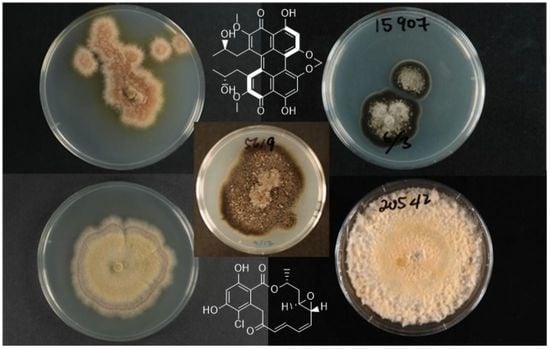
Antimicrobial Natural Products from Plant Pathogenic Fungi
Abstract
1. Introduction
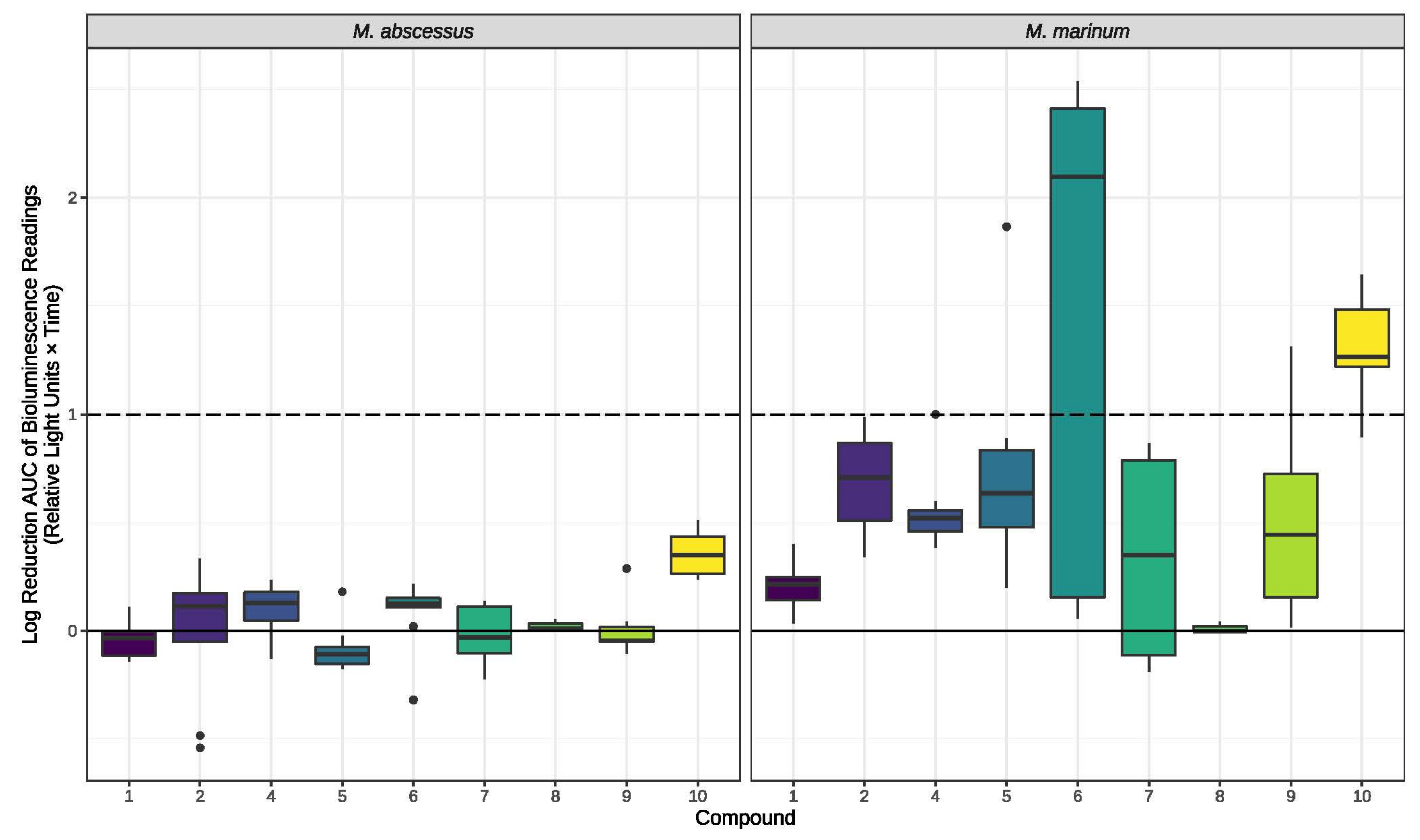
2. Results
3. Discussion
| Fungus | ICMP | Compounds Isolated | Bioactivity | References |
|---|---|---|---|---|
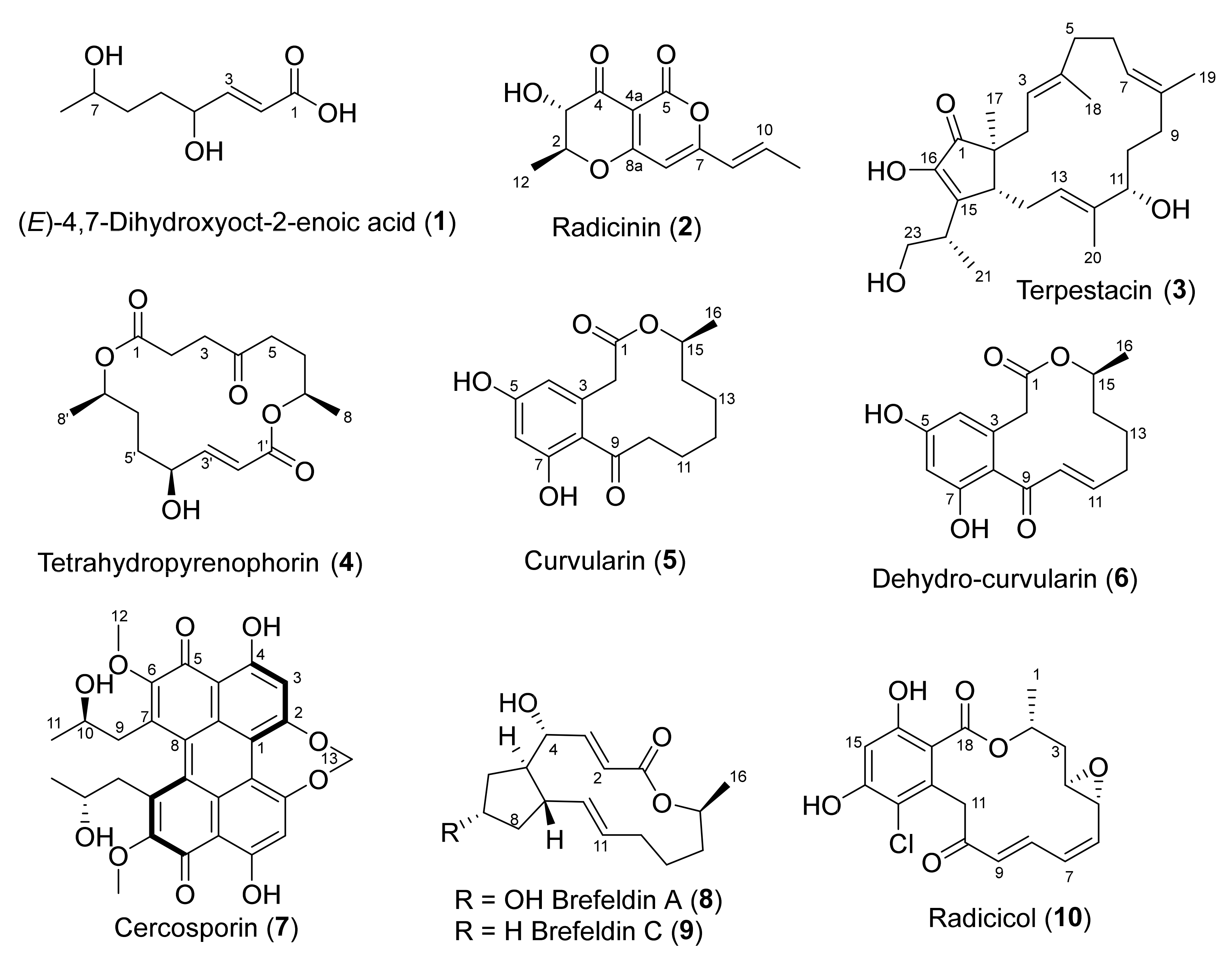
| Alternaria radicina | 5619 | (E)-4,7-dihydroxyoct-2-enoic acid (1) | None | |
| Radicinin (2) | Phytotoxic Antifungal Anti-proliferative Acetylcholinesterase inhibitor | [27,30,32,33,35] | ||
| Terpestacin (3) | Anti-proliferative | [41] | ||
| Tetrahydropyrenophorin (4) | Antibacterial Antifungal Algicidal | [36] | ||
| Curvularin (5) | Phytotoxic | [9,23,26,38,42] | ||
| Dehydrocurvularin (6) | Antifungal Phytotoxic Antimycobacterial | [9,23,26,42] | ||
| Cercospora beticola | 15907 | Cercosporin (7) | Photo-activated plant toxin | [22,43] |
| Dactylonectria macrodidyma | 16789 | Brefeldin A (8) (also known as Decumbin, Cyanein, Ascotoxin, Nectrolide, Synergisidin) | Anti-proliferative Antifungal | [19,38,44,45,46] |
| Brefeldin C (9) | [20] | |||
| Dactylonectria torresensis | 20542 | Brefeldin A (8) | Anti-proliferative | [19,38,44,45,46] |
| Ilyonectria europaea | 16794 | Radicicol (10) (also known as Monorden) | HSP90 inhibitor Antifungal Antimalarial | [24,25,34,39,47] |
| Ilyonectria liriodendra | 16795 | Radicicol (10) | HSP90 inhibitor Antifungal Antimalarial | [24,25,34,39,47] |
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Fungal Material
4.2.1. ICMP 5619—Alternaria radicina
4.2.2. ICMP 15907—Cercospora beticola
4.2.3. ICMP 16789—Dactylonectria macrodidyma
4.2.4. ICMP 20542—Dactylonectria torresensis
4.2.5. ICMP 16794—Ilyonectria europaea
4.2.6. ICMP 16795—Ilyonectria liriodendri
4.3. Fermentation, Extraction and Isolation
4.3.1. ICMP 5619—Alternaria radicina
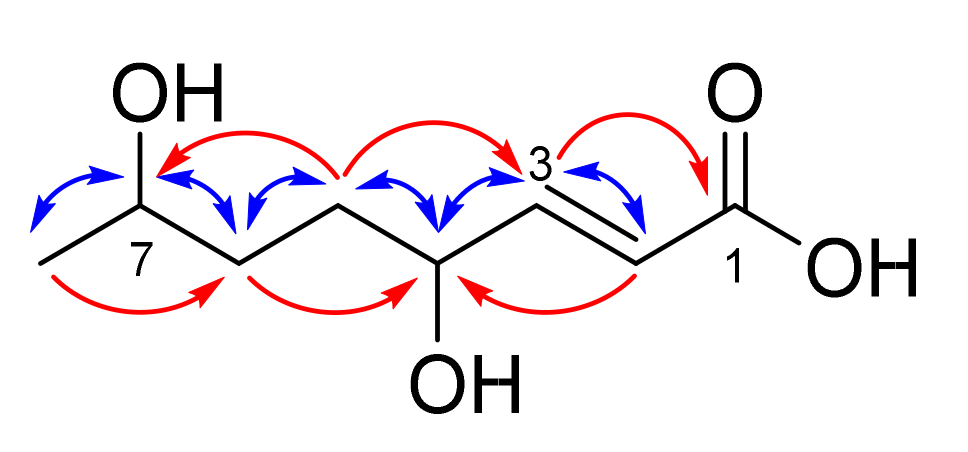
(E)-4,7-Dihydroxyoct-2-enoic acid (1)
Radicinin (2)
Terpestacin (3)
Tetrahydropyrenophorin (4)
Curvularin (5)
Dehydro-curvularin (6)
4.3.2. ICMP 15907—Cercospora beticola
Cercosporin (7)
4.3.3. ICMP 16789—Dactylonectria macrodidyma
Brefeldin A (8)
Brefeldin C (9)
4.3.4. ICMP 20542—Dactylonectria torresensis
4.3.5. ICMP 16794—Ilyonectria europaea
Radicicol (10)
4.3.6. ICMP 16795—Ilyonectria liriodendri
4.4. Antimicrobial Testing of Fungal Cultures
4.5. Antimicrobial Assays of Pure Compounds
