Water-Soluble Polyoxometal Clusters of Molybdenum (V) with Pyrazole and Triazole: Synthesis and Study of Cytotoxicity and Antiviral Activity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis of POMC
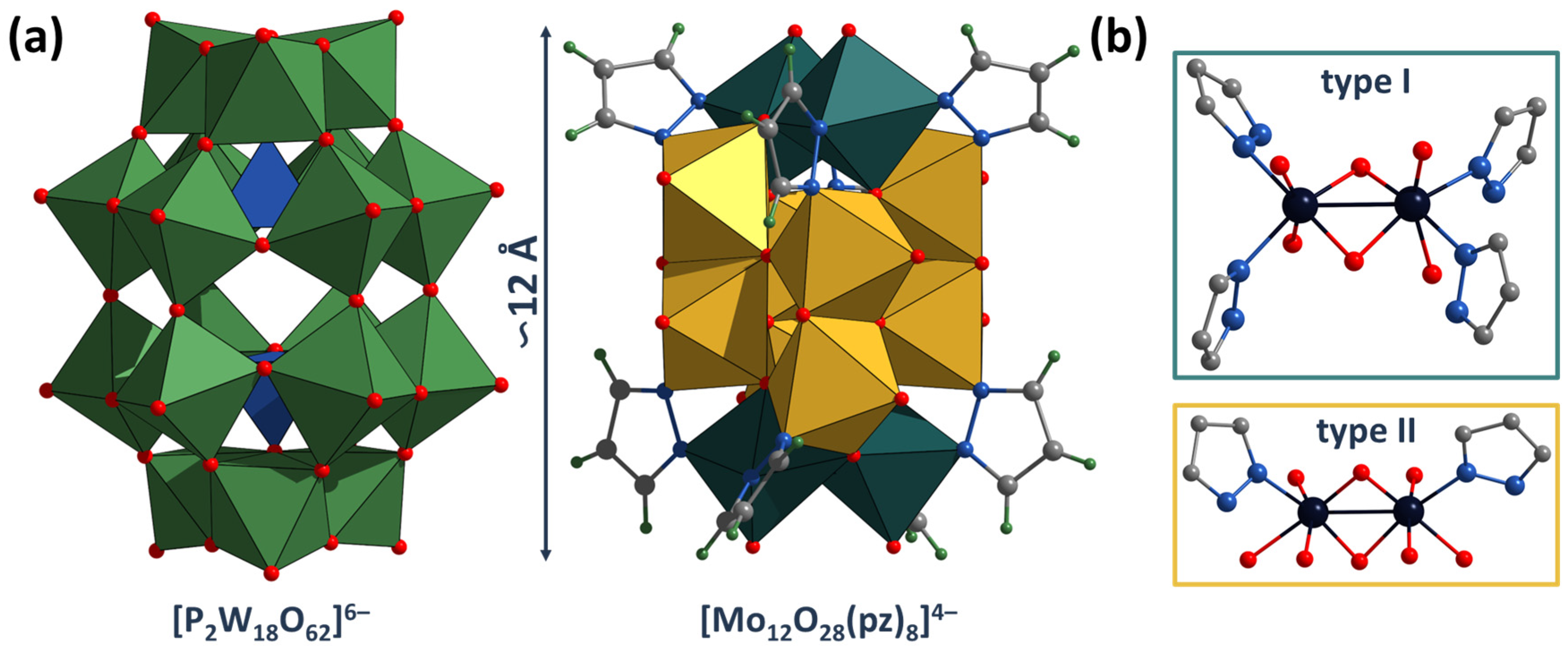
2.2. Crystal Structure
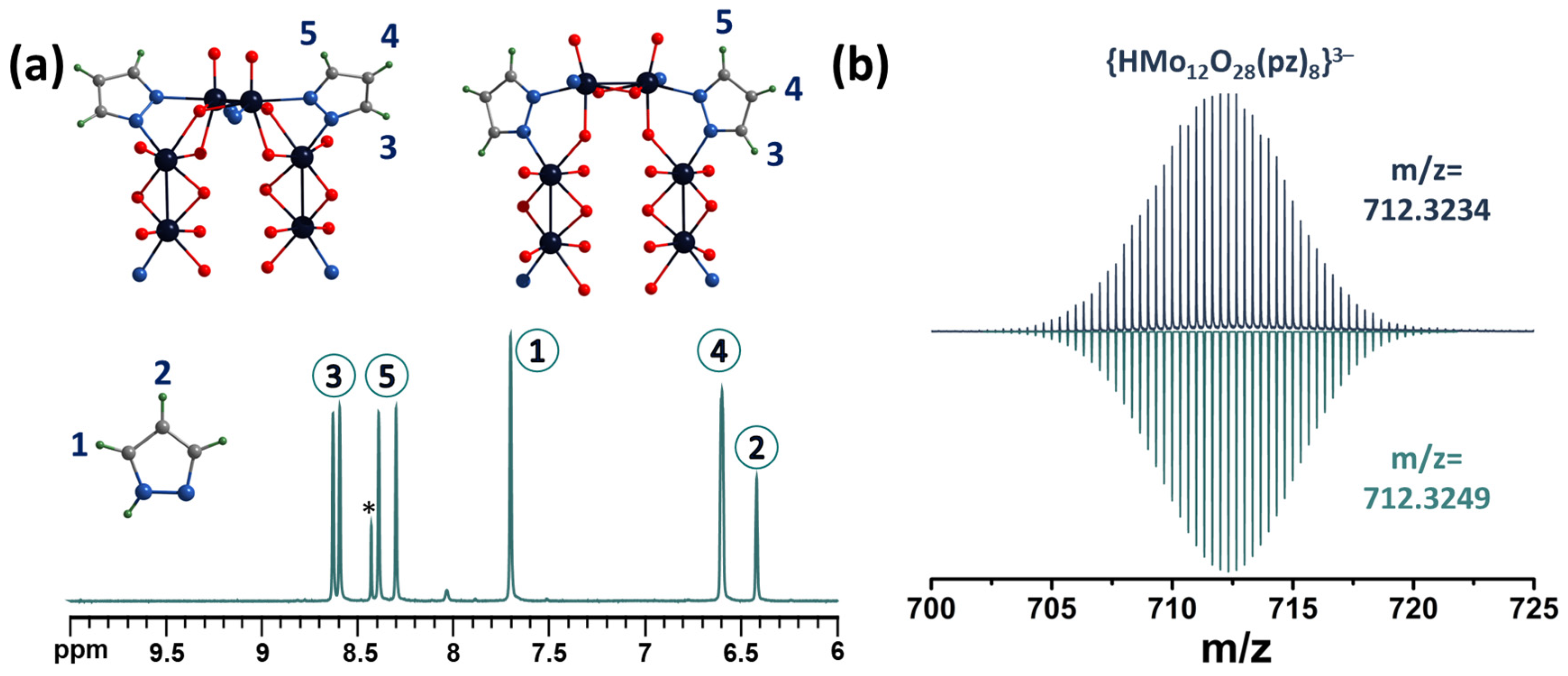
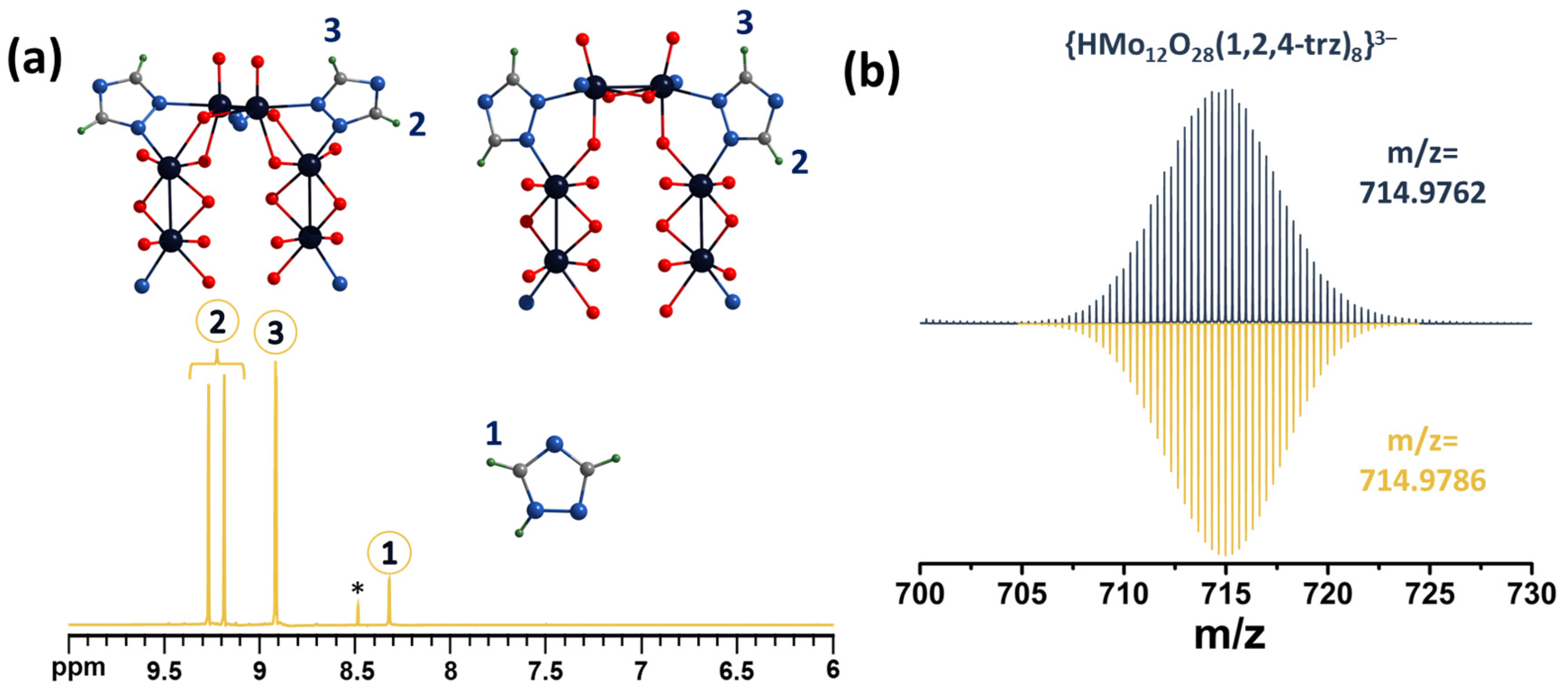
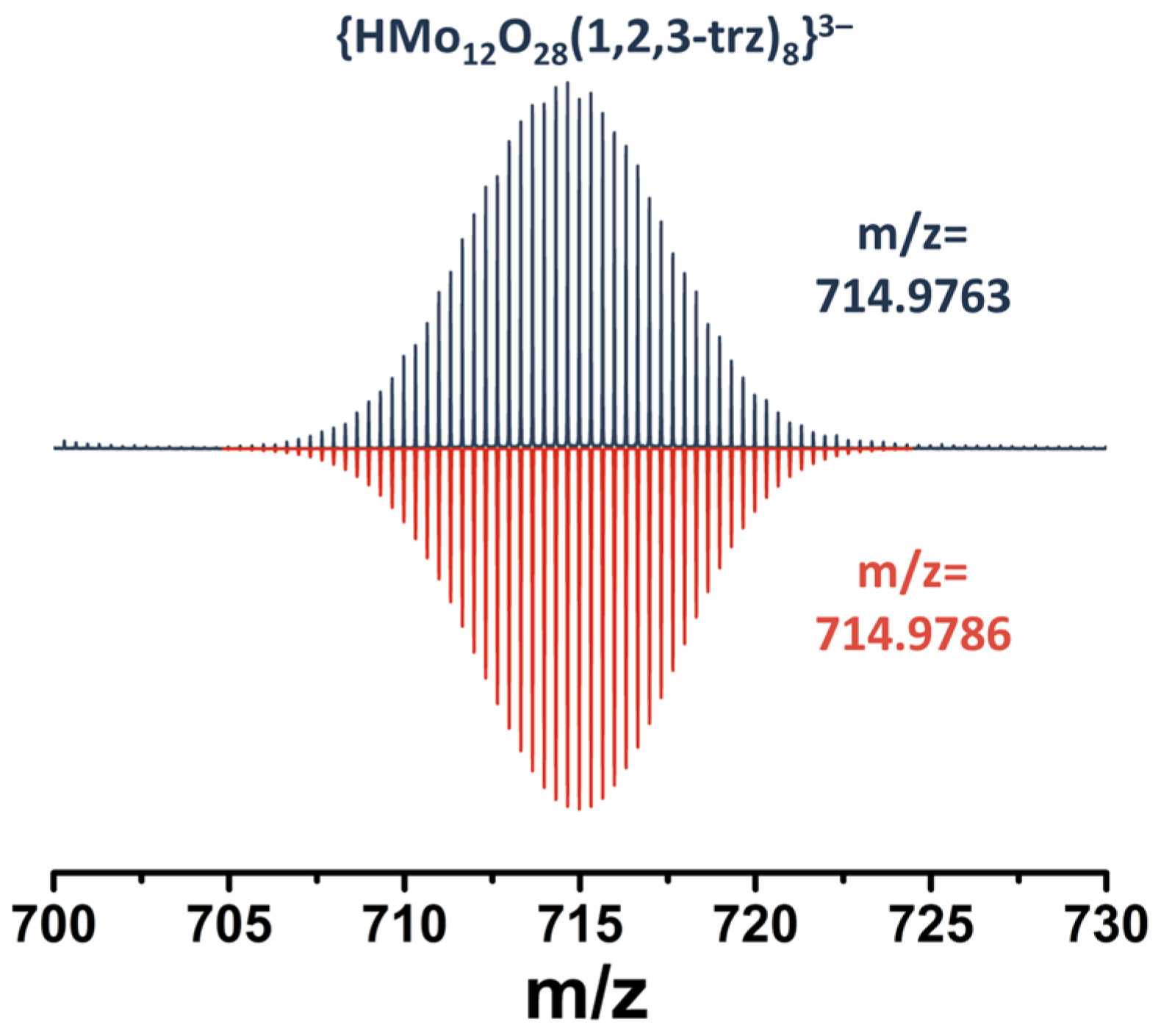
2.3. NMR Spectroscopy and Mass Spectrometry
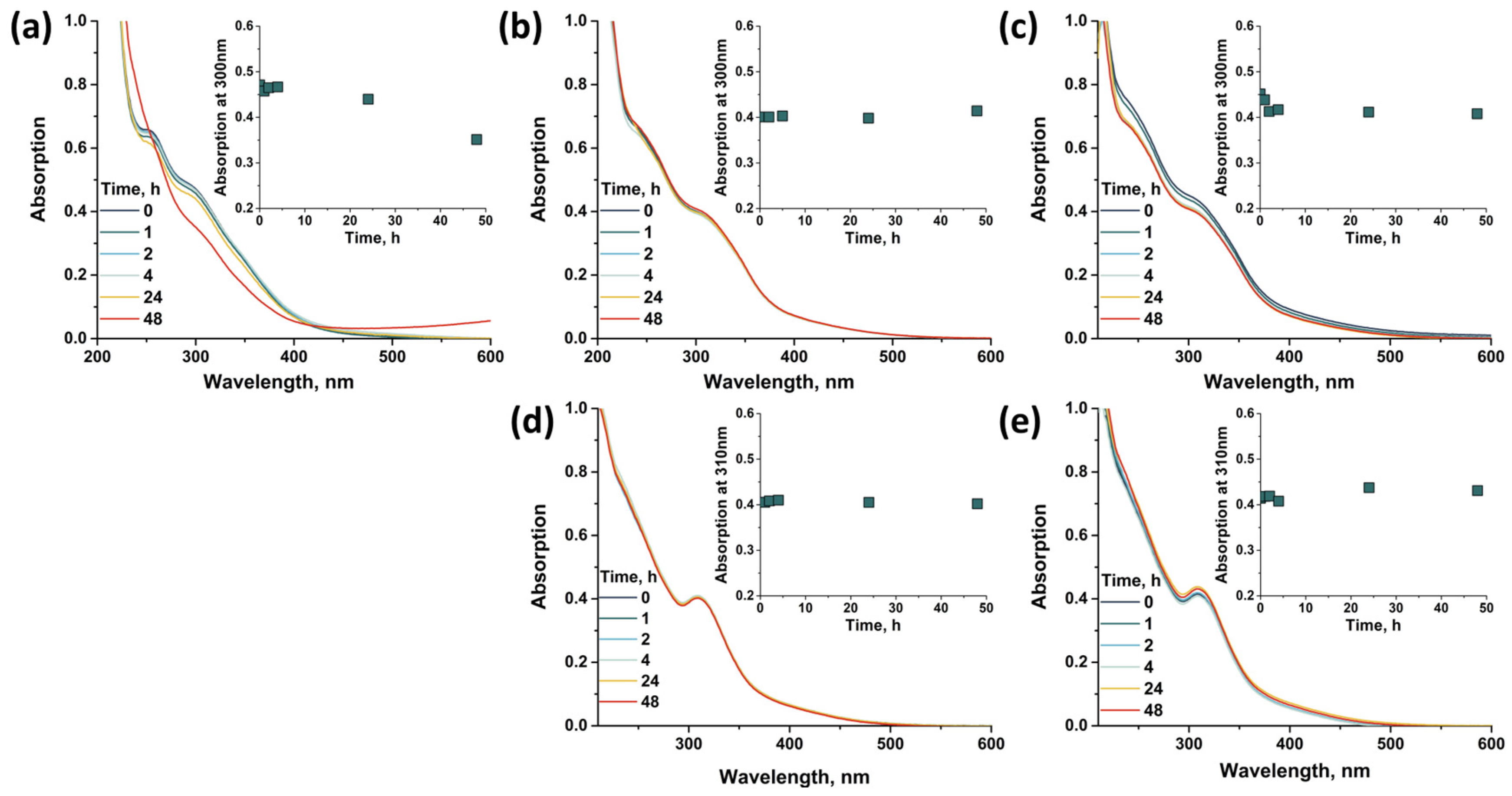
2.4. Stability in Water Solution
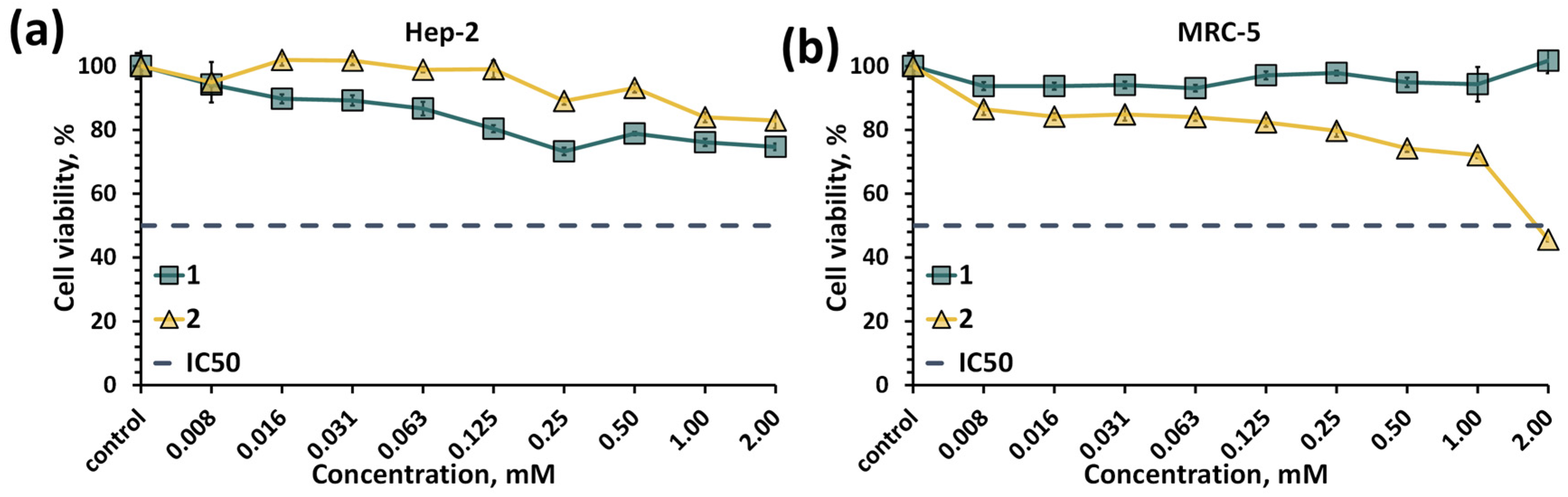
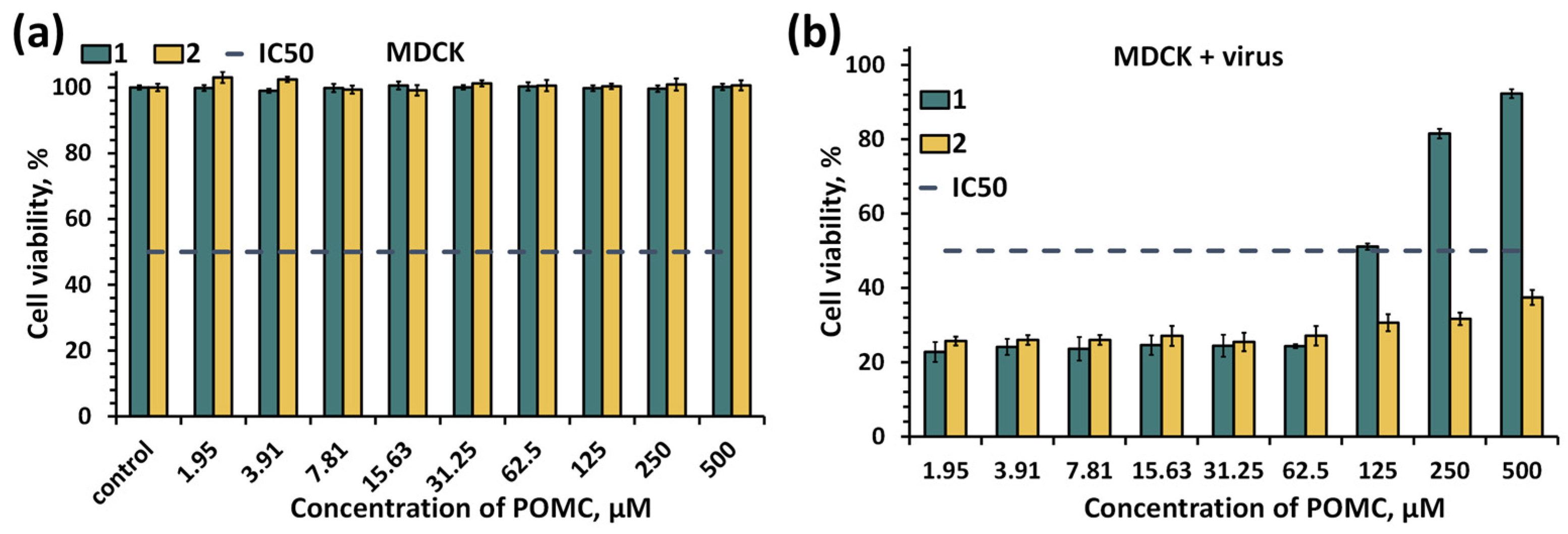
2.5. Biological Properties
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Syntheses
3.2.1. (NH4)4[Mo12O28(μ-pz)8]·1.5pzH·4.5H2O (Denoted as 1)
3.2.2. (NH4)4[Mo12O28(μ-1,2,4-trz)8]·1.5(1,2,4-trzH)·3H2O (Denoted as 2)
3.2.3. (NH4)4[Mo12O28(μ-1,2,3-trz)8]·4H2O (Denoted as 3)
3.3. Physical Methods
3.4. Crystallography
3.5. Cell Culture
3.6. The MTT Test
3.7. Inductively Coupled Plasma Atomic Emission Spectroscopy (ICP-AES)
3.8. The Antiviral Activity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hutin, M.; Rosnes, M.H.; Long, D.-L.; Cronin, L. 2.10—Polyoxometalates: Synthesis and structure—From building blocks to emergent materials. In Comprehensive Inorganic Chemistry II, 2nd ed.; Reedijk, J., Ed.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 241–269. [Google Scholar]
- Liu, J.-C.; Zhao, J.-W.; Streb, C.; Song, Y.-F. Recent advances on high-nuclear polyoxometalate clusters. Coord. Chem. Rev. 2022, 471, 214734. [Google Scholar] [CrossRef]
- Ribó, E.G.; Bell, N.L.; Long, D.-L.; Cronin, L. Engineering highly reduced molybdenum polyoxometalates via the incorporation of d and f block metal ions. Angew. Chem. Int. Ed. 2022, 61, e202201672. [Google Scholar] [CrossRef] [PubMed]
