Abstract
Fungi from the genus Diaporthe have been reported as plant pathogens, endophytes, and saprophytes on a wide range of host plants worldwide. Their precise identification is problematic since many Diaporthe species can colonize a single host plant, whereas the same Diaporthe species can inhabit many hosts. Recently, Diaporthe has been proven to be a rich source of bioactive secondary metabolites. In our initial study, 40 Diaporthe isolates were analyzed for their metabolite production. A total of 153 compounds were identified based on their spectroscopic properties—Ultraviolet-visible and mass spectrometry. From these, 43 fungal metabolites were recognized as potential chemotaxonomic markers, mostly belonging to the drimane sesquiterpenoid-phthalide hybrid class. This group included mainly phytotoxic compounds such as cyclopaldic acid, altiloxin A, B, and their derivatives. To the best of our knowledge, this is the first report on the metabolomic studies on Diaporthe eres species complex from fruit trees in the South-Eastern Poland. The results from our study may provide the basis for the future research on the isolation of identified metabolites and on their bioactive potential for agricultural applications as biopesticides or biofertilizers.
1. Introduction
The genus Diaporthe Nitschke belongs to the family Diaporthaceae, (Diaporthales, Diaporthomycetidae, Sordariomycetes, Pezizomycotina, Ascomycota; (MycoBank. 2022; Species Fungorum. 2022; accessed on 21 December 2022), with the anamorph known as Phomopsis. According to the implementation of “one fungus one name (1F:1N) nomen- clature”, Diaporthe has been adopted over Phomopsis because it was first introduced as this, is encountered commonly in the literature, and represents most species [1]. Diaporthe are present as plant pathogens, endophytes, and saprophytes in a wide range of hosts worldwide [2,3,4,5]. Some pathogenic Diaporthe are responsible for several serious diseases of economically important crops, including fruit plants [4,6,7]. Both sexual and asexual morphs of Diaporthe have been associated with cankers, shoot diebacks, bud and shoot blights, and leave spots of peach caused by Phomopsis amygdali [8], apple by P. mali [9], pear by D. eres [10], plum by D. perniciosa [11], grape by P. viticola, P. fukushii, D. eres [12,13], blueberry by D. australafricana, D. ambigua, D. neotheicola, D. passiflorae [14], and many others. Currently, 1177 names of Diaporthe and 984 of Phomopsis are listed in Index Fungorum (http://www.indexfungorum.org/; accessed on 20 September 2022). Identification of Diaporthe species is complicated and was initially based on morphological features, cultural characteristics, and host affiliation leading to a proliferation of names based on the hosts from which they were isolated [15]. It has been observed that the same Diaporthe species colonizes different hosts, and the co-occurrence of different species is commonly reported in the same host [4,16,17,18,19]. Thus, the identification and description of species based on host association are unreliable within Diaporthe [3,20,21]. Moreover, the identification of Diaporthe based only on morphological features such as the size and shape of ascomata [2] and conidiomata [16] also proved insufficient due to their variability under changing environmental conditions [3]. Currently, the taxonomy of Diaporthe is actively changing, with numerous species being described each year, primarily based on molecular data combined with morphological characterization and host associations [3,10,20,21,22,23,24].
In recent years the genus Diaporthe has been widely used in secondary metabolite study due to their production of a variety of unique low- and high-molecular-weight metabolites with different bioactivities which were recently summarized in the extensive review by Xu et al. [25]. Researchers focused mainly on the endophytic species of Diaporthe, which is reported as one of the most frequently isolated genera among endophytic fungi. Probably the same compounds can be produced by endophytic, saprotrophic, and pathogenic species [26]. Over the past decade, 335 bioactive secondary metabolites have been obtained from known Diaporthe species and from those for which only a generic name has been assigned. [25,27,28] Among bioactive compounds 246 were isolated from Phomopsis, 106 from Diaporthe, and 17 from both species [25,29]. The metabolites produced by this genus include terpenoids, steroids, macrolides, ten-membered lactones, alkaloids, flavonoids, fatty acids, and polyketides, being the main structural type [25].
Although endophytic Diaporthe species have been extensively screened in bioassays for metabolite production, no such information is available for fungi belonging to D. eres species complex isolated from fruit trees in Poland. The literature indicates that the same species of the genus Diaporthe can occur on one or different hosts with different lifestyles [2]. Some Diaporthe species described as endophytes include latent phytopathogens, which asymptomatically colonize various host plants [30]. An example is D. eres, which as a pathogen infects many crops, including orchards, and is often the main cause of serious economic losses worldwide [3,20,21,31]. In Poland, however, the endophytic form of D. eres on Prunus domestica was recorded [32]. In the era of global warming and climate change, we must remember that many species may switch their lifestyles or spread into new regions, where they will come into contact with new potential hosts and will become a dangerous cause of diseases [33,34]. Therefore, the additional data such as metabolite profiles of such important fungi like Diaporthe may be crucial to understand their pathogenicity and switching life mode triggers in future research. Since Diaporthe species are a valuable source of bioactive metabolites, it would be worthwhile to further explore the genus for novel compounds that have a biotechnological potential.
The main objective of our study was metabolite profiling of Diaporthe isolates from various orchard plants of south-eastern Poland. To fully delineate the secondary metabolite profile of any fungus is an ambitious undertaking. Thus, this work is just an initial step toward the further exploration of the novel compounds from Diaporthe and their agricultural or pharmaceutical bioactivities.
2. Results and Discussion
2.1. ITS-Based Fungal Identification
The sequences of the ITS regions were used to identify Diaporthe strains. Closely related species have been received by comparing the obtained sequence data with the NCBI database (https://www.ncbi.nlm.nih.gov; accessed on 19 December 2022). The results indicated that 34 of the tested strains were closely related to D. eres species complex with 100% of similarity, 5 strains with 99.8%, and one strain with 99.6% of similarity (Table 1).

Table 1.
Fungi most closely related to Diaporthe based on ITS sequences using BLASTn analysis.
2.2. Chemical Characterization of Fungal Metabolites
Our study revealed that the D. eres species complex isolated from fruit trees in south-eastern Poland showed high biodiversity in the secondary metabolite production. A total of 153 compounds were found as a result of screening of forty isolates belonging to the Diaporthe eres species complex, based on their spectroscopic properties—UV-Vis and mass spectrometry (Table 2). The identified metabolites mainly included polyketides, pyrones, fatty acids/oxylipins, chromones, sesquiterpenoids, phthalides, and numerous derivatives and hybrids belonging to the preceding groups of compounds. The metabolite profile of the studied isolates belonging to the Diaporthe eres species complex is unique and most of the detected compounds have not been described for Diaporthe species before. Furthermore, as far as we know, our research on the characterization of the metabolite profile of D. eres species complex isolated from orchard plants is pioneering and has not been conducted in Europe or other parts of the world before. There are few publications on Diaporthe (=Phomopsis) from fruit plants, but they focus mainly either on Diaporthe from the one host plant or on the selected group of metabolites produced by Diaporthe [35,36].

Table 2.
Annotation of specific metabolites in the studied Diaporthe isolates using UHPLC-qTOF-MS/MS in the negative (NI) and positive ionization (PI) modes.
2.2.1. Polyketides
The UHPLC-HRESIMS analysis of extracts from Diaporthe isolates led to the annotation of several compounds from the polyketide group—pyranones such as, dihydrohydroxyphomopsolide B isomers I–III (17, 23, 32), dihydrophomopsolide A (49), dihydrohydroxyphomopsolide A (26), and furanones such as phomopsolidone B (37) and dihydrohydroxyphomopsolidone B isomers I and II (15, 16) (Figure 1). These compounds were tentatively identified based on the high-resolution mass of the precursor ions and the fragments generated via common fragmentation pathways in positive ionization mode. Namely, the loss of one or two water molecules (−18 Da or 36 Da), followed by the loss of a tiglic acid (2-methylbut-2-enoic acid) residue (-C5H8O2), giving intense fragment ions with m/z 179 or 197 for compounds 17, 23, and 32 ion at m/z 177 for compounds 26 and 49, and m/z 181 for compounds 15, 16, and 37, was observed (Table 2). Phomopsolides are common secondary metabolites derived from Diaporthe [25]. They were initially isolated from Phomopsis oblonga, a fungus that provided some protection against elm bark beetle infestations [37]. They have been proved for their antibacterial activity against Staphylococcus aureus [38]. Moreover, phomopsolide A/C (62), from the endophytic Diaporthe sp. AC1 from Artemisia argyi, was proved to inhibit the growth of Fusarium graminearum, F. moniliforme, Botrytis cinerea, and Verticillium dahliae, indicating that the compound may have a broad spectrum of antifungal activity [29].