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Gudesblat, G.E.; Torres, P.S.; Vojno, A.A. Stomata and Pathogens. Plant Signal. Behav. 2009, 4, 1114–1116. [Google Scholar] [CrossRef] [PubMed]
- Demoor, A.; Silar, P.; Brun, S. Appressorium: The Breakthrough in Dikarya. J. Fungi 2019, 5, 72. [Google Scholar] [CrossRef] [PubMed]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal Effectors and Plant Susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Pusztahelyi, T.; Holb, I.; Pócsi, I. Secondary Metabolites in Fungus-Plant Interactions. Front. Plant Sci. 2015, 6, 573. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, A.S.; Moffat, C.S.; Lopez-Ruiz, F.J.; Gibberd, M.R.; Hamblin, J.; Zerihun, A. Host–Multi-Pathogen Warfare: Pathogen Interactions in Co-Infected Plants. Front. Plant Sci. 2017, 8, 1806. [Google Scholar] [CrossRef]
- Chan, J.; Jamison, T.F. Synthesis of (−)-Terpestacin via Catalytic, Stereoselective Fragment Coupling: Siccanol Is Terpestacin, Not 11-epi- Terpestacin. J. Am. Chem. Soc. 2003, 125, 11514–11515. [Google Scholar] [CrossRef]
- Wu, Y.; Shen, X.; Yang, Y.-Q.; Hu, Q.; Huang, J.-H. An Aldol Approach to the Total Synthesis of (+)-Brefeldin A. Tetrahedron Lett. 2004, 45, 199–202. [Google Scholar] [CrossRef]
- Barluenga, S.; Moulin, E.; Lopez, P.; Winssinger, N. Solution- and Solid-Phase Synthesis of Radicicol (Monorden) and Pochonin C. Chem. A Eur. J. 2005, 11, 4935–4952. [Google Scholar] [CrossRef]
- Xie, L.W.; Ouyang, Y.C.; Zou, K.; Wang, G.H.; Chen, M.J.; Sun, H.M.; Dai, S.K.; Li, X. Isolation and Difference in Anti-Staphylococcus Aureus Bioactivity of Curvularin Derivates from Fungus Eupenicillium sp. Appl. Biochem. Biotechnol. 2009, 159, 284–293. [Google Scholar] [CrossRef]
- Morgan, B.J.; Mulrooney, C.A.; Kozlowski, M.C. Perylenequinone Natural Products: Evolution of the Total Synthesis of Cercosporin. J. Org. Chem. 2010, 75, 44–56. [Google Scholar] [CrossRef]
- Archambaud, S.; Legrand, F.; Aphecetche-Julienne, K.; Collet, S.; Guingant, A.; Evain, M. Total Synthesis of (+)-Brefeldin C, (+)-nor-Me Brefeldin A and (+)-4- Epi -nor-Me Brefeldin A. Eur. J. Org. Chem. 2010, 2010, 1364–1380. [Google Scholar] [CrossRef]
- Shao, M.; Wang, N.; Lin, F.; Zhang, Y. Identification and Herbicidal Active Ingredients of DN02, a Fungus Residing in a Ceriagrion Sp. Gut. ActanPhytophylacica Sin. 2014, 41, 98–102. [Google Scholar]
