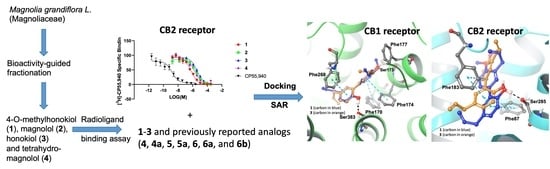
In Vitro and In Silico Studies of Neolignans from Magnolia grandiflora L. Seeds against Human Cannabinoids and Opioid Receptors
Abstract
1. Introduction
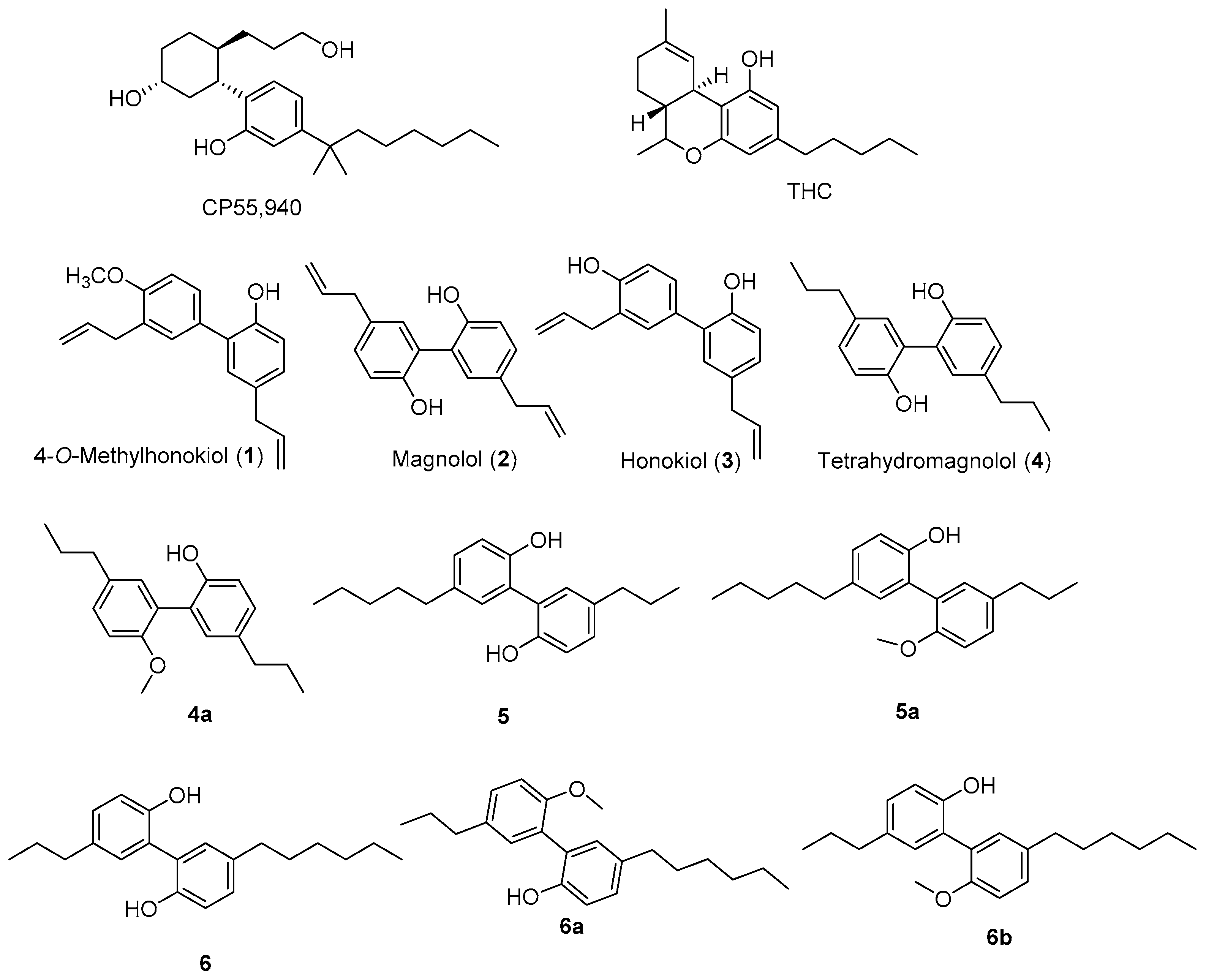
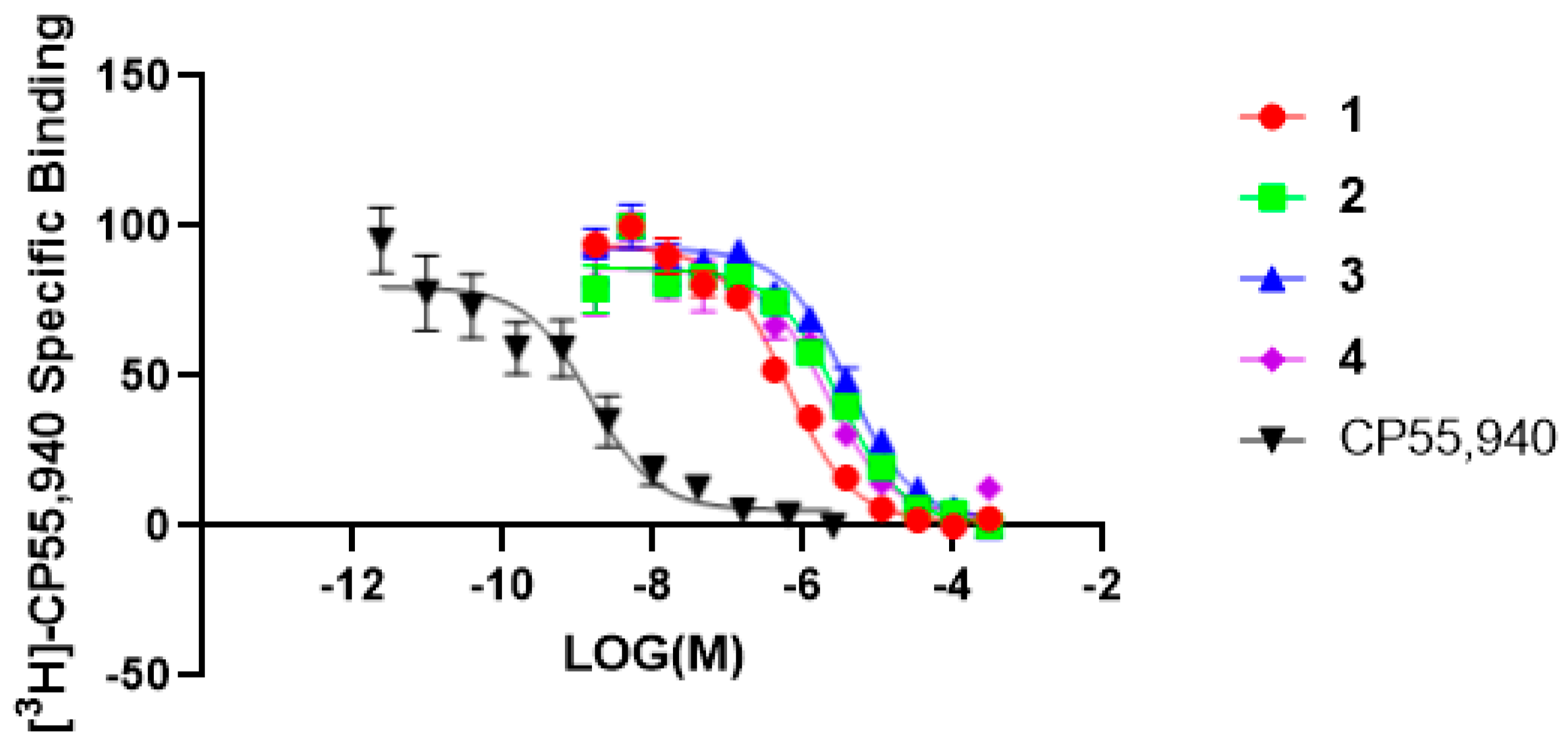
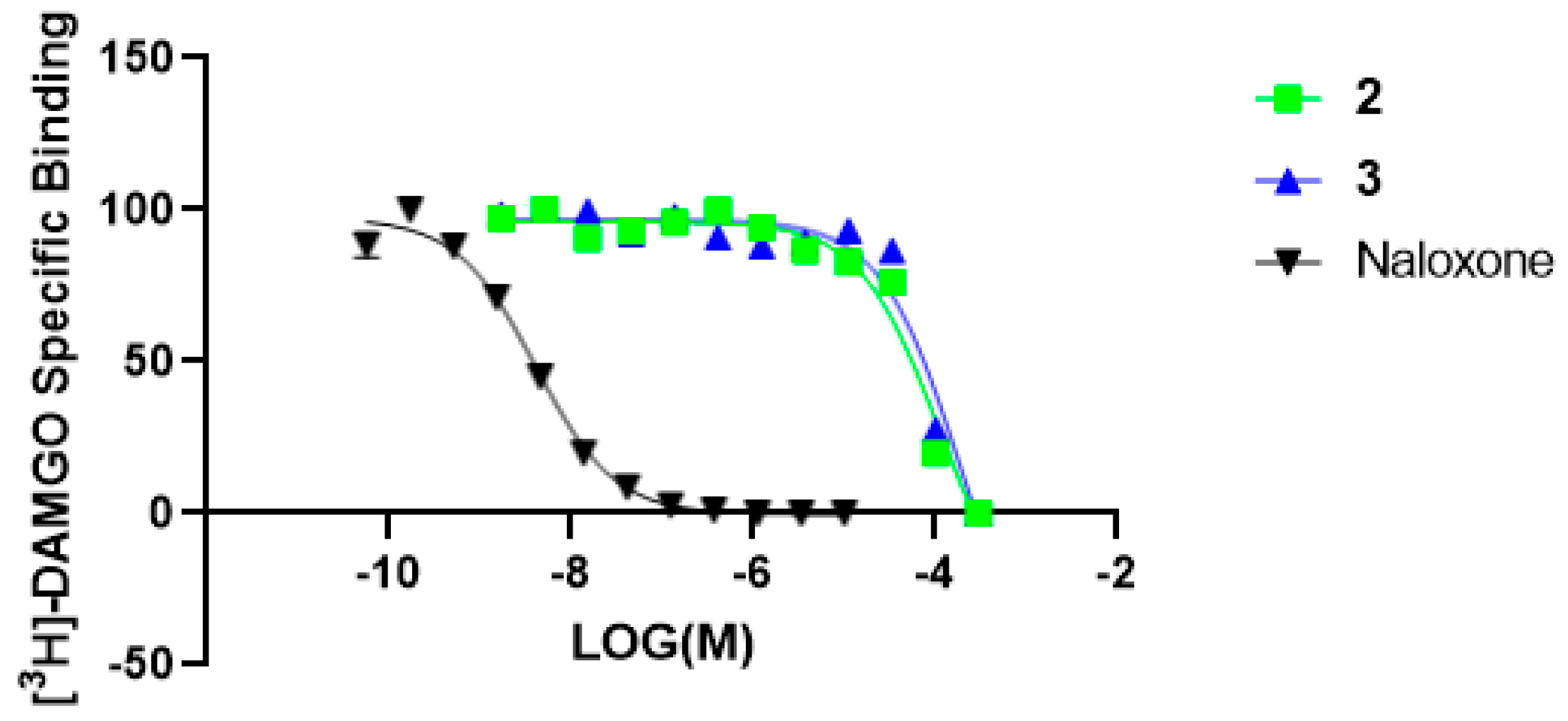
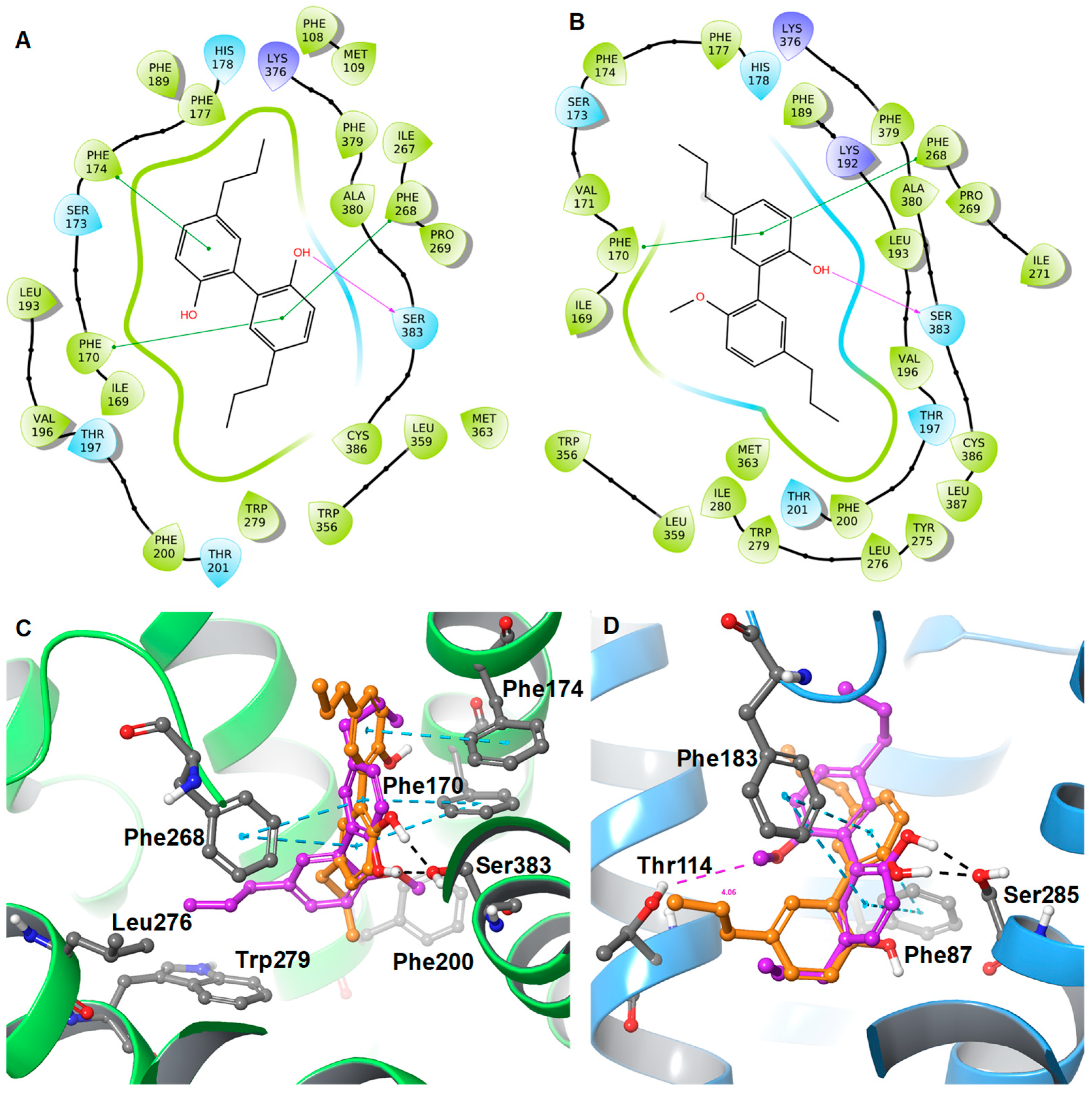
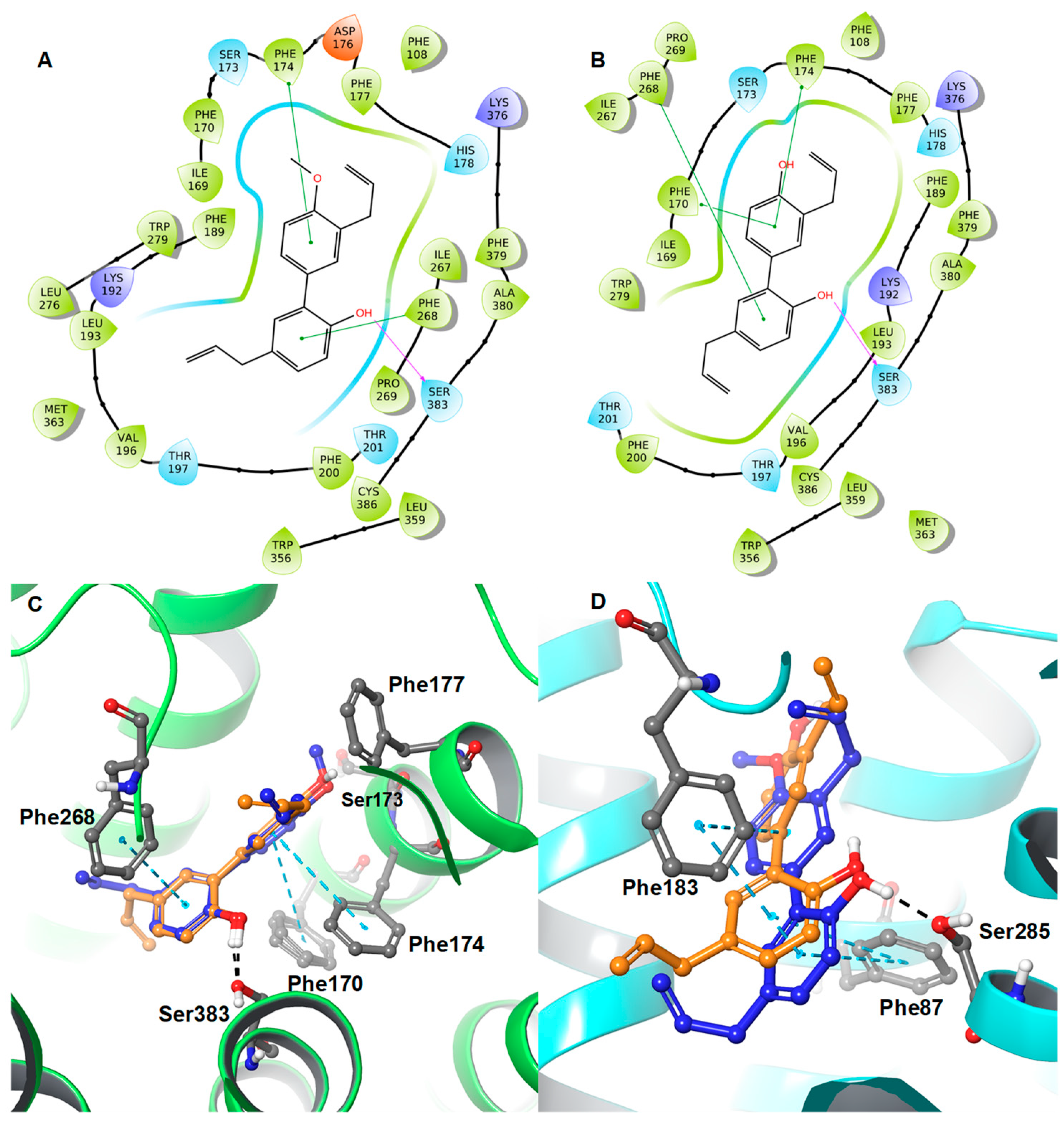
2. Results and Discussion
Protein-Ligand Interaction Study
3. Materials and Methods
3.1. Extraction and Bioassay-Guided Isolation of Compounds
3.2. Cannabinoid and Opioid Receptor Binding Assay
3.2.1. Reagents
3.2.2. Cell Culture and Membrane Preparation
3.2.3. Competitive Radioligand Binding Assays
3.3. Computational Method
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schiöth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2009, 16, 829. [Google Scholar] [CrossRef]
- Quarta, C.; Cota, D. Anti-obesity therapy with peripheral CB1 blockers: From promise to safe(?) practice. Int. J. Obes. 2020, 44, 2179–2193. [Google Scholar] [CrossRef] [PubMed]
- Kunos, G.; Tam, J. The case for peripheral CB(1) receptor blockade in the treatment of visceral obesity and its cardiometabolic complications. Br. J. Pharmacol. 2011, 163, 1423–1431. [Google Scholar] [CrossRef] [PubMed]