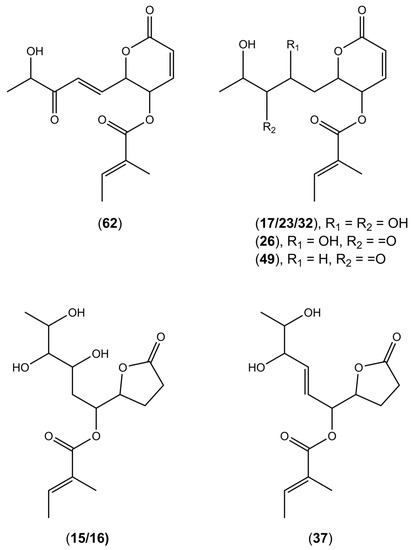
Figure 1.
Putative structures of polyketides found in the tested Diaporthe isolates.
2.2.2. Pyrones
Pyrones represent a class of oxygen-based heterocyclic compounds that naturally occur in two isomeric forms as either 2-pyrone (α-pyrone) or 4-pyrone (γ-pyrone). The number 2/4 is assigned based on the position of the carbonyl group relative to the oxygen atom within the ring system [39]. In our study, Diaporthe spp. isolated from fruit trees produced phomopsinone A (31) and pyrenocine P (3) (Figure 2), which belong to the α-pyrones. Their fragmentation spectra showed mainly water (−18 Da) and/or CO losses (−28 Da). However, characteristic UV maxima at around 280 nm indicated α-pyrone structures (Table 2). Previously, phomopsinone A and pyrenocine J-M have been isolated from the endophytic fungus Phomopsis sp. and have shown antifungal, antibacterial, and antialgal activity [27,28]. Phomopsinone A showed very strong antifungal activity against Botrytis cinerea, Pyricularia oryzae, and Septoria tritici. Pyrenocine J-M had strong antibacterial activity especially against the gram-negative bacterium E. coli, since gram-negative bacteria are usually difficult to inhibit. Similarly, all mentioned compounds showed algicidal activity against Chlorella fusca [27,28].
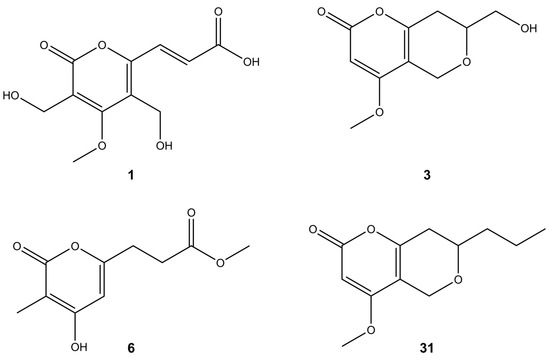
Figure 2.
Putative structures of pyrones found in the tested Diaporthe isolates.
The studied Diaporthe isolates, apart from the metabolite characteristics for the genus Diaporthe, also produced several new bioactive compounds usually present in the other species of fungi but not in Diaporthe [39]. For example, islandic acid-II (1), originally isolated from Penicillium islandicum, in the literature was reported as showing the complete growth inhibition of Yoshida sarcoma tumor cells [40]. Another compound produced by the tested Diaporthe isolates was scirpyrone K (6) (Figure 2). Its fragmentation pathway was very similar to that of compound 3. Previously, it had been isolated from a marine fungus identified as Phialocephala sp. strain FL30r. This compound exhibited weak radical scavenging activity with no cytotoxic activities reported [41].
2.2.3. Oxylipins
Oxylipins constitute a large family of oxidized fatty acids and their derivatives. Bioactive lipid production is widespread among many organisms including filamentous fungi [42]. In many cases, oxylipins have a role in both organismal development and communication with the host on a cellular basis [43,44]. The literature showed that fungal oxylipins are involved in influencing processes in infected host tissues, presumably by mimicking endogenous signal molecules [45,46]. Fungi have the ability to use the host plant’s oxylipin to achieve their own benefits. For example, by increasing the production of toxins, they improve their virulence [45,46], and by increasing sporulation they can accelerate reproduction in the tissues of the host plant [47]. Additional functions of fungal oxypilins have also been reported. They are related to fungal development regulation, metabolism, and host-pathogen interaction [42,48,49]. The synthesis of oxylipins proceeds due to substrates released by phospholipids and acylglycerides such as: oleic, linoleic, linolenic, and arachidonic acids [50,51]. Various reactions occurring in an oxidizing environment, in combination with enzymatic activity, contribute to the formation of various oxylipins from a given fatty acid. [52]. In our study we have tentatively identified thirty-seven oxylipins of predominantly C18 chain (Table 2); among them, trihydroxyoctadecenoic acid isomers I-VII (56, 69, 71, 74, 81, 83, 85) have been found in the tested Diaporthe isolates. It should be mentioned that the differences in fragmentation patterns between structural isomers were minimal and did not allow us to determine the position of double bonds or hydroxyl groups in the analyzed compounds. Previously, similar metabolites have been produced in the tubers of taro (Colocasia antiquorum) as a defense response to inoculation with black rot fungus (Ceratocystis fimbriata) [53]. They were isolated for the first time from the Chinese truffle Tuber indicum [54]. It has been proven, for example, that (9S,12S,13S)-tri-hydroxyoctadeca-10E-enoic acid had antifungal activities against Magnaporthe grisea causing rice blast disease [55], and (13S)-hydroxy-9,11-octadecadienoic acid had nematocidal properties [56].
2.2.4. Chromones
Chromones are naturally occurring phenolic derivatives of chromone (1,4-benzopyrone or 4H-chromen-4-one) and are isomers of coumarin. They are produced abundantly by many genera of plants, being a part of a normal healthy diet and by fungi. This class of compounds is mainly associated with antioxidant, antimicrobial, anticancer, and anti-inflammatory activities [57]. In our study, Diaporthe spp. produced phomochromone A (45) (Figure 3), which can exhibit an antifungal, antibacterial, and algicidal activities, which is supported by the literature. For example, two new chromones, phomochromone A and B, have been isolated from the endophytic fungus Phomopsis spp. from Cistus monspeliensis which showed good antifungal, antibacterial, and algicidal properties towards Septoria tritici, Microbotryum violaceum, Botrytis cinerea, E. coli, Bacillus megaterium, and Chlorella fusca [58]. Amycolachromone E (52) (Figure 3) and the series of other chromone derivatives were isolated from the deep-sea marine actinomycete Amycolatopsis sp. [59].

Figure 3.
Putative structures of chromones found in the tested Diaporthe isolates.
2.2.5. Sesquiterpenoids
Drimane-type sesquiterpenoids are a large group of compounds that have been found in plants and fungi, exhibiting various biological activities [60,61]. During the research conducted by Zang et al. [62] and Chen et al. [63], a variety of new drimane-type metabolites, including diaporols B–I (104), Q, and R, have been isolated from the mangrove endophytic Diaporthe sp. [62,63]. Furthermore, two drimane-type sesquiterpenoids, named altiloxins A (65) and B (77) (Figure 4), showing phytotoxic activity on the lettuce seedlings were obtained from Phoma asparagi [64]. Considering the fragmentation spectra of compounds 65, 77, and 104, in the Diaporthe isolates studied, we determined the presence of a number of their derivatives—dihydro-altiloxin A (54), dihydro-altiloxin B (72), hydroxy-altiloxin A isomers I and II (19, 22), hydroxy-altiloxin B isomers I and II (33, 50), and deoxy-altiloxin A (102).
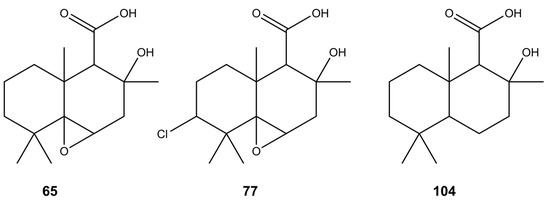
Figure 4.
Putative structures of sesquiterpenoids found in the tested Diaporthe isolates.
2.2.6. Phthalides
Phthalides are natural substances used in traditional medicine in Asia, Europe, and North America, which can be found both in plants and fungi [65,66,67,68,69]. In our study Diaporthe spp. produced the convolvulanic acid A isomers I–II (9,13), which was previously reported from Phomopsis convolvulus, a host-specific pathogen of field bindweed (Convolvulus arvensis) [66]. This metabolite showed phytotoxic activity against C. arvensis, proving that it could be used as an herbicide to control this weed effectively [66].