- Clarke, D.D.; Nord, F.F.; Bhacca, N.S. Radicinin: Revision of Its Structure Obtained from NMR Measurements. Arch. Biochem. Biophys. 1963, 102, 473–474. [Google Scholar] [CrossRef]
- Iimura, S.; Oka, M.; Narita, Y.; Konishi, M.; Kakisawa, H.; Gao, Q.; Oki, T. Terpestacin, a Novel Syncytium Formation Inhibitor, Isolated from Arthrinium Species. Tetrahedron Lett. 1993, 34, 493–496. [Google Scholar] [CrossRef]
- Nozoe, S.; Hirai, K.; Tsuda, K.; Ishibashi, K.; Shirasaka, M.; Frederick Grove, J. The Structure of Pyrenophorin. Tetrahedron Lett. 1965, 6, 4675–4677. [Google Scholar] [CrossRef]
- Musgrave, O.C. Curvularin. Part I. Isolation and Partial Characterisation of a Metabolic Product from a New Species of Curvularia. J. Chem. Soc. 1956, 4301–4305. [Google Scholar] [CrossRef]
- Starratt, A.N.; White, G.A. Identification of Some Metabolites of Alternaria cucumerina (E. & E.) Ell. Phytochemistry 1968, 7, 1883–1884. [Google Scholar] [CrossRef]
- Kuyama, S.; Tamura, T. Cercosporin. A Pigment of Cercosporina Kikuchii Matsumoto et Tomoyasu. I. Cultivation of Fungus, Isolation and Purification of Pigment. J. Am. Chem. Soc. 1957, 79, 5725–5726. [Google Scholar] [CrossRef]
- Singleton, V.L.; Bohonos, N.; Ullstrup, A.J. Decumbin, a New Compound from a Species of Penicillium. Nature 1958, 181, 1072–1073. [Google Scholar] [CrossRef]
- Sunagawa, M.; Ohta, T.; Nozoe, S. Isolation and Structure of Brefeldin C. Heterocycles 1979, 13, 267–270. [Google Scholar]
- Mirrington, R.N.; Ritchie, E.; Shoppee, C.W.; Taylor, W.C.; Sternhell, S. The Constitution of Radicicol. Tetrahedron Lett. 1964, 5, 365–370. [Google Scholar] [CrossRef]
- Fajola, A.O. Cercosporin, a Phytotoxin from Cercospora spp. Physiol. Plant Pathol. 1978, 13, 157–164. [Google Scholar] [CrossRef]
- Robeson, D.J.; Strobel, G.A.; Strange, R.N. The Identification of a Major Phytotoxic Component from Alternaria macrospora as Aβ-Dehydrocurvularin. J. Nat. Prod. 1985, 48, 139–141. [Google Scholar] [CrossRef]
- Shimada, Y.; Ogawa, T.; Sato, A.; Kaneko, I.; Tsujita, Y. Induction of Differentiation of HL-60 Cells by the Anti-Fungal Antibiotic, Radicicol. J. Antibiot. 1995, 48, 824–830. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Shiomi, K.; Kamei, K.; Sugoh-Hagino, M.; Enomoto, Y.; Fang, F.; Yamaguchi, Y.; Masuma, R.; Zhang, C.G.; Zhang, X.W.; et al. Antimalarial Activity of Radicicol. Heptelidic Acid and Other Fungal Metabolites. J. Antibiot. 1998, 51, 153–160. [Google Scholar] [CrossRef]
