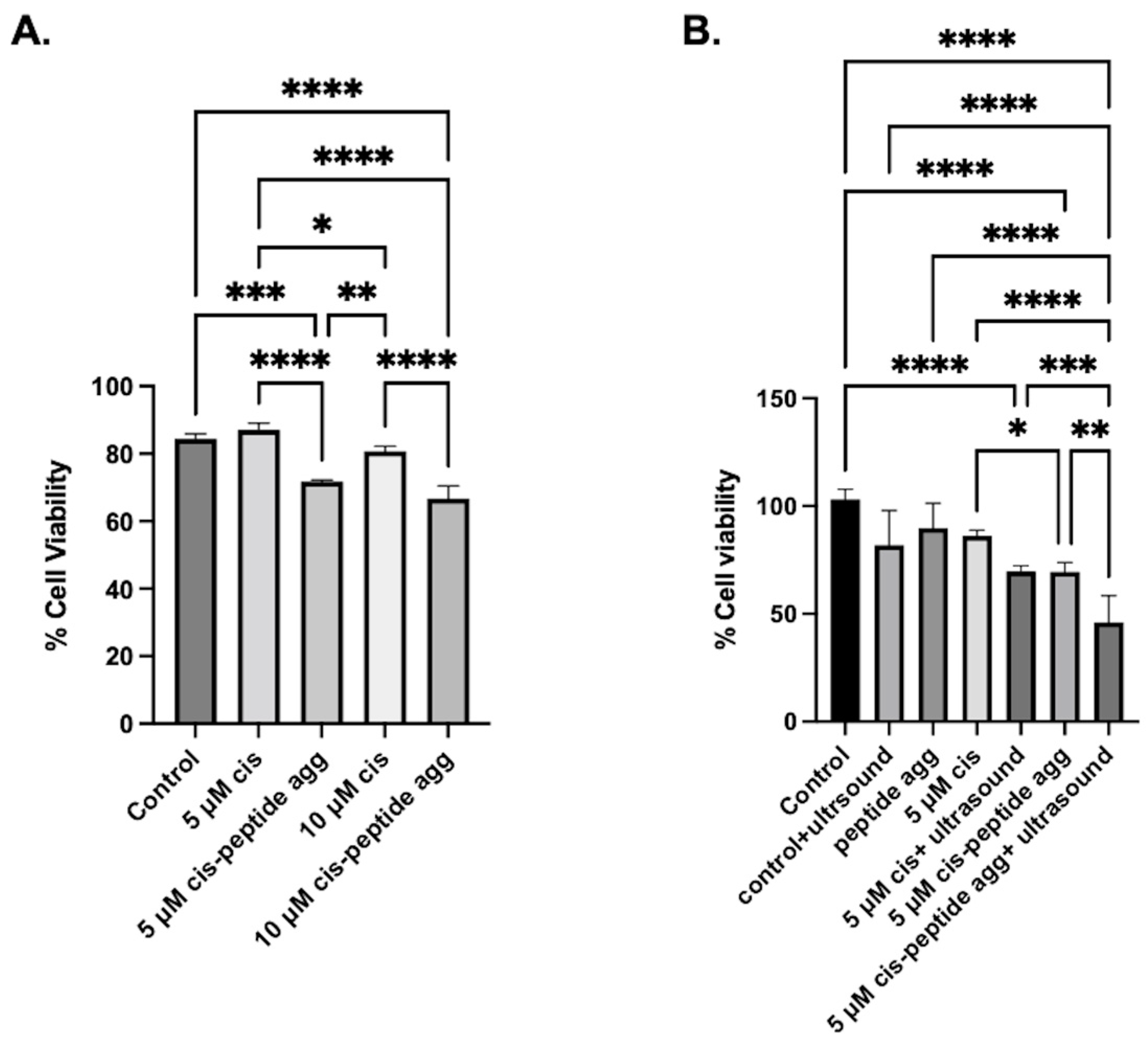
2. Results
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Aggregation Formation
4.3. ThT Fluorescence and Proteostat Assay
4.4. Turbidity
4.5. Transmission Electron Microscopy (TEM)
4.6. Mechanical Characterization
4.7. Cell Toxicity (alamarBlue) and ROS Assay
4.8. TNF Alpha Assay
4.9. LDH Toxicity Assay
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Furukawa, Y.; Nukina, N. Functional diversity of protein fibrillar aggregates from physiology to RNA granules to neurodegenerative diseases. Biochim. Biophys. Acta 2013, 1832, 1271–1278. [Google Scholar] [CrossRef]
- Hafner Bratkovic, I. Prions, prionoid complexes and amyloids: The bad, the good and something in between. Swiss Med. Wkly 2017, 147, w14424. [Google Scholar]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef]
- Wang, M.; Audas, T.E.; Lee, S. Disentangling a Bad Reputation: Changing Perceptions of Amyloids. Trends Cell Biol. 2017, 27, 465–467. [Google Scholar] [CrossRef]
- Chiesa, G.; Kiriakov, S.; Khalil, A.S. Protein assembly systems in natural and synthetic biology. BMC Biol. 2020, 18, 35. [Google Scholar] [CrossRef]
- Fassler, J.S.; Skuodas, S.; Weeks, D.L.; Phillips, B.T. Protein Aggregation and Disaggregation in Cells and Development. J. Mol. Biol. 2021, 433, 167215. [Google Scholar] [CrossRef] [PubMed]
- Merlini, G.; Bellotti, V.; Andreola, A.; Palladini, G.; Obici, L.; Casarini, S.; Perfetti, V. Protein aggregation. Clin. Chem. Lab. Med. 2001, 39, 1065–1075. [Google Scholar] [CrossRef] [PubMed]
- Fink, A.L. Protein aggregation: Folding aggregates, inclusion bodies and amyloid. Fold Des. 1998, 3, R9–R23. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, H.; Nishitoh, H.; Urano, F.; Sadamitsu, C.; Matsuzawa, A.; Takeda, K.; Masutani, H.; Yodoi, J.; Urano, Y.; Nagano, T.; et al. Amyloid beta induces neuronal cell death through ROS-mediated ASK1 activation. Cell Death Differ. 2005, 12, 19–24. [Google Scholar] [CrossRef] [PubMed]
- van Dam, L.; Dansen, T.B. Cross-talk between redox signalling and protein aggregation. Biochem. Soc. Trans. 2020, 48, 379–397. [Google Scholar] [CrossRef]
- Cenini, G.; Cecchi, C.; Pensalfini, A.; Bonini, S.A.; Ferrari-Toninelli, G.; Liguri, G.; Memo, M.; Uberti, D. Generation of reactive oxygen species by beta amyloid fibrils and oligomers involves different intra/extracellular pathways. Amino Acids 2010, 38, 1101–1106. [Google Scholar] [CrossRef]
- Peng, C.; Trojanowski, J.Q.; Lee, V.M. Protein transmission in neurodegenerative disease. Nat. Rev. Neurol. 2020, 16, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Spires-Jones, T.L.; Attems, J.; Thal, D.R. Interactions of pathological proteins in neurodegenerative diseases. Acta Neuropathol. 2017, 134, 187–205. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; McQuade, T.; Siemer, A.B.; Napetschnig, J.; Moriwaki, K.; Hsiao, Y.S.; Damko, E.; Moquin, D.; Walz, T.; McDermott, A.; et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 2012, 150, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Olrichs, N.K.; Gadella, B.M.; Kaloyanova, D.V.; Helms, J.B. Regulation of Functional Protein Aggregation by Multiple Factors: Implications for the Amyloidogenic Behavior of the CAP Superfamily Proteins. Int. J. Mol. Sci. 2020, 21, 6530. [Google Scholar] [CrossRef]