- Anyushin, A.V.; Kondinski, A.; Parac-Vogt, T.N. Hybrid polyoxometalates as post-functionalization platforms: From fundamentals to emerging applications. Chem. Soc. Rev. 2020, 49, 382–432. [Google Scholar] [CrossRef]
- Nohra, B.; El Moll, H.; Rodriguez Albelo, L.M.; Mialane, P.; Marrot, J.; Mellot-Draznieks, C.; O’Keeffe, M.; Ngo Biboum, R.; Lemaire, J.; Keita, B.; et al. Polyoxometalate-based metal organic frameworks (POMOFs): Structural trends, energetics, and high electrocatalytic efficiency for hydrogen evolution reaction. J. Am. Chem. Soc. 2011, 133, 13363–13374. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhou, Z.; Qin, L.; Zhang, D.; Wang, H.; Wang, S.; Yang, L. Structural regulation of two polyoxometalate-based metal–organic frameworks for the heterogeneous catalysis of quinazolinones. Inorg. Chem. 2023, 62, 5565–5575. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chang, S.; An, H.; Li, Y.; Zhu, Q.; Luo, H.; Huang, Y. Two polymorphic polyoxometalate-based metal–organic frameworks for the efficient synthesis of functionalized sulfoxides and detoxification of mustard gas simulants. ACS Sustain. Chem. Eng. 2021, 9, 15683–15693. [Google Scholar] [CrossRef]
- Falaise, C.; Moussawi, M.A.; Floquet, S.; Abramov, P.A.; Sokolov, M.N.; Haouas, M.; Cadot, E. Probing dynamic library of metal-oxo building blocks with γ-cyclodextrin. J. Am. Chem. Soc. 2018, 140, 11198. [Google Scholar] [CrossRef]
- Wu, Y.; Shi, R.; Wu, Y.-L.; Holcroft, J.M.; Liu, Z.; Frasconi, M.; Wasielewski, M.R.; Li, H.; Stoddart, J.F. Complexation of polyoxometalates with cyclodextrins. J. Am. Chem. Soc. 2015, 137, 4111–4118. [Google Scholar] [CrossRef]