2.2.7. Hybrid Compounds
HRESIMS analysis revealed a molecular formula of C27H33ClO10 ([M − H]− at m/z 551.1692) for compound 135, suggesting close structural analogy to pestalotiopene A [68]. The structural similarity of both compounds was further corroborated by detecting the same mass fragment at m/z 301.1206 with the characteristic chlorine isotope splitting, corresponding to the altiloxin B part of pestalotiopene A. Previously, drimane sesquiterpene-cyclopaldic acids hybrids, pestalotiopens A and B, were isolated from the mangrove-derived fungus Pestalotiopsis sp. obtained from leaves of the Chinese mangrove Rhizophora mucronate [68]. Pestalotiopen A (135), an altiloxin B—O-methylcyclopaldic acid hybrid, showed moderate antibacterial activity against Enterococcus faecalis [68]. Cyclopaldic acid was also produced by Seiridium cupressi, the pathogen of a canker disease of cypress, showing phytotoxic and antifungal activity [67], and by Coccomyces strobi isolated from needles of Pinus strobus, showing moderate growth inhibition of Microbotryum violaceum (=Ustilago violacea) and weak antibiotic activity against Bacillus subtilis, with no inhibition observed against E. coli at the highest tested concentration [69]. In the search for natural products as an alternative to synthetic pesticides, cyclopaldic acid has been reported to possess insecticidal [70], fungicidal [71], as well as herbicidal [72] activities. Recently, Samperna et al. [73], during the investigation of the effects of cyclopaldic acid in Arabidopsis thaliana plants and protoplasts, showed that this metabolite induced leaf chlorosis, ion leakage, membrane-lipid peroxidation, hydrogen peroxide production, and inhibited root proton extrusion in vivo and plasma membrane H+-ATPase activity in vitro. In our study, we report the presence of over twenty-five compounds, ethers of altiloxin A and its derivatives with cyclopolic acid (51, 88, 103, 109, 126, and 127), and ethers of altiloxin B and its derivatives with either (iso)cyclopaldic acid, cyclopolic acid, or salfredins A7/C3 (57, 63, 66, 70, 76, 79, 87, 90, 91, 93, 95, 98, 101, 106, 107, 112–115, 117, 122, and 135) (Figure 5). The identity of these compounds was tentatively established by the similarity of fragmentation spectra to those of compound 135 (Table 2).
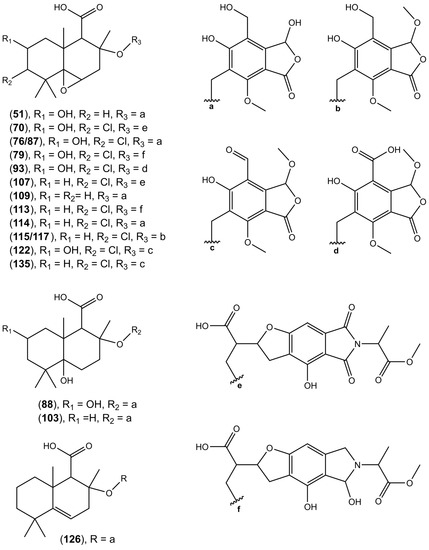
Figure 5.
Putative structures of hybrid sesquiterpenoids-phthalides found in the tested Diaporthe isolates.
2.3. Metabolite-Based Chemotaxonomy
As a preliminary step in multivariate statistical analysis, PCA analysis provided an unsupervised overview of LC-MS fingerprints obtained in both ionization modes (NI and PI). Both NI and PI PCA score plots revealed a close clustering of the QC samples (Figure 6A), indicating that the separation, observed between fungal isolates into two distinct chemotypes was mainly due to biological reasons.
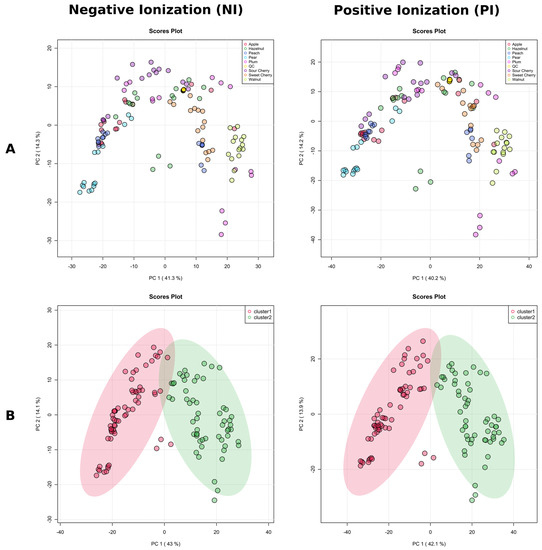
Figure 6.
The score plots of principal component analysis (PCA) in negative ionization mode and positive ionization mode LC-MS data of the tested Diaporthe isolates, where each point represents a single isolate. (A) PCA colored by the host plant. (B) Isolates in the PCA are colored by k–means clustering cluster assignments from negative ionization mode; the elliptic areas represent the 95% confidence regions.
To avoid biased group assignment of the PCA plots, samples were statistically assigned into 2 clusters (chemotypes) based on the k–means clustering algorithm in NI mode, and the groups generated by the k–means clustering algorithm in the negative mode were assigned to the positive mode (Figure 6B). The clustering of the data was easily visualized in both ionization modes, and confirmed by clusters obtained separately by HCA (Figure 7A,B). The first five PCs explained 75.1% of the variance in NI and 75.4% in PI modes, and 57.1% of the total variance was projected in the first two PCs in NI, while 56.0% in PI, which suggested the similar quality of data obtained in both ionization modes. Indeed, the PCA score plots showed similar patterns with specific host plants (understood as metadata) grouped together (pear, sweet cherry, and walnut), while the rest were much more dispersed, and there were no clear associations between the metadata and the groups in the PCA. We decided to use NI mode for further work due to the lower complexity of LC-MS data (high amount of in-source collision-induced dissociation in PI).
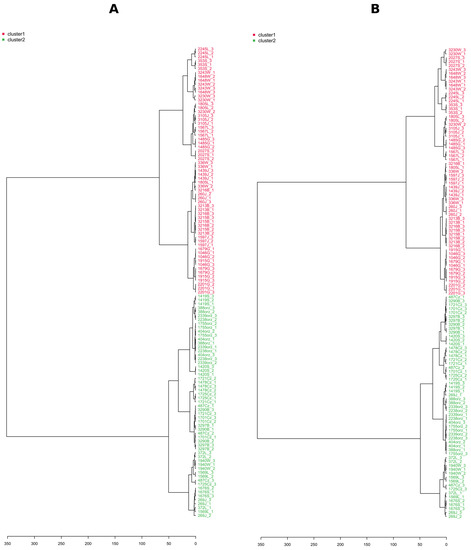
Figure 7.
LC-MS-based hierarchical cluster analyses (HCA, Pearson distance, and Ward’s linkage rule) show the tested Diaporthe isolates following differentiation in negative ionization mode (A) and positive ionization mode (B). Letters in isolate names refer to the host plant: J = Apple; L = Hazelnut; B = Peach; G = Pear; S = Plum; W = Sour Cherry; Cz = Sweet Cherry; and orz = Walnut. Numbers 1, 2, and 3 after the underscore refer to individual biological repetitions.
To validate the k–means/HCA model and to identify the features responsible for the classification, we performed a supervised PLS-DA analysis, and overall, 52.1% of the total variance was displayed on the first two principal component axes of the PLS-DA score plot (Figure 8A), with R2X = 0.946 and Q2 = 0.924 calculated from the first three components via a 10-fold cross-validation method, with Q2 as the measured performance. Since PLS-DA tends to overfit data, the model was validated to understand whether the separation is statistically significant or is due to random noise. This hypothesis was tested using the permutation test—separation distance (B/W), with 100 permutations with observed statistics having a p < 0.01 (Figure 8C).
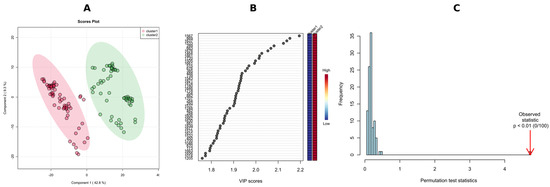
Figure 8.
PLS-DA score plots of clusters in NI-mode-based k–means clustering of the tested Diaporthe isolates; the elliptic areas represent the 95% confidence regions (A); the top 50 features ranked based on scores of VIP, features are numbered based on MS-DIAL ID (see Table 2) (B); and permutation test results of the PLS-DA model (statistical test: separation distance (B/W)), the number of permutations set at 100 (C).
A p-value below 0.01 in 100 permutations means that not even once (<0.01 × 100) did the permutated data yield a better performance (higher B/W) than the original label, suggesting the significant difference between these two clusters.