- Jacob, R.S.; Das, S.; Ghosh, S.; Anoop, A.; Jha, N.N.; Khan, T.; Singru, P.; Kumar, A.; Maji, S.K. Amyloid formation of growth hormone in presence of zinc: Relevance to its storage in secretory granules. Sci. Rep. 2016, 6, 23370. [Google Scholar] [CrossRef] [PubMed]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.; Simon, R.; Schubert, D.; et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef]
- Jang, H.; Arce, F.T.; Mustata, M.; Ramachandran, S.; Capone, R.; Nussinov, R.; Lal, R. Antimicrobial protegrin-1 forms amyloid-like fibrils with rapid kinetics suggesting a functional link. Biophys. J. 2011, 100, 1775–1783. [Google Scholar] [CrossRef]
- Li, J.; Zhang, F. Amyloids as Building Blocks for Macroscopic Functional Materials: Designs, Applications and Challenges. Int. J. Mol. Sci. 2021, 22, 10698. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, T.; Lei, T.; Zhang, D.; Du, S.; Girani, L.; Qi, D.; Lin, C.; Tong, R.; Wang, Y. RIP1/RIP3-regulated necroptosis as a target for multifaceted disease therapy (Review). Int. J. Mol. Med. 2019, 44, 771–786. [Google Scholar] [CrossRef]
- Li, D.; Liu, C. Better Together: A Hybrid Amyloid Signals Necroptosis. Cell 2018, 173, 1068–1070. [Google Scholar] [CrossRef] [PubMed]
- Vandenabeele, P.; Declercq, W.; Van Herreweghe, F.; Vanden Berghe, T. The role of the kinases RIP1 and RIP3 in TNF-induced necrosis. Sci. Signal. 2010, 3, re4. [Google Scholar] [CrossRef]
- Meylan, E.; Tschopp, J. The RIP kinases: Crucial integrators of cellular stress. Trends Biochem. Sci. 2005, 30, 151–159. [Google Scholar] [CrossRef]
- Zhang, D.; Lin, J.; Han, J. Receptor-interacting protein (RIP) kinase family. Cell Mol. Immunol. 2010, 7, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Feoktistova, M.; Leverkus, M. Programmed necrosis and necroptosis signalling. FEBS J. 2015, 282, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Festjens, N.; Vanden Berghe, T.; Cornelis, S.; Vandenabeele, P. RIP1, a kinase on the crossroads of a cell’s decision to live or die. Cell Death Differ. 2007, 14, 400–410. [Google Scholar] [CrossRef]
- Jang, T.H.; Zheng, C.; Li, J.; Richards, C.; Hsiao, Y.S.; Walz, T.; Wu, H.; Park, H.H. Structural study of the RIPoptosome core reveals a helical assembly for kinase recruitment. Biochemistry 2014, 53, 5424–5431. [Google Scholar] [CrossRef]
- Moquin, D.; Chan, F.K. The molecular regulation of programmed necrotic cell injury. Trends Biochem. Sci. 2010, 35, 434–441. [Google Scholar] [CrossRef]
- Stanger, B.Z.; Leder, P.; Lee, T.H.; Kim, E.; Seed, B. RIP: A novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell 1995, 81, 513–523. [Google Scholar] [CrossRef]