- Khlifi, S.; Marrot, J.; Haouas, M.; Shepard, W.E.; Falaise, C.; Cadot, E. Chaotropic effect as an assembly motif to construct supramolecular cyclodextrin–polyoxometalate-based frameworks. J. Am. Chem. Soc. 2022, 144, 4469–4477. [Google Scholar] [CrossRef]
- Gumerova, N.I.; Rompel, A. Synthesis, structures and applications of electron-rich polyoxometalates. Nat. Rev. Chem. 2018, 2, 0112. [Google Scholar] [CrossRef]
- Cao, Y.; Chen, Q.; Shen, C.; He, L. Polyoxometalate-based catalysts for CO2 conversion. Molecules 2019, 24, 2069. [Google Scholar] [CrossRef] [PubMed]
- Aureliano, M. The future is bright for polyoxometalates. BioChem 2022, 2, 8–26. [Google Scholar] [CrossRef]
- Woźniak Budych, M.J.; Staszak, K.; Bajek, A.; Pniewski, F.; Jastrząb, R.; Staszak, M.; Tylkowski, B.; Wieszczycka, K. The future of polyoxymetalates for biological and chemical apllications. Coord. Chem. Rev. 2023, 493, 215306. [Google Scholar] [CrossRef]
- Carvalho, F.; Aureliano, M. Polyoxometalates impact as anticancer agents. Int. J. Mol. Sci. 2023, 24, 5043. [Google Scholar] [CrossRef] [PubMed]
- Bijelic, A.; Aureliano, M.; Rompel, A. Polyoxometalates as potential next-generation metallodrugs in the combat against cancer. Angew. Chem. Int. Ed. 2018, 58, 2980–2999. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, J.; Wang, H.; Lu, S.; Song, Y.; Chen, H.; Ma, Y.; Wang, L.; Sun, T. Combinatorial discovery of Mo-based polyoxometalate clusters for tumor photothermal therapy and normal cell protection. Biomater. Sci. 2020, 8, 6017–6024. [Google Scholar] [CrossRef] [PubMed]
- Ehlert, M.K.; Rettig, S.J.; Storr, A.; Thompson, R.C.; Trotter, J. Octamolybdenum oxo-pyrazolate clusters. Syntheses, characterization, and crystal and molecular structures of the Mo(V)/Mo(VI) and Mo(VI) octamolybdenum clusters Mo8(pz)6O18(pzH)6 and Mo8(pz)6O21(pzH)6. Inorg. Chem. 1993, 32, 5176–5182. [Google Scholar] [CrossRef]