Potential variables to separate clusters 1 and 2 in the dendrogram were identified as potential biomarkers using VIP values which estimate the importance of each variable in the projection used in a PLS-DA model. The greater-than-one rule is usually considered for detecting the descriptors with the greatest importance in the projection. However, we decided, due to a large number of significant metabolites (>300), to use VIP scores > 1.8 (Figure 8B). The peak intensity ratios were also subjected to an unpaired non-parametric test (Wilcoxon rank-sum test, also known as the Mann–Whitney U test) within MetaboAnalyst, and false discovery rates (FDR < 0.05) were calculated to discover if those features are significantly different between cluster 1 and 2. A large fold change (FC > 10) between the two putative chemotypes was also considered a selection criterion, with FC > 100 indicating the presence/absence of the feature in question. As a result, 43 features meeting these conditions (VIP = 2.20–1.81, FDR adj. p-value = 3.23 × 10−19–4.71 × 10−18, FC = 348–19) were selected (Table 3) for receiver operating characteristic (ROC) analysis in order to assess their potential as chemotaxonomical biomarkers. ROC curves are used to evaluate classification and prediction models in bioinformatics. They are often summarized in a single metric known as area under the curve (AUC), where AUC = 1.0 indicates an excellent classifier and AUC = 0.5 means the classifier has no practical utility [74]. In this regard, we calculated the AUC for each selected candidate biomarker, and the AUC values obtained ranged from 0.972 to 1.000 (Table 3). Furthermore, to consider factors other than genetics, i.e., host plant, year of strain isolation or storage time, a combination of multiple individual markers must be considered into a single multivariate model, providing improved levels of discrimination and confidence. To this end, we applied the PLS-DA model to combine our 43 selected markers to obtain the AUC (Figure 9A), and predicted the classification probability into each chemotype (Figure 9B). The performance of this model was tested using a balanced Monte-Carlo cross-validation procedure, and as a result the average accuracy based on 100 cross-validations was 0.991.

Table 3.
The 43 top ranked features contributing to the group discrimination in PLS-DA and marked as potential biomarkers for the tested Diaporthe isolates from data generated in NI mode.
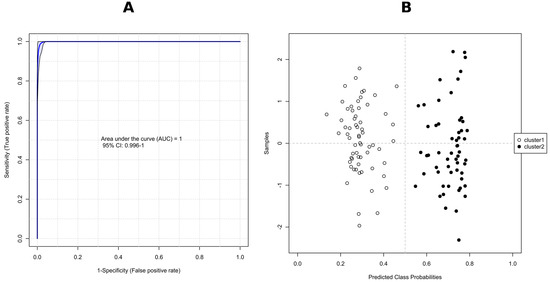
Figure 9.
ROC curve for combined biomarker models (set of 43 metabolites); 100 cross-validations were performed, and the results were averaged to generate the plot (A); The average of predicted class probabilities of each sample across the 100 cross-validations. As the algorithm uses a balanced sub-sampling approach, the classification boundary is located at the center (x = 0.5, the dotted line). The corresponding confusion matrix showed that all isolates were correctly classified in all cases (B).
Hierarchical clustering with a heat map is also shown to easily visualize the concentration variation of the top 100 tentatively identified metabolites (according to t-tests) expressed in the tested Diaporthe isolates (Figure 10). A sharp contrast of their accumulation is observed, while at the same time the samples are clearly grouped by their group membership, determined by HCA and k–means analyses.
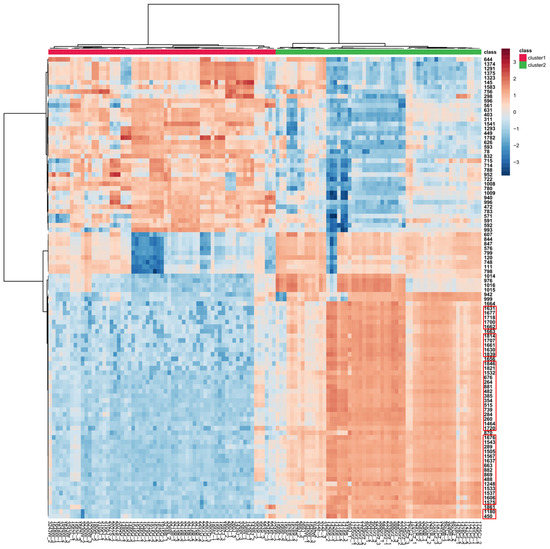
Figure 10.
Hierarchical clustering with the heat map generated from the top 100 tentatively identified metabolites present in the tested Diaporthe isolates, according to t-tests, using Pearson distance for similarity measure and Ward’s linkage algorithm for clustering. Clusters were grouped based on the HCA/k–means analyses shown in Figure 6B and Figure 7. Cell colors indicate relative concentration values as high (dark brown) or low (dark blue), with samples in columns and features (MS DIAL ID in NI) in rows. Features from Table 3 are enclosed in red rectangles.
The study on the utilization of metabolites as chemotaxonomic markers for species identification refers to the genus Penicillium, Aspergillus, Fusarium, Alternaria, and the Xylariaceae family [75,76]. However, in the case of Diaporthe, this type of research was limited. In the research conducted by Horn et al. [77,78] on endophytic Phomopsis (=Diaporthe) from woody host, three metabolites named phomodiol, phomopsolide B, and phomopsichalasin were indicated as potential chemotaxonomic markers for this fungi. In addition, Abreu et al. [79] showed that the production of secondary metabolites by Phomopsis and related Diaporthales may be species-specific, indicating the value of utilizing the metabolic analysis in taxonomic research on closely related species.
In our research, isolates belonging to the Diaporthe eres species complex isolated from fruit trees produced 153 metabolites from which 43 were recognized as potential chemotaxonomic markers, mostly belonging to the drimane sesquiterpenoid—phthalide hybrid class. This group included mainly phytotoxic compounds such as cyclopaldic acid and altiloxin A, B and their derivatives. It is noteworthy that during our investigation, the phytotoxic compound cyclopaldic acid was produced not only by the pathogenic Diaporthes species but also by the endophytic D. eres isolate 1420S, previously described by Abramczyk et al. [32] and used in the present study. Following the observations of Graniti et al. [67] and McMullin et al. [69], the production of phytotoxic cyclopaldic acid may be related to Diaporthe changing its lifestyle from endophytic to pathogenic, under favorable conditions. Thus, it is possible that endophytic D. eres isolate 1420S [32], is a weak opportunistic pathogen, switching from an endophytic to a pathogenic phase when the host tissue becomes weakened. This issue requires more advanced research in the future.
3. Materials and Methods
3.1. Chemicals and Reagents
Hypergrade for LC-MS acetonitrile (≥99.9%) and HPLC gradient-grade methanol (≥99.9%) were purchased from Merck (Darmstadt, Germany), LC-MS grade formic acid (98–100%) was purchased from Sigma Aldrich (Steinheim, Germany). A Milli-Q Simplicity 185 water purification system from Millipore (Milford, MA, USA) was used for preparation of ultrapure water (18.2 MΩ·cm).
3.2. Fungal Strains and Culture Conditions
We investigated 40 Diaporthe strains isolated during previous studies from different species of fruit trees growing in south-eastern Poland (Table 1) [4,31]. All axenic cultures were deposited at the Fungal Collection of Phytopathology and Mycology Subdepartment, University of Life Sciences in Lublin (Poland). Thirty-nine came from shoots with visible disease symptoms and one from healthy Prunus domestica as endophyte, described previously by Abramczyk et al. [32]. Diaporthe strains were isolated according to the methodology described by Król [80]. Healthy fragments of the tested plants were properly disinfected by rinsing several times, first in a 10% sodium hypochlorite solution, then in sterile distilled water. After drying, the plant fragments were placed on potato dextrose agar (PDA, Difco) and incubated for 5 days at 25°C, in the dark. When the fungus colonies appeared, pure cultures were prepared according to the methodology described previously [80].
3.3. DNA Extraction, Amplification and Sequencing
Strains were incubated on PDA at 25 °C for 7 days before to DNA extraction. The total genomic DNA was extracted using the FastDNA®SPIN Kit and the FastPrep®Instrument (Qbiogene, Inc., Carlsbad, CA, USA), according to the manufacturer’s protocol. All extracted DNA was stored at −20 °C until use.
The amplification of the fragment of the internal transcribed spacer region (ITS) of the nuclear ribosomal RNA gene, the universal primers ITS1: TCCGTAGGTGAACCTGCGG and ITS4: TCCTCCGCTTATTGATATGC were used [81]. For the amplification of ITS regions, 25 μL of the reaction mixture was prepared, which consisted of the following components: 1 μL of genomic DNA (5 ng/μL), DreamTaq™ Green PCR Master Mix (2×) (Thermo Scientific, Waltham, MA, USA) in a volume of 12.5 μL, primers (10 μM) in a volume of 1 μL each and purified water in a volume of 9.5 μL. The PCR reaction was run under the following conditions: 95 °C for 3 min, followed by 39 cycles of 95 °C for 30 sec, 55 °C for 50 sec, 72 °C for 1 min, and final extension at 72 °C for 10 min. Sequencing of the obtained PCR products was performed in the Genomed S.A. (Warsaw, Poland). The sequence data received were deposited in GenBank (Table 1). The Bionumerics 7.6 (Applied Maths NV., Sint-Martens-Latem, Belgium) and SEED v.2.1.05 (Institute of Microbiology CAS, Prague, Czech Republic) software was used for bioinformatic analyses.