- Soler-Cedeno, O.; Xi, Z.-X. Neutral CB1 Receptor Antagonists as Pharmacotherapies for Substance Use Disorders: Rationale, Evidence, and Challenge. Cells 2022, 11, 3262. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Roy, K.K.; Liu, H.; Ma, G.; Pettaway, S.; Alsharif, W.F.; Gadepalli, R.S.; Rimoldi, J.M.; McCurdy, C.R.; Cutler, S.J.; et al. Structure-based identification of potent natural product chemotypes as cannabinoid receptor 1 inverse agonists. Molecules 2018, 23, 2630. [Google Scholar] [CrossRef]
- GGaliegue, S.; Mary, S.; Marchand, J.; Dussossoy, D.; Carriere, D.; Carayon, P.; Bouaboula, M.; Shire, D.; Le Fur, G.; Casellas, P. Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur. J. Biochem. 1995, 232, 54–61. [Google Scholar] [CrossRef]
- Ashton, J.C.; Glass, M. The cannabinoid CB2 receptor as a target for inflammation-dependent neurodegeneration. Curr. Neuropharmacol. 2007, 5, 73–80. [Google Scholar] [CrossRef]
- Defer, N.; Wan, J.; Souktani, R.; Escoubet, B.; Perier, M.; Caramelle, P.; Manin, S.; Deveaux, V.; Bourin, M.C.; Zimmer, A.; et al. The cannabinoid receptor type 2 promotes cardiac myocyte and fibroblast survival and protects against ischemia/reperfusion-induced cardiomyopathy. FASEB J. 2009, 23, 2120–2130. [Google Scholar] [CrossRef]
- Han, S.; Thatte, J.; Buzard, D.J.; Jones, R.M. Therapeutic utility of cannabinoid receptor type 2 (CB(2)) selective agonists. J. Med. Chem. 2013, 56, 8224–8256. [Google Scholar] [CrossRef]
- Pressly, J.D.; Mustafa, S.M.; Adibi, A.H.; Alghamdi, S.; Pandey, P.; Roy, K.K.; Doerksen, R.J.; Moore, B.M., Jr.; Park, F. Selective cannabinoid 2 receptor stimulation reduces tubular epithelial cell damage after renal ischemia-reperfusion Injury. J. Pharmacol. Exp. Ther. 2018, 364, 287–299. [Google Scholar] [CrossRef]
- Trang, T.; Al-Hasani, R.; Salvemini, D.; Salter, M.W.; Gutstein, H.; Cahill, C.M. Pain and poppies: The good, the bad, and the ugly of opioid analgesics. J. Neurosci. 2015, 35, 13879–13888. [Google Scholar] [CrossRef] [PubMed]
- Abrams, D.I.; Couey, P.; Shade, S.B.; Kelly, M.E.; Benowitz, N.L. Cannabinoid-opioid interaction in chronic pain. Clin. Pharmacol. Ther. 2011, 90, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Singla, R.K.; Banerjee, S.; Sinha, B.; Shen, B.; Sharma, R. Role of Shankhpushpi (Convolvulus pluricaulis) in neurological disorders: An umbrella review covering evidence from ethnopharmacology to clinical studies. Neurosci. Biobehav. Rev. 2022, 140, 104795. [Google Scholar] [CrossRef]
- von Linnaeus, C. Magnolia grandiflora L. Syst. Nat. Ed. 1759, 10, 1082. [Google Scholar]
- Schühly, W.; Khan, S.I.; Fischer, N.H. Neolignans from North American Magnolia species with cyclooxygenase 2 inhibitory activity. Inflammopharmacology 2009, 17, 106–110. [Google Scholar] [CrossRef]
- Lee, Y.J.; Lee, Y.M.; Lee, C.K.; Jung, J.K.; Han, S.B.; Hong, J.T. Therapeutic applications of compounds in the Magnolia family. Pharmacol. Ther. 2011, 130, 157–176. [Google Scholar] [CrossRef] [PubMed]
- Schifano, F.; Guarino, V.; Papanti, D.G.; Baccarin, J.; Orsolini, L.; Corkery, J.M. Is there a potential of misuse for Magnolia officinalis compounds/metabolites? Hum. Psychopharmacol. 2017, 32, 2595. [Google Scholar] [CrossRef]
- Singh, T.; Katiyar, S.K. Honokiol, a phytochemical from Magnolia spp., inhibits breast cancer cell migration by targeting nitric oxide and cyclooxygenase-2. Int. J. Oncol. 