- Modec, B.; Brenčič, J.V.; Finn, R.C.; Rarig, R.S.; Zubieta, J. Structural isomerism among octanuclear oxomolybdenum(V) coordination compounds with pyridines. Two isomers of [Mo8O16(OCH3)8(R Py)4]. Inorg. Chim. Acta 2001, 1–2, 113–119. [Google Scholar] [CrossRef]
- Modec, B.; Brenčič, J.V.; Rotar, R.; Golic, L.; Prout, K. An oxomolybdenum(V) cluster, [Mo8O16(OCH3)8(C5H5N)4]. Acta Cryst. C 1998, C54, 1573–1575. [Google Scholar] [CrossRef]
- Modec, B.; Brenčič, J.V.; Golič, L.; Daniels, L.M. Oxomolybdenum coordination compounds with alkyl-substituted pyridines. Solvothermal syntheses and structural characterization of [Mo10O26(3,5-Lut)8]·2(3,5-Lut), [Mo10O26(3-MePy)8]·(3-MePy), [Mo10O26(3-MePy)8] and [Mo12O28(OCH3)2Cl2(3-MePy)8]. Polyhedron 2000, 19, 1407–1414. [Google Scholar] [CrossRef]
- Modec, B.; Brenčič, J.V.; Zubieta, J.; Hagrman, P.J. Synthesis and structure of a high-nuclearity oxomolybdenum(V) complex, [Mo12O28(OC2H5)4(C6H7N)8]. Inorg. Chem. Comm. 2001, 4, 537–540. [Google Scholar] [CrossRef]
- Modec, B.; Brenčič, J.V.; Giester, G. A decanuclear oxomolybdenum(V,VI) cluster with 4-isopropylpyridine. Acta Cryst. C 2001, C57, 246–247. [Google Scholar] [CrossRef]
- Zhao, J.; Xu, L. Solvothermal treatment of triangular molybdenum(IV) oxo species—A new approach for the synthesis of new molybdenum oxo clusters. Eur. J. Inorg. Chem. 2011, 2011, 4096–4102. [Google Scholar] [CrossRef]
- Deng, L.; Dong, X.; Zhou, Z.-H. Intrinsic molybdenum-based POMOFs with impressive gas adsorptions and photochromism. Chem. Eur. J. 2021, 27, 9643–9653. [Google Scholar] [CrossRef]
- Gumerova, N.I.; Rompel, A. Polyoxometalates in solution: Speciation under spotlight. Chem. Soc. Rev. 2020, 49, 7568–7601. [Google Scholar] [CrossRef] [PubMed]