The obtained sequences were blasted against the NCBIs GenBank nucleotide database to determine the closest related species.
3.4. Extraction of Fungal Metabolites
For metabolite extraction, 40 Diaporthe strains were three-point inoculated on 90 mm Petri plates containing PDA, and incubated for 28 days at 23 °C under a 12 h photoperiod, referring to the methodology of Abreu et al. [80], with modifications. Fungal discs (5-mm diameter) were collected in three individual biological repetitions each (n = 3). Each fungal culture (120 total) and three non-inoculated medium samples were freeze-dried (Christ Gamma 1–16 LSC, Martin Christ, Osterode am Harz, Germany) and subsequently ground with a mortar. Dried material (25 mg) was transferred to a 5 mL screw-capped centrifuge tube (Eppendorf, Hamburg, Germany) and added to 2.5 mL of extraction solvent mixture, MeOH/H2O 80:20 (v/v). Samples were then thoroughly vortex-mixed for 1 min and ultrasonicated for 20 min under 4 °C. Samples were centrifuged (18,000× g for 20 min under 4 °C), and the supernatants were transferred to separate vials and analyzed using UHPLC-QTOF HRMS. A QC (Quality Control) sample (aliquot of all samples) was also prepared and injected six times before randomized sample injection for column conditioning and at every forty samples to evaluate the performance of the LC-MS method during the detection.
3.5. UHPLC-QTOF HRMS Profiling
Ultrahigh-performance liquid chromatography-quadrupole time of flight-high-resolution MS (UHPLC-QTOF HRMS) analyses were performed on an Impact II HD mass spectrometer (Bruker, Billerica, USA) coupled to a U-HPLC Ultimate 3000 RSLC system (Thermo Fisher Scientfic, Hemel Hempstead, UK). Five-microliter injections of samples were fed from a thermostatted autosampler (8 °C) onto a CORTECS T3 column (150 mm × 2.1 mm i.d., 2.7 μm, Waters, Milford, USA), equipped with a guard column, and the column was kept at 35 °C. Mobile phases were (A) ultrapure water with 0.1% formic acid (FA), and (B) acetonitrile with 0.1% FA. The flow rate was set at 500 μL/min and the solvent gradient profile was as follows: 0.0–1.0 min, 5% B; 1.0–27.0 min, 5–99% B (concave-shaped gradient—Dionex gradient curve 6); 27.0–30.0 min, 99% B. Between the injections, the column was equilibrated with six volumes of 5% B. Mass detection was performed using an electrospray source in positive ionization (PI) and negative ionization (NI) modes. Ionization spray voltages were set to 4.0 kV (for PI) and 3.0 kV (for NI); dry gas flow was 6 l/min; the dry gas temperature was 200 °C; collision cell transfer time was 90 μs; and nebulizer pressure was 0.7 bar. MS1 and MS/MS data (range 80–1800 m/z) were collected using Bruker DataAnalysis 4.3 software in data-dependent acquisition (DDA) mode—after each full MS1 scan, the two most intense ions were fragmented with collision energies of 20 eV for PI and 30 eV for NI.
3.6. Data Processing and Metabolite Identification
LC-MS raw data were first converted into the ‘Analysis Base File’ (ABF) format [82] using Reifycs Abf (Analysis Base File) Converter (https://www.reifycs.com/AbfConverter/ (accessed on 25 May 2021)) and processed with MS-DIAL (RIKEN, version 4.90) [83]. MS1 and MS2 tolerances were set to 0.01 and 0.05 Da, respectively, in centroid mode for each data set (PI and NI). In PI and NI modes, automatic feature detection was performed between 3.0 and 27.0 min for mass range between 80 and 1800 Da. The minimum peak height intensity was set to 2000 for NI and 3000 for PI modes, respectively; linear-weighted moving average as the smoothing method using 5 scans and peak width 5 scans. Peaks were aligned on a QC reference file with an RT tolerance of 0.10 min and a mass tolerance of 0.015 Da and retained in the feature table if they appeared in at least 3 samples. All peaks detected from non-inoculated medium were removed from the generated matrix if their “Sample average/blank average” ratio was lower than 10, thus removing the background and contaminants and preserving the true biological mass signals from LC-MS data.
The kept significant features were exported to the MS-FINDER program (RIKEN, version 3.52) for in silico-based annotation using the hydrogen rearrangement rules (HRR) scoring system [84]. The MS1 and MS2 tolerances were set to 10 and 25 ppm, respectively, and the isotopic ratio tolerance set to 20%. The formulas were filtered to exclusively contain only C, H, O, N, P, S, and Cl atoms. Selected compounds were searched against the built-in database in the MS-FINDER system: NANPDB (Northern African Natural Products Database), KNApSAcK, COCONUT, T3DB (the toxin and toxin target database), and NPA (Natural Products Atlas), and only structures with a score above 5 were retained for thorough analysis. Fungal metabolites were tentatively identified by their high-resolution mass data, MS/MS fragmentation pattern analysis, UV data, and published literature.
3.7. Multivariate Statistical Analysis
The aligned data table was LOWESS (locally weighted scatterplot smoothing), normalized using the pooled QC samples and exported from MS-DIAL software to comma-separated value (CSV) format prior to analysis using MetaboAnalyst (version 5.0) [85]. The data were filtered by removing variables showing low repeatability among QC samples (RSD > 20%). Two data matrices were constructed, one in PI mode (120 isolates × 3557 metabolites) and the second in NI mode (120 isolates × 1759 metabolites). The samples were then normalized by the sum to account for the effects of sample dilution (different content of culture medium in the samples), data were log10-transformed to correct for heteroscedasticity and Pareto-scaled to reduce the influence of intense peaks, which transformed the data matrix into a more Gaussian-type distribution [86,87]. First, unsupervised principal component analysis (PCA) was used as an exploratory data analysis to provide an overview of LC-MS fingerprints. Unsupervised groups from the PCA were assigned by k–means clustering analysis and confirmed by hierarchical cluster analysis (HCA) performed to obtain a dendrogram of fungal strains according to metabolite profiling (Pearson distance measure, Ward’s clustering algorithm). On the clusters obtained, a partial least squares discriminant analysis (PLS-DA) was conducted using clusters as Y value, and their potential variables were selected based on variable importance in projection (VIP > 1.0) values and false discovery rate (FDR < 0.05) by Wilcoxon rank-sum test.
4. Conclusions
The results of our study demonstrated a rich diversity of metabolites secreted by the tested Diaporth eres species complex. The characterization of these compounds could be the basis for the future research on their isolation and bioactive potential for agricultural applications as biopesticides or biofertilizers.
Furthermore, the future research should include a larger population of Diaporthe from fruit plants from various areas of Poland. It would be worth determining their metabolic profile, then isolating more important compounds to confirm their structure and bioactive properties. In addition, the optimization of culture media and cultivation conditions for producing richer metabolite profiles are necessary for a more conclusive chemical classification of these fungi.
Although the bioactivity of cyclopaldic acid and altiloxins (the main components of the drimane sesquiterpenoid—phthalide hybrids) identified in the present study as potential biomarkers for species belonging to the Diaporthe eres complex is known, as described above, the genes involved in their biosynthesis have not yet been defined. In general, the eukaryotic genes involved in a single metabolic pathway are scattered throughout the genome, whereas the genes required for a fungus to produce a given secondary metabolite are very frequently clustered, adjacent to one another on the chromosome [88]. Such clusters are found in the majority of filamentous fungi and may range from only a few to more than 20 genes [89]. Thus, identifying a biosynthetic gene cluster for the main compounds reported as biomarkers for species from Diaporthe eres complex, could be the next step to supplement the current research by the results relied on the genetic methods used on a larger Diaporthe population.
Over the last decade, multi-locus DNA sequence data and morphological characterization have been extensively used to identify Diaporthe on a species level [3,7,10,20,21,90,91,92]. The gene regions most commonly used for this purpose in Diaporthe are the internal transcribed spacer (ITS), together with translation elongation factor-1α (EF-1α), β-tubulin, partial histone H3 (HIS), and calmodulin (CAL) [3,6,20,21,93,94]. However, they are still limited to those species for which the comparative sequence data have been deposited in the public database. Nevertheless, a multi-locus sequencing should always be used for identification of Diaporthe species [6]. In agreement with the study of Abreu et al. [79] and Horn et al. [77], the metabolite profiling may support phenotypic species recognition in Diaporthe. Thus, when studying closely related species in the Diaporthe eres complex, a holistic approach combining morphological characterization, metabolic profile and multi-locus sequencing for species identification is certainly worth considering [79].