- de Souza, A.O.; Galetti, F.C.S.; Silva, C.L.; Bicalho, B.; Parma, M.M.; Fonseca, S.F.; Marsaioli, A.J.; Trindade, A.C.L.B.; Gil, R.P.F.; Bezerra, F.S.; et al. Antimycobacterial and Cytotoxicity Activity of Synthetic and Natural Compounds. Quím. Nova 2007, 30, 1563–1566. [Google Scholar] [CrossRef]
- Zhang, Y.; Kong, L.; Jiang, D.; Yin, C.; Cai, Q.; Chen, Q.; Zheng, J. Phytotoxic and Antifungal Metabolites from Curvularia sp. FH01 Isolated from the Gut of Atractomorpha sinensis. Bioresour. Technol. 2011, 102, 3575–3577. [Google Scholar] [CrossRef]
- Bladt, T.; Frisvad, J.; Knudsen, P.; Larsen, T. Anticancer and Antifungal Compounds from Aspergillus, Penicillium and Other Filamentous Fungi. Molecules 2013, 18, 11338–11376. [Google Scholar] [CrossRef]
- Sztiller-Sikorska, M.; Koprowska, K.; Majchrzak, K.; Hartman, M.; Czyz, M. Natural Compounds’ Activity against Cancer Stem-Like or Fast-Cycling Melanoma Cells. PLoS ONE 2014, 9, e90783. [Google Scholar] [CrossRef]
- Li, S.; Shao, M.-W.; Lu, Y.-H.; Kong, L.-C.; Jiang, D.-H.; Zhang, Y.-L. Phytotoxic and Antibacterial Metabolites from Fusarium proliferatum ZS07 Isolated from the Gut of Long-Horned Grasshoppers. J. Agric. Food Chem. 2014, 62, 8997–9001. [Google Scholar] [CrossRef]
- Ye, X.; Anjum, K.; Song, T.; Wang, W.; Yu, S.; Huang, H.; Lian, X.-Y.; Zhang, Z. A New Curvularin Glycoside and Its Cytotoxic and Antibacterial Analogues from Marine Actinomycete Pseudonocardia sp. HS7. Nat. Prod. Res. 2016, 30, 1156–1161. [Google Scholar] [CrossRef]
- Piemontese, L.; Vitucci, G.; Catto, M.; Laghezza, A.; Perna, F.; Rullo, M.; Loiodice, F.; Capriati, V.; Solfrizzo, M. Natural Scaffolds with Multi-Target Activity for the Potential Treatment of Alzheimer’s Disease. Molecules 2018, 23, 2182. [Google Scholar] [CrossRef] [PubMed]
- Handayani, D.; Putri, R.A.; Ismed, F.; Hertiani, T.; Ariantari, N.P.; Proksch, P. Bioactive Metabolite from Marine Sponge-Derived Fungus Cochliobolus geniculatus WR12. RJC 2020, 13, 417–422. [Google Scholar] [CrossRef]
- Kuttikrishnan, S.; Prabhu, K.S.; Al Sharie, A.H.; Al Zu’bi, Y.O.; Alali, F.Q.; Oberlies, N.H.; Ahmad, A.; El-Elimat, T.; Uddin, S. Natural Resorcylic Acid Lactones: A Chemical Biology Approach for Anticancer Activity. Drug Discov. Today 2022, 27, 547–557. [Google Scholar] [CrossRef]