- Sun, X.; Lee, J.; Navas, T.; Baldwin, D.T.; Stewart, T.A.; Dixit, V.M. RIP3, a novel apoptosis-inducing kinase. J. Biol. Chem. 1999, 274, 16871–16875. [Google Scholar] [CrossRef]
- Moriwaki, K.; Chan, F.K. RIP3: A molecular switch for necrosis and inflammation. Genes Dev. 2013, 27, 1640–1649. [Google Scholar] [CrossRef]
- Hu, H.; Wu, X.; Wu, G.; Nan, N.; Zhang, J.; Zhu, X.; Zhang, Y.; Shu, Z.; Liu, J.; Liu, X.; et al. RIP3-mediated necroptosis is regulated by inter-filament assembly of RIP homotypic interaction motif. Cell Death Differ. 2021, 28, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.N.; Yang, Z.H.; Wang, X.K.; Zhang, Y.; Wan, H.; Song, Y.; Chen, X.; Shao, J.; Han, J. Distinct roles of RIP1-RIP3 hetero- and RIP3-RIP3 homo-interaction in mediating necroptosis. Cell Death Differ. 2014, 21, 1709–1720. [Google Scholar] [CrossRef] [PubMed]
- Della Torre, L.; Nebbioso, A.; Stunnenberg, H.G.; Martens, J.H.A.; Carafa, V.; Altucci, L. The Role of Necroptosis: Biological Relevance and Its Involvement in Cancer. Cancers 2021, 13, 684. [Google Scholar] [CrossRef] [PubMed]
- Mompean, M.; Li, W.; Li, J.; Laage, S.; Siemer, A.B.; Bozkurt, G.; Wu, H.; McDermott, A.E. The Structure of the Necrosome RIPK1-RIPK3 Core, a Human Hetero-Amyloid Signaling Complex. Cell 2018, 173, 1244–1253 e10. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Yin, J.; Starovasnik, M.A.; Fairbrother, W.J.; Dixit, V.M. Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. J. Biol. Chem. 2002, 277, 9505–9511. [Google Scholar] [CrossRef] [PubMed]
- Dalgediene, I.; Luciunaite, A.; Zvirbliene, A. Activation of Macrophages by Oligomeric Proteins of Different Size and Origin. Mediat. Inflamm. 2018, 2018, 7501985. [Google Scholar] [CrossRef]
- Polumuri, S.K.; Haile, L.A.; Ireland, D.D.C.; Verthelyi, D. Aggregates of IVIG or Avastin, but not HSA, modify the response to model innate immune response modulating impurities. Sci. Rep. 2018, 8, 11477. [Google Scholar] [CrossRef]
- Vasconcelos, B.; Stancu, I.C.; Buist, A.; Bird, M.; Wang, P.; Vanoosthuyse, A.; Van Kolen, K.; Verheyen, A.; Kienlen-Campard, P.; Octave, J.N.; et al. Heterotypic seeding of Tau fibrillization by pre-aggregated Abeta provides potent seeds for prion-like seeding and propagation of Tau-pathology in vivo. Acta Neuropathol. 2016, 131, 549–569. [Google Scholar] [CrossRef]
- Krol, S.; Osterlund, N.; Vosough, F.; Jarvet, J.; Warmlander, S.; Barth, A.; Ilag, L.L.; Magzoub, M.; Graslund, A.; Morman, C. The amyloid-inhibiting NCAM-PrP peptide targets Abeta peptide aggregation in membrane-mimetic environments. iScience 2021, 24, 102852. [Google Scholar] [CrossRef]
- Varache, M.; Bezverkhyy, I.; Weber, G.; Saviot, L.; Chassagnon, R.; Baras, F.; Bouyer, F. Loading of Cisplatin into Mesoporous Silica Nanoparticles: Effect of Surface Functionalization. Langmuir 2019, 35, 8984–8995. [Google Scholar] [CrossRef]