Characterizing metabolites biosynthesized by Diaporthe infecting shoots of fruit trees is vital for the phytotoxic properties and chemotaxonomy. It is also essential to better understand the conditions under which the fungi start producing the toxins and switch their lifestyle from endophytic to pathogenic.
Finally, it is hoped that the results from our initial research will enrich the biodiversity of the chemical compounds of species from Diaporthe eres complex and provide a series of new information for this genus.
Author Contributions
Conceptualization, B.A. and Ł.P.; methodology, B.A., Ł.P. and M.K.; validation, B.A. and Ł.P.; formal analysis, B.A., Ł.P., S.K. and A.M.-G.; investigation, B.A., Ł.P. and S.K.; resources, B.A. and Ł.P.; data curation, B.A. and Ł.P.; writing—original draft preparation, B.A. and Ł.P.; writing—review and editing, B.A., Ł.P., S.K. and M.K.; visualization, B.A. and Ł.P.; supervision, E.K., A.G. and W.O.; project administration, B.A.; funding acquisition, B.A.; All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science Centre (NCN), Poland, the grant number 2016/21/N/NZ9/01526.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in Zenodo at https://doi.org/10.5281/zenodo.7371706 (accessed on 28 November 2022).
Acknowledgments
The Diaporthe isolates used in the present work came from the Fungal Collection of Phytopathology and Mycology Subdepartment, University of Life Sciences in Lublin (Poland).
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Not applicable.
References
- Rossman, A.Y.; Adams, G.C.; Cannon, P.F.; Castlebury, L.A.; Crous, P.W.; Gryzenhout, M.; Jaklitsch, W.M.; Mejia, L.C.; Stoykov, D.; Udayanga, D. Recommendations of generic names in Diaporthales competing for protection or use. IMA Fungus 2015, 6, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Udayanga, D.; Liu, X.; McKenzie, E.H.C.; Chukeatirote, E.; Bahkali, A.H.A.; Hyde, K.D. The genus Phomopsis: Biology, applications, species concepts and names of common phytopathogens. Fungal Divers. 2011, 50, 189–225. [Google Scholar] [CrossRef]
- Gomes, R.R.; Glienke, C.; Videira, S.I.R.; Lombard, L.; Groenewald, J.Z.; Crous, P.W. Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 2013, 31, 1–41. [Google Scholar] [CrossRef]
- Król, E.D.; Abramczyk, B.A.; Zalewska, E.D.; Zimowska, B. Fungi inhabiting fruit tree shoots with special reference to the Diaporthe (Phomopsis) genus. Acta Sci. Pol. Hortorum Cultus 2017, 16, 113–126. [Google Scholar] [CrossRef]
- Sun, W.; Huang, S.; Xia, J.; Zhang, X.; Li, Z. Morphological and molecular identification of Diaporthe species in south-western China, with description of eight new species. MycoKeys 2021, 77, 65–95. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.; Alves, A.; Alves, R. Evaluating multi-locus phylogenies for species boundaries determination in the genus Diaporthe. PeerJ 2017, 5, e3120. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Groenewald, J.Z.; Woodhall, J.; Armengol, J.; Cinelli, T.; Eichmeier, A.; Ezra, D.; Fontaine, F.; Gramaje, D.; Gutierrez-Aguirregabiria, A. Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia 2018, 40, 135–153. [Google Scholar] [CrossRef]
- Farr, D.F.; Castlebury, L.A.; Pardo-Schultheiss, R.A. Phomopsis amygdali causes peach shoot blight of cultivated peach trees in the southeastern United States. Mycologia 1999, 91, 1008–1015. [Google Scholar] [CrossRef]
- Karaoglanidis, G.S.; Bardas, G. First report of Phomopsis fruit decay on apple caused by Phomopsis mali in Greece. Plant Dis. 2006, 90, 375. [Google Scholar] [CrossRef]
- Guo, Y.S.; Crous, P.W.; Bai, Q.; Fu, M.; Yang, M.M.; Wang, X.H.; Du, Y.M.; Hong, N.; Xu, W.X.; Wang, G.P. High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia 2020, 45, 132–162. [Google Scholar] [CrossRef]
- Harris, D.C. Diaporthe perniciosa associated with plum dieback. Plant Pathol. 1988, 37, 604–606. [Google Scholar] [CrossRef]
- Król, E. Identification and differentiation of Phomopsis spp. isolates from grapevine and some other plant species. Phytopathol. Pol. 2005, 35, 151–156. [Google Scholar]
- Baumgartner, K.; Fujiyoshi, P.T.; Travadon, R.; Castlebury, L.A.; Wilcox, W.F.; Rolshausen, P.E. Characterization of species of Diaporthe from wood cankers of grapes in Eastern North American vineyards. Plant Dis. 2013, 97, 912–920. [Google Scholar] [CrossRef]
- Elfar, K.; Torres, R.; Díaz, G.A.; Latorre, B.A. Characterization of Diaporthe australafricana and Diaporthe spp. associated with stem canker of blueberry in Chile. Plant Dis. 2013, 97, 1042–1050. [Google Scholar] [CrossRef]
- Uecker, F.A. A world list of Phomopsis names with notes on nomenclature, morphology and biology. Mycol. Mem. 1988, 13, 1–231. [Google Scholar]
- Rehner, S.A.; Uecker, F.A. Nuclear ribosomal internal transcribed spacer phylogeny and host diversity in the coelomycete Phomopsis. Can. J. Bot. 1994, 72, 1666–1674. [Google Scholar] [CrossRef]
- Mostert, L.; Crous, P.W.; Kang, J.C.; Phillips, A.J.L. Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: Morphological, cultural, molecular and pathological characterization. Mycologia 2001, 93, 146–167. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Vitale, A.; Cirvilleri, G.; Aiello, D.; Susca, A.; Epifani, F.; Perrone, G.; Polizzi, G. Characterisation and pathogenicity of fungal species associated with branch cankers and stem-end rot of avocado in Italy. Eur. J. Plant Pathol. 2016, 146, 963–976. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Crous, P.W. Emerging citrus diseases in Europe caused by species of Diaporthe. IMA Fungus 2017, 8, 317–334. [Google Scholar] [CrossRef]
- Udayanga, D.; Castlebury, L.A.; Rossman, A.Y.; Chukeatirote, E.; Hyde, K.D. Insights into the genus Diaporthe: Phylogenetic species delimitation in the D. eres species complex. Fungal Divers. 2014, 67, 203–229. [Google Scholar] [CrossRef]
- Udayanga, D.; Castlebury, L.A.; Rossman, A.Y.; Hyde, K.D. Species limits in Diaporthe: Molecular re-assessment of D. citri, D. cytosporella, D. foeniculina and D. rudis. Persoonia 2014, 32, 83–101. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Udayanga, D.; Manamgoda, D.S.; Tedersoo, L.; Larsson, E.; Abarenkov, K.; Bertrand, Y.J.K.; Oxelman, B.; Hartmann, M.; Kauserud, H.; et al. Incorporating molecular data in fungal systematics: A guide for aspiring researchers. Curr. Res. Environ. Appl. Mycol. 2013, 3, 1–32. [Google Scholar] [CrossRef]
- Norphanphoun, C.; Gentekaki, E.; Hongsanan, S.; Jayawardena, R.; Senanayake, I.C.; Manawasinghe, I.S.; Abeywickrama, P.D.; Bhunjun, C.S.; Hyde, K.D. Diaporthe: Formalizing the species-group concept. Mycosphere 2022, 13, 752–819. [Google Scholar] [CrossRef]
- Bhunjun, C.S.; Niskanen, T.; Suwannarach, N.; Wannathes, N.; Chen, Y.-J.; McKenzie, E.; Maharachchikumbura, S.; Buyck, B.; Zhao, C.-L.; Fan, Y.-G.; et al. The numbers of fungi: Are the most speciose genera truly diverse? Fungal Divers. 2022, 114, 387–462. [Google Scholar] [CrossRef]