2011, 38, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Jung, J.K.; Han, S.B.; Nam, S.Y.; Oh, K.W.; Hong, J.T. Anxiolytic-like effects of 4-O-methylhonokiol isolated from Magnolia officinalis through enhancement of GABAergic transmission and chloride influx. J. Med. Food. 2011, 14, 724–731. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, P.P.; Hu, K.L.; Li, L.N.; Yu, X.; Lu, Y.; Chang, H.S. Antidepressant-Like effect and mechanism of action of honokiol on the mouse lipopolysaccharide (LPS) depression model. Molecules 2019, 24, 2035. [Google Scholar] [CrossRef]
- Singha, S.K.; Muhammad, I.; Ibrahim, M.A.; Wang, M.; Ashpole, N.M.; Shariat-Madar, Z. 4-O-Methylhonokiol influences normal cardiovascular development in medaka embryo. Molecules 2019, 24, 475. [Google Scholar] [CrossRef] [PubMed]
- Kuo, W.L.; Chung, C.Y.; Hwang, T.L.; Chen, J.J. Biphenyl-type neolignans from Magnolia officinalis and their anti-inflammatory activities. Phytochemistry 2013, 85, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Khalid, S.; Ullah, M.Z.; Khan, A.U.; Afridi, R.; Rasheed, H.; Khan, A.; Ali, H.; Kim, Y.S.; Khan, S. Antihyperalgesic properties of honokiol in inflammatory pain models by targeting of NF-κB and Nrf2 signaling. Front. Pharmacol. 2018, 9, 140. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, A.; Baur, R.; Schoeder, C.; Sigel, E.; Müller, C.E. Structural analogues of the natural products magnolol and honokiol as potent allosteric potentiators of GABA(A) receptors. Bioorg. Med. Chem. 2014, 22, 6908–6917. [Google Scholar] [CrossRef]
- Stout, K.; Bernaskova, M.; Miller, G.W.; Hufner, A.; Schuehly, W. Bioinspired honokiol analogs and their evaluation for activity on the norepinephrine transporter. Molecules 2018, 23, 2536. [Google Scholar] [CrossRef]
- Kong, L.D.; Cheng, C.H.; Tan, R.X. Inhibition of MAO A and B by some plant-derived alkaloids, phenols and anthraquinones. J. Ethnopharmacol. 2004, 91, 351–355. [Google Scholar] [CrossRef]
- Rempel, V.; Fuchs, A.; Hinz, S.; Karcz, T.; Lehr, M.; Koetter, U.; Müller, C.E. Magnolia extract, magnolol, and metabolites: Activation of cannabinoid CB2 receptors and blockade of the related GPR55. ACS Med. Chem. Lett. 2013, 4, 41–45. [Google Scholar] [CrossRef]
- Anavi-Goffer, S.; Gertsch, J. CB2 Receptor Ligands for the Treatment of Psychiatric Disorders. Google Patents 1498379858369718886-09486419, 8 November 2016. Available online: storage.googleapis.com (accessed on 4 January 2022).