- Gumerova, N.I.; Rompel, A. Speciation atlas of polyoxometalates in aqueous solutions. Sci. Adv. 2023, 9, eadi081. [Google Scholar] [CrossRef] [PubMed]
- Majumder, S.; Pasayat, S.; Panda, A.K.; Dash, S.P.; Roy, S.; Biswas, A.; Varma, M.E.; Joshi, B.N.; Garribba, E.; Kausar, C.; et al. Monomeric and dimeric oxidomolybdenum(V and VI) complexes, cytotoxicity, and DNA interaction studies: Molybdenum assisted C=N bond cleavage of salophen ligands. Inorg. Chem. 2017, 56, 11190–11210. [Google Scholar] [CrossRef]
- Gretarsdottir, J.M.; Lambert, I.H.; Sturup, S.; Suman, S.G. In vitro characterization of a threonine-ligated molybdenyl–sulfide cluster as a putative cyanide poisoning antidote; intracellular distribution, effects on organic osmolyte homeostasis, and induction of cell death. ACS Pharmacol. Transl. Sci. 2022, 5, 907–918. [Google Scholar] [CrossRef]
- Fuior, A.; Cebotari, D.; Garbuz, O.; Calancea, S.; Gulea, A.; Floquet, S. Biological properties of a new class of [Mo2O2S2]-based thiosemicarbazone coordination complexes. Inorg. Chim. Acta 2023, 548, 121372. [Google Scholar] [CrossRef]
- Fuior, A.; Hijazi, A.; Garbuz, O.; Bulimaga, V.; Zosim, L.; Cebotari, D.; Haouas, M.; Toderaş, I.; Gulea, A.; Floquet, S. Screening of biological properties of MoV2O2S2- and MoV2O4-based coordination complexes: Investigation of antibacterial, antifungal, antioxidative and antitumoral activities versus growing of Spirulina platensis biomass. J. Inorg. Biochem. 2022, 226, 111627. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Hashemi, S.A.; Mazraedoost, S.; Chiang, W.-H.; Yousefi, K.; Arjmand, O.; Ghahramani, Y.; Gholami, A.; Omidifar, N.; Rumjit, N.P.; et al. Anticancer, antimicrobial and biomedical features of polyoxometalate as advanced materials: A review study. Inorg. Chem. Commun. 2022, 146, 110074. [Google Scholar] [CrossRef]