- Hussain, H.; Kliche-Spory, C.; Al-Harrasi, A.; Al-Rawahi, A.; Abbas, G.; Green, I.R.; Schulz, B.; Krohn, K.; Shah, A. Antimicrobial Constituents from Three Endophytic Fungi. Asian Pac. J. Trop. Med. 2014, 7, S224–S227. [Google Scholar] [CrossRef]
- Zhang, W.; Krohn, K.; Egold, H.; Draeger, S.; Schulz, B. Diversity of Antimicrobial Pyrenophorol Derivatives from an Endophytic Fungus, Phoma sp. Eur. J. Org. Chem. 2008, 2008, 4320–4328. [Google Scholar] [CrossRef]
- Rutala, W.A.; Weber, D.J. Infection Control: The Role of Disinfection and Sterilization. J. Hosp. Infect. 1999, 43, S43–S55. [Google Scholar] [CrossRef]
- Betina, V.; Mičeková, D. Antimicrobial Properties of Fungal Macrolide Antibiotics. Z. Allg. Mikrobiol. 1972, 12, 355–364. [Google Scholar] [CrossRef]
- Wicklow, D.T.; Joshi, B.K.; Gamble, W.R.; Gloer, J.B.; Dowd, P.F. Antifungal Metabolites (Monorden, Monocillin IV, and Cerebrosides) from Humicola fuscoatra Traaen NRRL 22980, a Mycoparasite of Aspergillus flavus Sclerotia. Appl Env. Microbiol 1998, 64, 4482–4484. [Google Scholar] [CrossRef]
- Trisuwan, K.; Rukachaisirikul, V.; Sukpondma, Y.; Phongpaichit, S.; Preedanon, S.; Sakayaroj, J. Lactone Derivatives from the Marine-Derived Fungus Penicillium sp. PSU-F44. Chem. Pharm. Bull. 2009, 57, 1100–1102. [Google Scholar] [CrossRef]
- Jung, H.J.; Shim, J.S.; Lee, J.; Song, Y.M.; Park, K.C.; Choi, S.H.; Kim, N.D.; Yoon, J.H.; Mungai, P.T.; Schumacker, P.T.; et al. Terpestacin Inhibits Tumor Angiogenesis by Targeting UQCRB of Mitochondrial Complex III and Suppressing Hypoxia-Induced Reactive Oxygen Species Production and Cellular Oxygen Sensing*. J. Biol. Chem. 2010, 285, 11584–11595. [Google Scholar] [CrossRef] [PubMed]
- Caputo, O.; Viola, F. Isolation of α, β–Dehydrocurvularin from Aspergillus aureofulgens. Planta Med. 1977, 31, 31–32. [Google Scholar] [CrossRef] [PubMed]
- Daub, M.E.; Ehrenshaft, M. The Photoactivated Cercospora Toxin Cercosporin: Contributions to Plant Disease and Fundamental Biology. Annu. Rev. Phytopathol. 2000, 38, 461–490. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Tanaka, H.; Aoki, H.; Tamura, T. Ascotoxin (Decumbin), a Metabolite of Ascochyta imperfecta PECK. Agric. Biol. Chem. 1970, 34, 395–413. [Google Scholar] [CrossRef]
- Hutchinson, C.R.; Shu-Wen, L.; McInnes, A.G.; Walter, J.A. Comparative Biochemistry of Fatty Acid and Macrolide Antibiotic (Brefeldin a). Formation in Penicillium brefeldianum. Tetrahedron 1983, 39, 3507–3513. [Google Scholar] [CrossRef]