- Fuchs, A.; Rempel, V.; Müller, C.E. The natural product magnolol as a lead structure for the development of potent cannabinoid receptor agonists. PLoS ONE 2013, 8, e77739. [Google Scholar] [CrossRef]
- Schuehly, W.; Paredes, J.M.; Kleyer, J.; Huefner, A.; Anavi-Goffer, S.; Raduner, S.; Altmann, K.H.; Gertsch, J. Mechanisms of osteoclastogenesis inhibition by a novel class of biphenyl-type cannabinoid CB(2) receptor inverse agonists. Chem. Biol. 2011, 18, 1053–1064. [Google Scholar] [CrossRef]
- Sarrica, A.; Kirika, N.; Romeo, M.; Salmona, M.; Diomede, L. Safety and toxicology of magnolol and honokiol. Planta Med. 2018, 84, 1151–1164. [Google Scholar] [CrossRef]
- Li, X.; Hua, T.; Vemuri, K.; Ho, J.H.; Wu, Y.; Wu, L.; Popov, P.; Benchama, O.; Zvonok, N.; Locke, K.; et al. Crystal structure of the human cannabinoid receptor CB2. Cell 2019, 176, 459–467.e13. [Google Scholar] [CrossRef]
- Hua, T.; Vemuri, K.; Nikas, S.P.; Laprairie, R.B.; Wu, Y.; Qu, L.; Pu, M.; Korde, A.; Jiang, S.; Ho, J.H.; et al. Crystal structures of agonist-bound human cannabinoid receptor CB1. Nature 2017, 547, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tang, Y.; Wei, S.; Yu, H.; Lv, H.; Ge, H. Isolation and purification of phenolic compounds from magnoliae officinalis by preparative high-performance liquid chromatography. J. Liq. Chromatogr. Relat. Technol. 2010, 33, 431–440. [Google Scholar] [CrossRef]
- Chen, Y.H.; Lu, M.H.; Guo, D.S.; Zhai, Y.Y.; Miao, D.; Yue, J.Y.; Yuan, C.H.; Zhao, M.M.; An, D.R. An antifungal effect of magnolol and honokiol from Magnolia officinalis on Alternaria alternata causing Tobacco Brown Spot. Molecules 2019, 24, 2140. [Google Scholar] [CrossRef] [PubMed]
- Husni, A.S.; McCurdy, C.R.; Radwan, M.M.; Ahmed, S.A.; Slade, D.; Ross, S.A.; ElSohly, M.A.; Cutler, S.J. Evaluation of phytocannabinoids from high potency Cannabis sativa using in vitro bioassays to determine structure-activity relationships for cannabinoid receptor 1 and cannabinoid receptor 2. Med. Chem. Res. 2014, 23, 4295–4300. [Google Scholar] [CrossRef] [PubMed]
- Tarawneh, A.H.; Pandey, P.; Lo’ay, A.; Gadetskaya, A.V.; Abu-Orabi, S.T.; Doerksen, R.J.; Cutler, S.J. 1, 2, 3-Triazole derivatives as highly selective cannabinoid receptor type 2 (CB2) agonists. Arab J. Chem. 2022, 15, 103545. [Google Scholar] [CrossRef]
- Giacometti, R.D.; Duchek, J.; Werner, L.; Husni, A.S.; McCurdy, C.R.; Cutler, S.J.; Cox, D.P.; Hudlicky, T. Heteroatom analogues of hydrocodone: Synthesis and biological activity. J. Org. Chem. 2013, 78, 2914–2925. [Google Scholar] [CrossRef]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aid. Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.P.; Friesner, R.A.; Xiang, Z.; Honig, B. On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 2002, 320, 597–608. [Google Scholar] [CrossRef] [PubMed]



| 10 μM Compounds or 10 μg/mL Extracts * | Cannabinoid Receptors (% Displacement) | Opioid Receptors (% Displacement) | |||
|---|---|---|---|---|---|
| CB1 | CB2 | Δ | κ | µ | |
| n-Hexanes extract | 63.8 | 73.7 | 93.0 | 60.8 | 84.5 |
| Ethanol extract | 55.4 | 94.7 | 76.2 | 23.1 | 72.6 |
| 4-O-Methylhonokiol (1) | 91.3 | 82.2 | 31.4 | 18.3 | 46.2 |