- Zhang, W.; Tung, C.H. Cisplatin Cross-Linked Multifunctional Nanodrugplexes for Combination Therapy. ACS Appl. Mater Interfaces 2017, 9, 8547–8555. [Google Scholar] [CrossRef]
- Naiki, H.; Higuchi, K.; Hosokawa, M.; Takeda, T. Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Anal. Biochem. 1989, 177, 244–249. [Google Scholar] [CrossRef] [PubMed]
- LeVine, H., 3rd; Thioflavine, T. interaction with synthetic Alzheimer’s disease beta-amyloid peptides: Detection of amyloid aggregation in solution. Protein Sci. 1993, 2, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.; Coleman, J.; Chan, E.; Nicholson, T.P.; Dai, L.; Sheppard, P.W.; Patton, W.F. Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell Biochem. Biophys. 2011, 60, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Oshinbolu, S.; Shah, R.; Finka, G.; Molloy, M.; Uden, M.; Bracewell, D.G. Evaluation of fluorescent dyes to measure protein aggregation within mammalian cell culture supernatants. J. Chem. Technol. Biotechnol. 2018, 93, 909–917. [Google Scholar] [CrossRef]
- Zhao, R.; So, M.; Maat, H.; Ray, N.J.; Arisaka, F.; Goto, Y.; Carver, J.A.; Hall, D. Measurement of amyloid formation by turbidity assay-seeing through the cloud. Biophys. Rev. 2016, 8, 445–471. [Google Scholar] [CrossRef] [PubMed]
- Samluk, L.; Ostapczuk, P.; Dziembowska, M. Long-term mitochondrial stress induces early steps of Tau aggregation by increasing reactive oxygen species levels and affecting cellular proteostasis. Mol. Biol. Cell 2022, 33, ar67. [Google Scholar] [CrossRef]
- Vasconcellos, L.R.; Dutra, F.F.; Siqueira, M.S.; Paula-Neto, H.A.; Dahan, J.; Kiarely, E.; Carneiro, L.A.; Bozza, M.T.; Travassos, L.H. Protein aggregation as a cellular response to oxidative stress induced by heme and iron. Proc. Natl. Acad. Sci. USA 2016, 113, E7474–E7482. [Google Scholar] [CrossRef]
- Kaja, S.; Payne, A.J.; Naumchuk, Y.; Koulen, P. Quantification of Lactate Dehydrogenase for Cell Viability Testing Using Cell Lines and Primary Cultured Astrocytes. Curr. Protoc. Toxicol. 2017, 72, 2.26.1–2.26.10. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ismail, M.; Kanapathipillai, M. Amyloid-like RIP1/RIP3 RHIM Fragments’ Characterization and Application as a Drug Depot. Molecules 2023, 28, 1480. https://doi.org/10.3390/molecules28031480
Ismail M, Kanapathipillai M. Amyloid-like RIP1/RIP3 RHIM Fragments’ Characterization and Application as a Drug Depot. Molecules. 2023; 28(3):1480. https://doi.org/10.3390/molecules28031480
Chicago/Turabian StyleIsmail, Maytham, and Mathumai Kanapathipillai. 2023. "Amyloid-like RIP1/RIP3 RHIM Fragments’ Characterization and Application as a Drug Depot" Molecules 28, no. 3: 1480. https://doi.org/10.3390/molecules28031480
APA StyleIsmail, M., & Kanapathipillai, M. (2023). Amyloid-like RIP1/RIP3 RHIM Fragments’ Characterization and Application as a Drug Depot. Molecules, 28(3), 1480. https://doi.org/10.3390/molecules28031480