- Anadu, N.O.; Davisson, V.J.; Cushman, M. Synthesis and Anticancer Activity of Brefeldin A Ester Derivatives. J. Med. Chem. 2006, 49, 3897–3905. [Google Scholar] [CrossRef]
- Moulin, E.; Zoete, V.; Barluenga, S.; Karplus, M.; Winssinger, N. Design, Synthesis, and Biological Evaluation of HSP90 Inhibitors Based on Conformational Analysis of Radicicol and Its Analogues. J. Am. Chem. Soc. 2005, 127, 6999–7004. [Google Scholar] [CrossRef]
- Trivedi, R.S.; Hampton, J.G.; Townshend, J.M.; Jaspers, M.V.; Ridgway, H.J. First Report of Alternaria carotiincultae on Carrot Seed Produced in New Zealand. Plant Dis. 2010, 94, 1168. [Google Scholar] [CrossRef]
- Specimen Details. Available online: https://scd.landcareresearch.co.nz/Specimen/ICMP_5619 (accessed on 5 July 2022).
- Cercospora beticola Sacc. 1876—Biota of NZ. Available online: https://biotanz.landcareresearch.co.nz/scientific-names/1cb180db-36b9-11d5-9548-00d0592d548c (accessed on 15 July 2022).
- Specimen Details. Available online: https://scd.landcareresearch.co.nz/Specimen/ICMP%2015907 (accessed on 25 October 2022).
- Lombard, L.; van der Merwe, A.; Groenewald, J.Z.; Crous, P.W. Lineages in Nectriaceae: Re-Evaluating the Generic Status of Ilyonectria and Allied Genera. Phytopathol. Mediterr. 2014, 53, 515–532. [Google Scholar]
- Specimen Details. Available online: https://scd.landcareresearch.co.nz/Specimen/ICMP%2016789 (accessed on 25 October 2022).
- Cabral, A.; Rego, C.; Nascimento, T.; Oliveira, H.; Groenewald, J.Z.; Crous, P.W. Multi-Gene Analysis and Morphology Reveal Novel Ilyonectria Species Associated with Black Foot Disease of Grapevines. Fungal Biol. 2012, 116, 62–80. [Google Scholar] [CrossRef]
- Specimen Details. Available online: https://scd.landcareresearch.co.nz/Specimen/ICMP%2020542 (accessed on 25 October 2022).
- Specimen Details. Available online: https://scd.landcareresearch.co.nz/Specimen/ICMP%2016794 (accessed on 25 October 2022).
- Probst, C.M.; Ridgway, H.J.; Jaspers, M.V.; Eirian Jones, E. Pathogenicity of Ilyonectria liriodendri and Dactylonectria macrodidyma Propagules in Grapevines. Eur. J. Plant Pathol. 2019, 154, 405–421. [Google Scholar] [CrossRef]
- Specimen Details. Available online: https://scd.landcareresearch.co.nz/Specimen/ICMP%2016795 (accessed on 25 October 2022).