- Qi, Y.; Han, L.; Qi, Y.; Jin, X.; Zhang, B.; Niu, J.; Zhong, J.; Xu, Y. Anti-flavivirus activity of polyoxometalate. Antivir. Res. 2020, 179, 104813. [Google Scholar] [CrossRef]
- European Food Safety Authority, European Centre for Disease Prevention and Control, European Union Reference Laboratory for Avian Influenza; Adlhoch, C.; Fusaro, A.; Gonzales, J.L.; Kuiken, T.; Mirinaviciute, G.; Niqueux, É.; Stahl, K.; Staubach, C.; Terregino, C.; et al. Avian influenza overview March–April 2023. EFSA J. 2023, 21, e08039. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, R.; Saad, M.D.; Davis, C.T.; Swayne, D.E.; Wang, D.; Wong, F.Y.K.; McCauley, J.W.; Peiris, J.S.M.; Webby, R.J.; Fouchier, R.A.M.; et al. Pandemic potential of highly pathogenic avian influenza clade 2.3.4.4 A(H5) viruses. Rev. Med. Virol. 2020, 30, e2099. [Google Scholar] [CrossRef] [PubMed]
- Lugovtsev, V.Y.; Melnyk, D.; Weir, J.P. Heterogeneity of the MDCK cell line and its applicability for influenza virus research. PLoS ONE 2013, 8, e75014. [Google Scholar] [CrossRef]
- Kongsomros, S.; Manopwisedjaroen, S.; Chaopreecha, J.; Wang, S.-F.; Borwornpinyo, S.; Thitithanyanont, A. Rapid and efficient cell-to-cell transmission of avian influenza H5N1 virus in MDCK cells is achieved by trogocytosis. Pathogens 2021, 10, 483. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Y.; Xu, K.; Qi, Y.; Zhong, J.; Zhang, K.; Li, J.; Wang, E.; Wu, Z.; Kang, Z. Broad-spectrum antiviral property of polyoxometalate localized on a cell surface. ACS Appl. Mater. Interfaces 2014, 6, 9785–9789. [Google Scholar] [CrossRef] [PubMed]
- Aoki, F.Y.; Doucette, K.E. Oseltamivir: A clinical and pharmacological perspective. Expert Opin. Pharmacother. 2001, 2, 1671–1683. [Google Scholar] [CrossRef]