- Xu, T.-C.; Lu, Y.-H.; Wang, J.-F.; Song, Z.-Q.; Hou, Y.-G.; Liu, S.-S.; Liu, C.-S.; Wu, S.-H. Bioactive Secondary Metabolites of the Genus Diaporthe and Anamorph Phomopsis from Terrestrial and Marine Habitats and Endophytes: 2010–2019. Microorganisms 2021, 9, 217. [Google Scholar] [CrossRef]
- Chepkirui, C.; Stadler, M. The genus Diaporthe: A rich source of diverse and bioactive metabolites. Mycol. Prog. 2017, 16, 477–494. [Google Scholar] [CrossRef]
- Hussain, H.; Ahmed, I.; Schulz, B.; Draeger, S.; Krohn, K. Pyrenocines J–M: Four new pyrenocines from the endophytic fungus, Phomopsis sp. Fitoterapia 2012, 83, 523–526. [Google Scholar] [CrossRef]
- Hussain, H.; Krohn, K.; Ahmed, I.; Draeger, S.; Schulz, B.; Di Pietro, S.; Pescitelli, G. Phomopsinones A–D: Four New Pyrenocines from Endophytic Fungus Phomopsis sp. Eur. J. Org. Chem. 2012, 9, 1783–1789. [Google Scholar] [CrossRef]
- Gu, H.; Zhang, S.; Liu, L.; Yang, Z.; Zhao, F.; Tian, Y. Antimicrobial Potential of Endophytic Fungi From Artemisia argyi and Bioactive Metabolites From Diaporthe sp. AC1. Front. Microbiol. 2022, 13, 908836. [Google Scholar] [CrossRef]
- Sessa, L.; Abreo, E.; Lupo, S. Diversity of fungal latent pathogens and true endophytes associated with fruit trees in Uruguay. J. Phytopathol. 2018, 166, 633–647. [Google Scholar] [CrossRef]
- Abramczyk, B.A.; Król, E.D.; Zalewska, E.D.; Zimowska, B. Morphological characteristics and pathogenicity of Diaporthe eres isolates to the fruit tree shoots. Acta Sci. Pol. Hortorum Cultus 2018, 17, 125–133. [Google Scholar] [CrossRef]
- Abramczyk, B.; Marzec-Grządziel, A.; Grządziel, J.; Król, E.; Gałązka, A.; Oleszek, W. Biocontrol Potential and Catabolic Profile of Endophytic Diaporthe eres Strain 1420S from Prunus domestica L. in Poland A Preliminary Study. Agronomy 2022, 12, 165. [Google Scholar] [CrossRef]
- Nnadi, N.E.; Carter, D.A. Climate change and the emergence of fungal pathogens. PLoS Pathog. 2021, 17, e1009503. [Google Scholar] [CrossRef] [PubMed]
- Elad, Y.; Pertot, I. Climate Change Impacts on Plant Pathogens and Plant Diseases. J. Crop Improv. 2014, 28, 139–199. [Google Scholar] [CrossRef]
- Goddard, M.L.; Mottier, N.; Jeanneret-Gris, J.; Christen, D.; Tabacchi, R.; Abou-Mansour, E. Differential production of phytotoxins from Phomopsis sp. from grapevine plants showing esca symptoms. J. Agric. Food Chem. 2014, 62, 8602–8607. [Google Scholar] [CrossRef]
- Reveglia, P.; Pacetti, A.; Masi, M.; Cimmino, A.; Carella, G.; Marchi, G.; Mugnai, L.; Evidente, A. Phytotoxic metabolites produced by Diaporthe eres involved in cane blight of grapevine in Italy. Nat. Prod. Res. 2021, 35, 2872–2880. [Google Scholar] [CrossRef] [PubMed]
- Grove, J.F.J. Metabolic products of Phomopsis oblonga. Part 2. Phomopsolides A and B, tiglic esters of two 6-substituted 5,6-dihydo-5-hydroxypyran-2-ones. J. Chem. Soc. Perkin Trans. 1985, 1, 865–869. [Google Scholar] [CrossRef]
- Stierle, D.B.; Stierle, A.A.; Ganser, B. New phomopsolides from a Penicillium sp. J. Nat. Prod. 1997, 60, 1207–1209. [Google Scholar] [CrossRef]
- Bhat, Z.S.; Rather, M.A.; Maqbool, M.; Lah, H.U.; Yousuf, S.K.; Ahmad, Z. α-pyrones: Small molecules with versatile structural diversity reflected in multiple pharmacological activities-an update. Biomed. Pharmacother. 2017, 91, 265–277. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Kihara, T.; Isono, K.; Tsunoda, H.; Tatsuno, T.; Matsumoto, K.; Hirokawa, H. Studies on the Biological Activities of Islandic Acid and Related Compounds. Chem. Pharm. Bull. 1984, 32, 1583–1586. [Google Scholar] [CrossRef]
- Zhang, Z.; He, X.; Che, Q.; Zhang, G.; Zhu, T.; Gu, Q.; Li, D. Sorbicillasins AB and Scirpyrone K from a Deep-See-Derived Fungus, Phialocephala sp. FL30r. Mar. Drugs 2018, 16, 245. [Google Scholar] [CrossRef]
- Brodhun, F.; Feussner, I. Oxylipins in fungi. FEBS J. 2011, 278, 2609–2610. [Google Scholar] [CrossRef]
- Noverr, M.C.; Erb-Downward, J.R.; Huffnagle, G.B. Production of eicosanoids and other oxylipins by pathogenic eukaryotic microbes. Clin. Microbiol. Rev. 2003, 16, 517–533. [Google Scholar] [CrossRef]
- Kock, J.L.; Strauss, C.J.; Pohl, C.H.; Nigam, S. The distribution of 3-hydroxy oxylipins in fungi. Prostaglandins Other Lipid Mediat. 2003, 71, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Brodhagen, M.; Tsitsigiannis, D.I.; Hornung, E.; Goebel, C.; Feussner, I.; Keller, N.P. Reciprocal oxylipin-mediated cross-talk in the Aspergillus–seed pathosystem. Mol. Microbiol. 2008, 67, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Burow, G.B.; Gardner, H.W.; Keller, N.P. A peanut seed lipoxygenase responsive to Aspergillus colonization. Plant. Mol. Biol. 2000, 42, 689–701. [Google Scholar] [CrossRef]
- Scarpari, M.; Punelli, M.; Scala, V.; Zaccaria, M.; Nobili, C.; Ludovici, M.; Camera, E.; Fabbri, A.A.; Reverberi, M.; Fanelli, C. Lipids in Aspergillus flavus-maize interaction. Front. Microbiol. 2014, 5, 74. [Google Scholar] [CrossRef] [PubMed]
- Reverberi, M.; Punelli, F.; Scarpari, M.; Camera, E.; Zjalic, S.; Ricelli, A.; Fanelli, C.; Fabbri, A.A. Lipoperoxidation affects ochratoxin A biosynthesis in Aspergillus ochraceus and its interaction with wheat seeds. Appl. Microbiol. Biotechnol. 2010, 85, 1935–1946. [Google Scholar] [CrossRef]
- Christensen, S.A.; Kolomiets, M.V. The lipid language of plant-fungal interactions. Fungal Genet. Biol. 2011, 48, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Sakuradani, E.; Ando, A.; Ogawa, J.; Shimizu, S. Improved production of various polyunsaturated fatty acids through filamentous fungus Mortierella alpina breeding. Appl. Microbiol. Biotechnol. 2009, 84, 1–10. [Google Scholar] [CrossRef]
- Beccaccioli, M.; Reverberi, M.; Scala, V. Fungal lipids: Biosynthesis and signalling during plant-pathogen interaction. Front. Biosci. 2019, 24, 172–185. [Google Scholar] [CrossRef]
- Beccaccioli, M.; Pucci, N.; Salustri, M.; Scortichini, M.; Zaccaria, M.; Momeni, B.; Loreti, S.; Reverberi, M.; Scala, V. Fungal and bacterial oxylipins are signals for intra- and inter-cellular communication within plant disease. Front. Plant Sci. 2022, 13, 823233. [Google Scholar] [CrossRef] [PubMed]
- Masui, H.; Kondo, T.; Kojima, M. An antifungal compound, 9,12,13-trihydroxy-(E)-10-octadecenoicacid, from Colocasia antiquorum inoculated with Ceratocystis fimbriata. Phytochemistry 1989, 28, 2613–2615. [Google Scholar] [CrossRef]
- Gao, J.M.; Wang, C.Y.; Zhang, A.L.; Liu, J.K. A new trihydroxy fatty acid from the ascomycete, Chinese truffle Tuber indicum. Lipids 2001, 36, 1365–1370. [Google Scholar] [CrossRef]
- Kato, T.; Yamaguchi, Y.; Abe, N.; Uyehara, T.; Namai, T.; Kodama, M.; Shiobara, Y. Structure and Synthesis of Unsaturated Trihydroxy C18 Fatty Acids in Rice Plant Suffering from Rice Blast Disease. Tetrahedron Lett. 1985, 26, 2357–2360. [Google Scholar] [CrossRef]
- Stadler, M.; Mayer, A.; Anke, H.; Sterner, O. FattyAcids and Other Compounds with Nematicidal Activity from Cultures of Basidiomycetes. Planta Med. 1994, 60, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Semwal, R.B.; Semwal, D.K.; Combrinck, S.; Vilioen, A. Health benefits of chromones: Common ingredients of our daily diet. Phytochem. Rev. 2020, 19, 761–785. [Google Scholar] [CrossRef]