- Hansen, O.R. Stemphylone, a Root-Killing Substance from Stemphylium radicinum. Acta Chem. Scand. 1954, 8, 1332–1334. [Google Scholar] [CrossRef]
- Chan, J.; Jamison, T.F. Enantioselective Synthesis of (−)-Terpestacin and Structural Revision of Siccanol Using Catalytic Stereoselective Fragment Couplings and Macrocyclizations. J. Am. Chem. Soc. 2004, 126, 10682–10691. [Google Scholar] [CrossRef]
- Bicalho, B.; Gonçalves, R.A.C.; Zibordi, A.P.M.; Manfio, G.P.; Marsaioli, A.J. Antimicrobial Compounds of Fungi Vectored by Clusia spp. (Clusiaceae) Pollinating Bees. Z. Nat. C 2003, 58, 746–751. [Google Scholar] [CrossRef]
- Wiles, S.; Grey, A. Bioluminescence-Based 24 Well Plate Assay for Screening Fungi for Activity against Mycobacterium marinum. Available online: https://www.protocols.io/view/bioluminescence-based-24-well-plate-assay-for-scre-bvnbn5an (accessed on 12 December 2022).
- Cadelis, M.M.; Gordon, H.; Grey, A.; Geese, S.; Mulholland, D.R.; Weir, B.S.; Copp, B.R.; Wiles, S. Isolation of a Novel Polyketide from Neodidymelliopsis Sp. Molecules 2021, 26, 3235. [Google Scholar] [CrossRef]
- Dalton, J.P.; Uy, B.; Okuda, K.S.; Hall, C.J.; Denny, W.A.; Crosier, P.S.; Swift, S.; Wiles, S. Screening of Anti-Mycobacterial Compounds in a Naturally Infected Zebrafish Larvae Model. J. Antimicrob. Chemother. 2017, 72, 421–427. [Google Scholar] [CrossRef]
- Blaskovich, M.A.T.; Zuegg, J.; Elliott, A.G.; Cooper, M.A. Helping Chemists Discover New Antibiotics. ACS Infect. Dis. 2015, 1, 285–287. [Google Scholar] [CrossRef]
- Cadelis, M.M.; Geese, S.; Gris, L.; Weir, B.S.; Copp, B.R.; Wiles, S. A Revised Structure and Assigned Absolute Configuration of Theissenolactone A. Molecules 2020, 11, 4823. [Google Scholar] [CrossRef]
- Cadelis, M.M.; Geese, S.; Uy, B.B.; Mulholland, D.R.; van de Pas, S.J.; Grey, A.; Weir, B.S.; Copp, B.R.; Wiles, S. Antimicrobial Metabolites against Methicillin-Resistant Staphylococcus aureus from the Endophytic Fungus Neofusicoccum australe. Molecules 2021, 26, 1094. [Google Scholar] [CrossRef]
- Grey, A.B.J.; Cadelis, M.M.; Diao, Y.; Park, D.; Lumley, T.; Weir, B.S.; Copp, B.R.; Wiles, S. Screening of Fungi for Antimycobacterial Activity Using a Medium-Throughput Bioluminescence-Based Assay. Front. Microbiol. 2021, 12, 2525. [Google Scholar] [CrossRef]