- Sidwell, R.W.; Huffman, J.H.; Barnard, D.L.; Bailey, K.W.; Wong, M.-H.; Morrison, A.; Syndergaard, T.; Kim, C.U. Inhibition of influenza virus infections in mice by GS4104, an orally effective influenza virus neuraminidase inhibitor. Antivir. Res. 1998, 37, 107–120. [Google Scholar] [CrossRef]
- Mitchell, P.C.H. The co-ordination chemistry of molybdenum—II(1): Properties of the oxalato complexes of molybdenum (V). J. Inorg. Nucl. Chem. 1964, 26, 1967–1976. [Google Scholar] [CrossRef]
- Bruker. APEX2 (Version 1.08), SAINT (Version 07.03), SADABS (Version 02.11), SHELXTL (Version 06.12); Bruker AXS Inc.: Madison, WI, USA, 2004. [Google Scholar]
- Kärber, G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Archiv Experiment. Pathol. Pharmakol. 1931, 162, 480–483. [Google Scholar] [CrossRef]
- Spearman, C. The method of ‘right and wrong cases’ (‘constant stimuli’) without gauss’s formulae. Br. J. Psychol. 1908, 2, 227–242. [Google Scholar] [CrossRef]
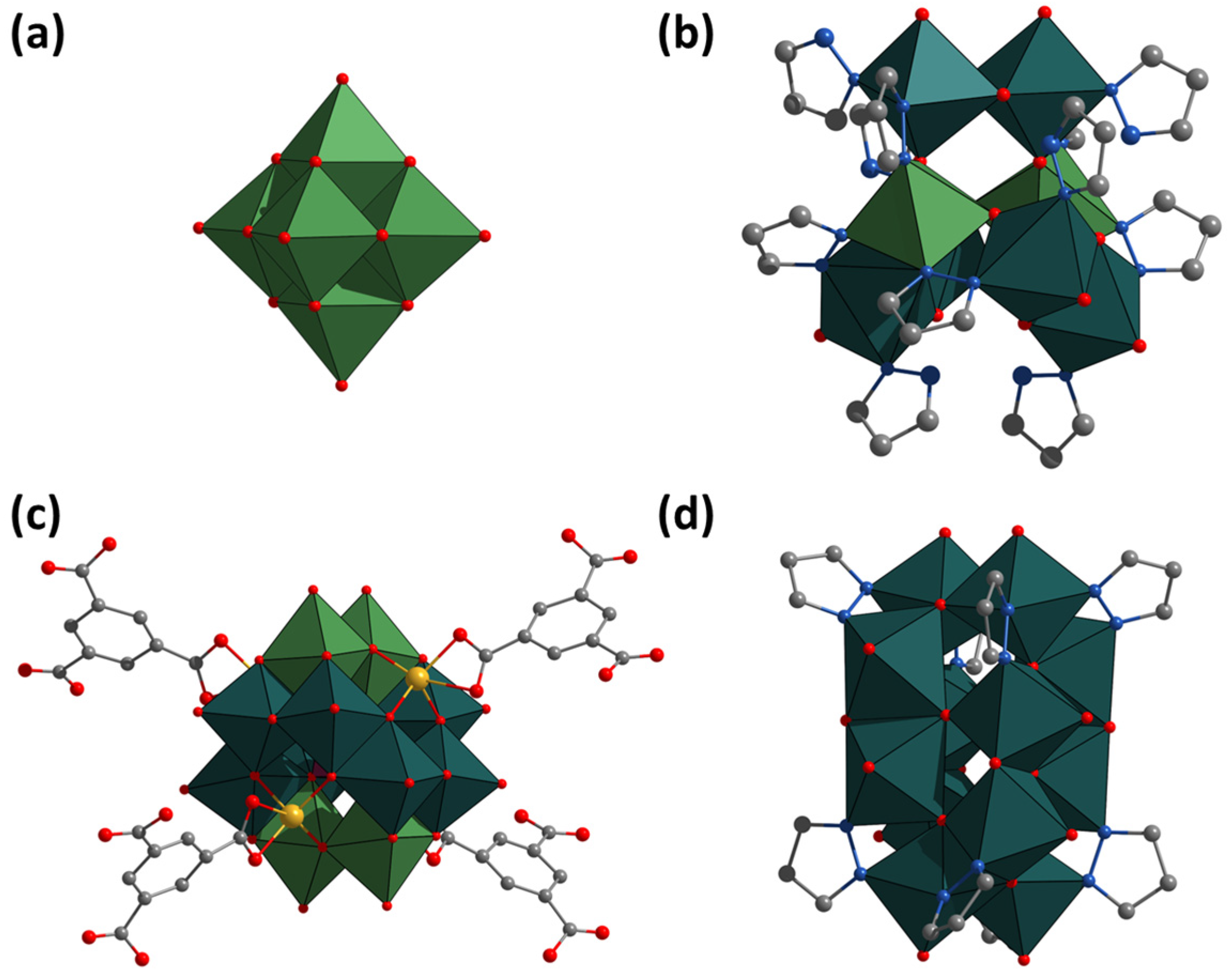
| Compound | Starting Compound | Yield | Reference |
|---|---|---|---|
| [MoV6MoVI2(μ-pz)6O18(pzH)6] (pzH = pyrazole) (Figure 1b) | Molybdenum blue | 29% | [18] |
| [MoV8O16(OCH3)8L4] (L = 4-methylpyridine (4-Me-Py), pyridine (Py)) | (PyH)2[MoVOCl5] | few crystals | [19,20] |
| [Mo8VMo2VIO26L8] (L = 3,5-lutidine, 3-methylpyridine (3-Me-Py)) | (PyH)2[MoVOCl5]/(NH4)2[MoVOCl5(H2O)] | 9–30% | [21] |
| [MoV12O28(OCH3)2Cl2(3-Me-Py)8] | (PyH)2[MoVOCl5] | 28% | [21] |
| [MoV12O28(OC2H5)4(4-Me-Py)8] | (PyH)[MoVOBr4(H2O)] | 75% | [22] |
| [MoV8MoVI2O26(4-iPr-Py)8] | (PyH)2[MoVOCl5] | few crystals | [23] |
| [MoV8MoVI2O26(py)8] | K2[MoIV3O4(Hnta)3] | 20% | [24] |
| [MoIV6MoVI4O24(py)8] | (H2bipy)[MoIV3O4(C2O4)3(H2O)3] | 60% | [24] |