| Magnolol (2) | 52.7 | 74.7 | 7.3 | 10.7 | 52.9 |
| Honokiol (3) | 50.8 | 65.7 | NA | NA | 54.2 |
| 2 + 3 (1:1) | 99.8 | 91.0 | 93.3 | 59.2 | 74.0 |
| Tetrahydromagnolol (4) | 30.8 | 78.2 | NA | NA | NA |
| CP55,940 | 82.5 | 101.3 | NT | NT | NT |
| Naloxone | NT | NT | 97.0 | 100.2 | 99.8 |
| Compound | CB1R (μM) | CB2R (μM) | µ Opioid Receptor (μM) | |||
|---|---|---|---|---|---|---|
| IC50 | Ki ± SEM | IC50 | Ki ± SEM | IC50 | Ki ± SEM | |
| 1 | 7.69 | 3.85 ± 0.89 | 0.59 | 0.29 ± 0.02 | n/a | n/a |
| 2 | 35.64 | 17.82 ± 3.43 | 2.89 | 1.40 ± 0.14 | 106.20 * | 53.12 * |
| 3 | 29.11 | 14.55 ± 2.47 | 3.88 | 1.94 ± 0.16 | 91.06 * | 182.10 * |
| 4 | 38.17 | 19.08 ± 0.79 | 1.99 | 0.99 ± 0.14 | NA | NA |
| CP55,940 # | 0.006859 | 0.0033 ± 0.00128 | 0.00287 | 0.001439 ± 0.00027 | NT | NT |
| Naloxone ^ | NT | NT | NT | NT | 0.00409 | 0.002049 ± 0.000179 |
| Compound | CB1R Agonist Ki (nM) * | CB2R Agonist Ki (nM) * | CB1R | CB2R | ||
|---|---|---|---|---|---|---|
| GlideScore (kcal/mol) | ΔG (kcal/mol) | GlideScore (kcal/mol) | ΔG (kcal/mol) | |||
| CP55,940 | 1.28 | 1.42 | −12.045 | −85.71 | −12.156 | −81.21 |
| 2 (Magnolol) | 3150 | 1440 | −10.243 | −74.55 | −9.765 | −64.48 |
| 4 (Tetrahydromagnolol) | 2260 | 416 | −10.346 | −71.06 | −11.194 | −73.41 |
| 3 (Honokiol) | 6460 | 5610 | −10.854 | −71.17 | −8.989 | −59.73 |
| 1 (4-O-methylhonokiol) | 8340 ± 3200 | 43.3 ± 17.1 | −11.106 | −78.20 | −10.564 | −73.06 |
| 4a | 267 ± 58 | 221 ± 57 | −11.275 | −76.78 | −10.251 | −71.46 |
| 5 | 362 ± 113 | 37.5 ± 7.8 | −12.307 | −78.78 | −11.195 | −77.88 |
| 5a | 17.3 ± 1.4 | 31.0 ± 9.9 | −11.652 | −83.77 | −10.401 | −76.00 |
| 6 | 145 ± 48 | 29.4 ± 9.0 | −12.228 | −80.02 | −11.577 | −81.06 |
| 6a | 9.57 ± 5.43 | 23.8 ± 7.1 | −12.311 | −88.39 | −11.224 | −80.56 |
| 6b | 313 | 281 ± 101 | ND | ND | −10.944 | −69.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, P.; Kumarihamy, M.; Chaturvedi, K.; Ibrahim, M.A.M.; Lambert, J.A.; Godfrey, M.; Doerksen, R.J.; Muhammad, I. In Vitro and In Silico Studies of Neolignans from Magnolia grandiflora L. Seeds against Human Cannabinoids and Opioid Receptors. Molecules 2023, 28, 1253. https://doi.org/10.3390/molecules28031253
Pandey P, Kumarihamy M, Chaturvedi K, Ibrahim MAM, Lambert JA, Godfrey M, Doerksen RJ, Muhammad I. In Vitro and In Silico Studies of Neolignans from Magnolia grandiflora L. Seeds against Human Cannabinoids and Opioid Receptors. Molecules. 2023; 28(3):1253. https://doi.org/10.3390/molecules28031253
Chicago/Turabian StylePandey, Pankaj, Mallika Kumarihamy, Krishna Chaturvedi, Mohamed A. M. Ibrahim, Janet A. Lambert, Murrell Godfrey, Robert J. Doerksen, and Ilias Muhammad. 2023. "In Vitro and In Silico Studies of Neolignans from Magnolia grandiflora L. Seeds against Human Cannabinoids and Opioid Receptors" Molecules 28, no. 3: 1253. https://doi.org/10.3390/molecules28031253
APA StylePandey, P., Kumarihamy, M., Chaturvedi, K., Ibrahim, M. A. M., Lambert, J. A., Godfrey, M., Doerksen, R. J., & Muhammad, I. (2023). In Vitro and In Silico Studies of Neolignans from Magnolia grandiflora L. Seeds against Human Cannabinoids and Opioid Receptors. Molecules, 28(3), 1253. https://doi.org/10.3390/molecules28031253