- Ahmed, I.; Hussain, H.; Schulz, B.; Draeger, S.; Padula, D.; Pescitelli, G.; van Ree, T.; Krohn, K. Three new antimicrobial metabolites from the endophytic fungus Phomopsis sp. Eur. J. Org. Chem. 2011, 15, 2867–2873. [Google Scholar] [CrossRef]
- Chen, J.; Chen, J.; Wang, S.; Bao, X.; Li, S.; Wei, B.; Zhang, H.; Wang, H. Amycolachromones A–F, Isolated from a Streptomycin-Resistant Strain of the Deep-Sea Marine Actinomycete Amycolatopsis sp. WP1. Mar. Drugs 2022, 20, 162. [Google Scholar] [CrossRef]
- Jansen, B.J.M.; de Groot, A. Occurrence, biological activity and synthesis of drimane sesquiterpenoids Nat. Prod. Rep. 2004, 21, 449–477. [Google Scholar] [CrossRef]
- Du, W.; Yang, Q.; Xu, H.; Dong, L. Drimane-type sesquiterpenoids from fungi, Chin. J. Nat. Med. 2022, 20, 737–748. [Google Scholar] [CrossRef]
- Zang, L.Y.; Wei, W.; Guo, Y.; Wang, T.; Jiao, R.H.; Ng, S.W.; Tan, R.X.; Ge, H.M. Sesquiterpenoids from the mangrove-derived endophytic fungus Diaporthe sp. J. Nat. Prod. 2012, 75, 1744–1749. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Liu, X.X.; Zhang, W.J.; Zang, L.Y.; Wang, G.; Ng, S.W.; Tan, R.X.; Ge, H.M. Sesquiterpenoids isolated from an endophyte fungus Diaporthe sp. RSC Adv. 2015, 5, 17559–17565. [Google Scholar] [CrossRef]
- Ichihara, A.; Sawamura, S.; Sakamura, S. Structures of altiloxins A and B, phytotoxins from Phoma asparagi Sacc. Tetrahedron Lett. 1984, 25, 3209–3212. [Google Scholar] [CrossRef]
- León, A.; Del-Ángel, M.; Ávila, J.L.; Delgado, G. Phthalides: Distribution in Nature, Chemical Reactivity, Synthesis, and Biological Activity. Prog. Chem. Org. Nat. Prod. 2017, 104, 127–246. [Google Scholar] [CrossRef]
- Tsantrizos, Y.S.; Ogilvie, K.K.; Watson, A.K. Phytotoxic metabolites of Phomopsis convolvulus, a host-specific pathogen of field bindweed. Can. J. Chem. 1992, 70, 2276–2284. [Google Scholar] [CrossRef]
- Graniti, A.; Sparapano, L.; Evidente, A. Cyclopaldic acid, a major phytotoxic metabolite of Seiridium cupressi, the pathogen of a canker disease of cypress. Plant Pathol. 1992, 41, 563–568. [Google Scholar] [CrossRef]
- Hemberger, Y.; Xu, J.; Wray, V.; Proksch, P.; Wu, J.; Bringmann, G. Pestaliopens A and B: Stereochemically challenging flexible sesquiterpene-cyclopaldic acid hybrids from Pestalotiopsis sp. Chem. Eur. J. 2013, 19, 15556–15564. [Google Scholar] [CrossRef]
- McMullin, D.R.; Tanney, J.B.; McDonald, K.P.; Miller, J.D. Phthalides produced by Coccomyces strobi (Rhytismataceae, Rhytismatales) isolated from needles of Pinus strobus. Phytochem. Lett. 2019, 29, 17–24. [Google Scholar] [CrossRef]
- Aznar-Fernández, T.; Cimmino, A.; Masi, M.; Rubiales, D.; Evidente, A. Antifeedant activity of long-chain alcohols, and fungal and plant metabolites against pea aphid (Acyrthosiphon pisum) as potential biocontrol strategy. Nat. Prod. Res. 2019, 33, 2471–2479. [Google Scholar] [CrossRef]
- Barilli, E.; Cimmino, A.; Masi, M.; Evidente, M.; Rubiales, D.; Evidente, A. Inhibition of early development stages of rust fungi by the two fungal metabolites cyclopaldic acid and epi- epoformin. Pest Manag. Sci. 2017, 73, 1161–1168. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Fernandez-Aparicio, M.; Andolfi, A.; Basso, S.; Rubiales, D.; Evidente, A. Effect of fungal and plant metabolites on broomrapes (Orobanche and Phelipanche spp.) seed germination and radicle growth. J. Agric. Food Chem. 2014, 62, 10485–10492. [Google Scholar] [CrossRef] [PubMed]
- Samperna, S.; Masi, M.; Vurro, M.; Evidente, A.; Marra, M. Cyclopaldic Acid, the Main Phytotoxic Metabolite of Diplodia cupressi, Induces Programmed Cell Death and Autophagy in Arabidopsis thaliana. Toxins 2022, 14, 474. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metabolomics 2013, 9, 280–299. [Google Scholar] [CrossRef] [PubMed]
- Larsen, T.O.; Smedsgaard, J.; Nielsen, K.F.; Hansen, M.E.; Frisvad, J.C. Phenotypic taxonomy and metabolite profiling in microbial drug discovery. Nat. Prod. Rep. 2005, 22, 672–695. [Google Scholar] [CrossRef]
- Stadler, M.; Ju, Y.; Rogers, J.D. Chemotaxonomy of Entonaema, Rhopalostroma and other Xylariaceae. Mycol. Res. 2004, 108, 239–256. [Google Scholar] [CrossRef]
- Horn, W.S.; Schwartz, R.E.; Simmonds, M.S.J.; Blaney, W.M. Isolation and characterization of phomodiol, a new antifungal from Phomopsis. Tetrahedron Lett. 1994, 35, 6037–6040. [Google Scholar] [CrossRef]
- Horn, W.S.; Simmonds, M.S.J.; Schwartz, R.E.; Blaney, W.M. Phomopsichalasin, a novel antimicrobial agent from an endophytic Phomopsis sp. Tetrahedron 1995, 51, 3969–3978. [Google Scholar] [CrossRef]
- Abreu, L.M.; Costa, S.S.; Pfenning, L.H.; Takahashi, J.A.; Larsen, T.O.; Andersen, B. Chemical and molecular characterization of Phomopsis and Cytospora-like endophytes from different host plants in Brazil. Fungal Biol. 2012, 116, 249–260. [Google Scholar] [CrossRef]
- Król, E. Grzyby zasiedlające zdrowe łozy winorośli (Vitis spp.) w wybranych szkółkach. Acta Agrobot. 2006, 59, 163–173. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, J.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Tsugawa, H.; Kanazawa, M.; Ogiwara, A.; Arita, M. MRMPROBS suite for metabolomics using large-scale MRM assays. Bioinformatics 2014, 30, 2379–2380. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Kind, T.; Nakabayashi, R.; Yukihira, D.; Tanaka, W.; Cajka, T.; Saito, K.; Fiehn, O.; Arita, M. Hydrogen Rearrangement Rules: Computational MS/MS Fragmentation and Structure Elucidation Using MS-FINDER Software Anal. Chem. 2016, 88, 7946–7958. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Steuer, R.; Morgenthal, K.; Weckwerth, W.; Selbig, J. A gentle guide to the analysis of metabolomic data. Methods Mol. Biol. 2007, 358, 105–126. [Google Scholar] [CrossRef]
- Keller, N.P.; Hohn, T.M. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 1997, 21, 17–29. [Google Scholar] [CrossRef]
- Shwab, E.K.; Keller, N.P. Regulation of secondary metabolite production in filamentous ascomycetes. Mycol. Res. 2008, 112, 225–230. [Google Scholar] [CrossRef]
- Yang, Q.; Jiang, N.; Tian, C.M. Three new Diaporthe species from Shaanxi Province, China. MycoKeys 2020, 67, 1–18. [Google Scholar] [CrossRef]
- Manawasinghe, I.S.; Dissanayake, A.J.; Li, X.; Liu, M.; Wanasinghe, D.; Xu, J.; Zhao, W.; Zhang, W.; Zhou, Y.; Hyde, K.D.; et al. High genetic diversity and species complexity of Diaporthe associated with grapevine dieback in China. Front. Microbiol. 2019, 10, 1936. [Google Scholar] [CrossRef]
- Cao, L.; Luo, D.; Lin, W.; Yang, Q.; Deng, X. Four new species of Diaporthe (Diaporthaceae, Diaporthales) from forest plants in China. MycoKeys 2022, 91, 25–47. [Google Scholar] [CrossRef]
- Udayanga, D.; Xingzhong, L.; Crous, P.W.; McKenzie, E.H.C.; Chukeatirote, E.; Hyde, K.D. A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Divers. 2012, 56, 157–171. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, F.; Duan, W.; Crous, P.W.; Cai, L. Diaporthe is paraphyletic. Ima Fungus 2017, 8, 153–187. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).