| [Mo8O20(μ-1,2,3-trzH)8] (1,2,3-trzH = 1,2,3-triazole) | Na2MoVIO4 | 90% | [25] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konkova, A.V.; Savina, I.V.; Evtushok, D.V.; Pozmogova, T.N.; Solomatina, M.V.; Nokhova, A.R.; Alekseev, A.Y.; Kuratieva, N.V.; Eltsov, I.V.; Yanshole, V.V.; et al. Water-Soluble Polyoxometal Clusters of Molybdenum (V) with Pyrazole and Triazole: Synthesis and Study of Cytotoxicity and Antiviral Activity. Molecules 2023, 28, 8079. https://doi.org/10.3390/molecules28248079
Konkova AV, Savina IV, Evtushok DV, Pozmogova TN, Solomatina MV, Nokhova AR, Alekseev AY, Kuratieva NV, Eltsov IV, Yanshole VV, et al. Water-Soluble Polyoxometal Clusters of Molybdenum (V) with Pyrazole and Triazole: Synthesis and Study of Cytotoxicity and Antiviral Activity. Molecules. 2023; 28(24):8079. https://doi.org/10.3390/molecules28248079
Chicago/Turabian StyleKonkova, Anna V., Iulia V. Savina, Darya V. Evtushok, Tatiana N. Pozmogova, Maria V. Solomatina, Alina R. Nokhova, Alexander Y. Alekseev, Natalia V. Kuratieva, Ilia V. Eltsov, Vadim V. Yanshole, and et al. 2023. "Water-Soluble Polyoxometal Clusters of Molybdenum (V) with Pyrazole and Triazole: Synthesis and Study of Cytotoxicity and Antiviral Activity" Molecules 28, no. 24: 8079. https://doi.org/10.3390/molecules28248079
APA StyleKonkova, A. V., Savina, I. V., Evtushok, D. V., Pozmogova, T. N., Solomatina, M. V., Nokhova, A. R., Alekseev, A. Y., Kuratieva, N. V., Eltsov, I. V., Yanshole, V. V., Shestopalov, A. M., Ivanov, A. A., & Shestopalov, M. A. (2023). Water-Soluble Polyoxometal Clusters of Molybdenum (V) with Pyrazole and Triazole: Synthesis and Study of Cytotoxicity and Antiviral Activity. Molecules, 28(24), 8079. https://doi.org/10.3390/molecules28248079