- Dalton, J.P.; Uy, B.; Phummarin, N.; Copp, B.R.; Denny, W.A.; Swift, S.; Wiles, S. Effect of Common and Experimental Anti-Tuberculosis Treatments on Mycobacterium tuberculosis Growing as Biofilms. PeerJ 2016, 4, e2717. [Google Scholar] [CrossRef] [PubMed]
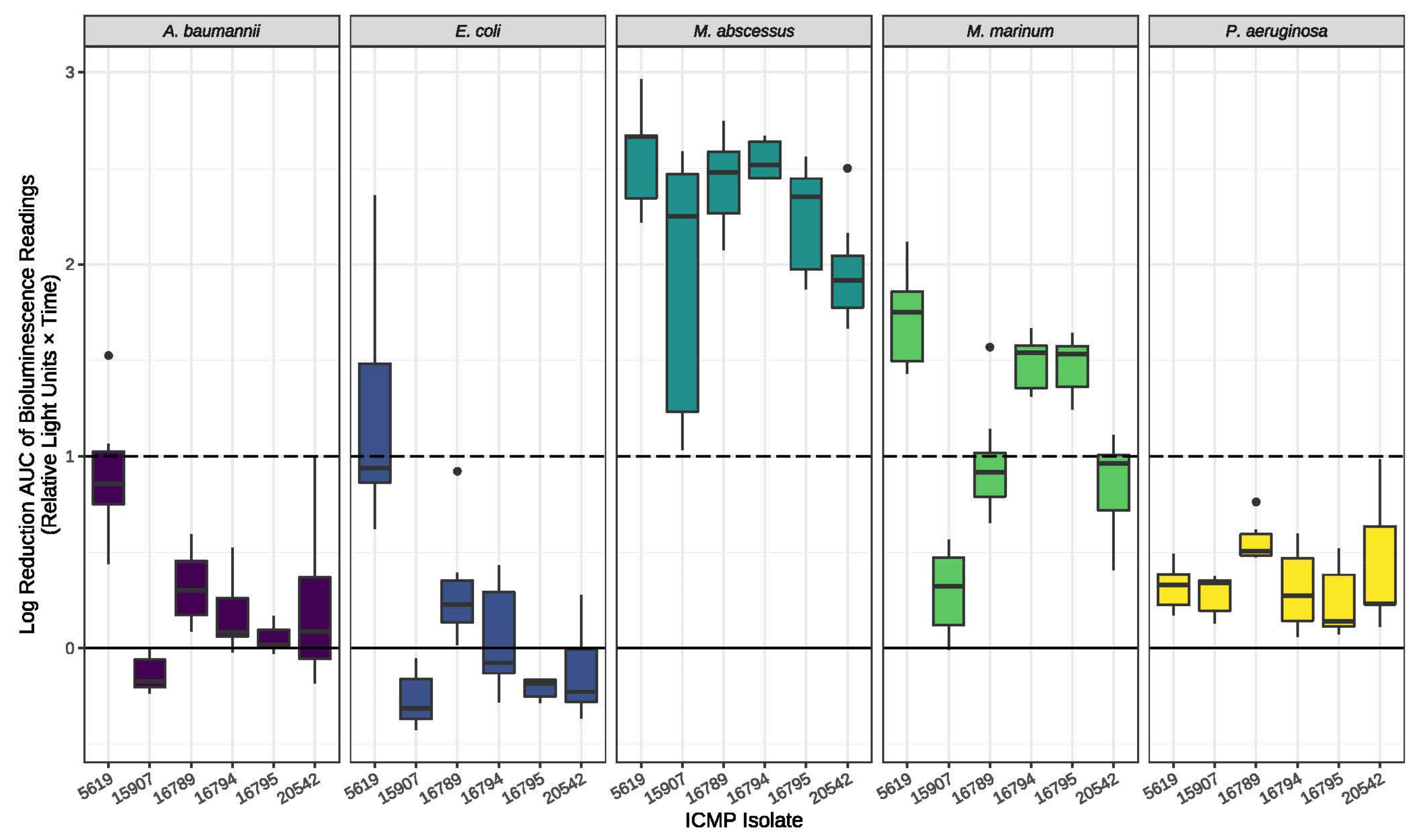
| MIC (µM) | HEK293 CC50 (µM) h | HC10 (µM) i | |||||||
|---|---|---|---|---|---|---|---|---|---|
| S. a a | E. c b | K. p c | P. a d | A. b e | C. a f | C. n g | |||
| 1 | >184 | >184 | >184 | >184 | >184 | >184 | >184 | n.t j | n.t j |
| 2 | >135 | >135 | >135 | >135 | >135 | >135 | >135 | n.t j | n.t j |
| 3 | >79 | >79 | >79 | >79 | >79 | >79 | >79 | n.t j | n.t j |
| 4 | >102 | >102 | >102 | >102 | >102 | >102 | >102 | n.t j | n.t j |
| 5 | >109 | >109 | >109 | >109 | >109 | >109 | >109 | n.t j | n.t j |
| 6 | >110 | >110 | >110 | >110 | >110 | >110 | >110 | 1.27 | >110 |
| 7 | ≤0.47 | >60 | >60 | >60 | >60 | >60 | >60 | 25.33 | >60 |
| 8 | >114 | >114 | >114 | >114 | >114 | 57 | >114 | 0.89 | >114 |
| 10 | >88 | >88 | >88 | >88 | >88 | 44 | >88 | >88 | >88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cadelis, M.M.; Li, S.A.; van de Pas, S.J.; Grey, A.; Mulholland, D.; Weir, B.S.; Copp, B.R.; Wiles, S. Antimicrobial Natural Products from Plant Pathogenic Fungi. Molecules 2023, 28, 1142. https://doi.org/10.3390/molecules28031142
Cadelis MM, Li SA, van de Pas SJ, Grey A, Mulholland D, Weir BS, Copp BR, Wiles S. Antimicrobial Natural Products from Plant Pathogenic Fungi. Molecules. 2023; 28(3):1142. https://doi.org/10.3390/molecules28031142
Chicago/Turabian StyleCadelis, Melissa M., Steven A. Li, Shara J. van de Pas, Alex Grey, Daniel Mulholland, Bevan S. Weir, Brent R. Copp, and Siouxsie Wiles. 2023. "Antimicrobial Natural Products from Plant Pathogenic Fungi" Molecules 28, no. 3: 1142. https://doi.org/10.3390/molecules28031142
APA StyleCadelis, M. M., Li, S. A., van de Pas, S. J., Grey, A., Mulholland, D., Weir, B. S., Copp, B. R., & Wiles, S. (2023). Antimicrobial Natural Products from Plant Pathogenic Fungi. Molecules, 28(3), 1142. https://doi.org/10.3390/molecules28031142