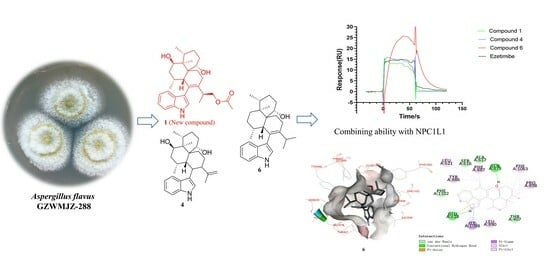
Indole Diterpene Derivatives from the Aspergillus flavus GZWMJZ-288, an Endophytic Fungus from Garcinia multiflora
Abstract
1. Introduction
2. Results and Discussion
2.1. Strain Fermentation and Secondary Metabolites’ Isolation
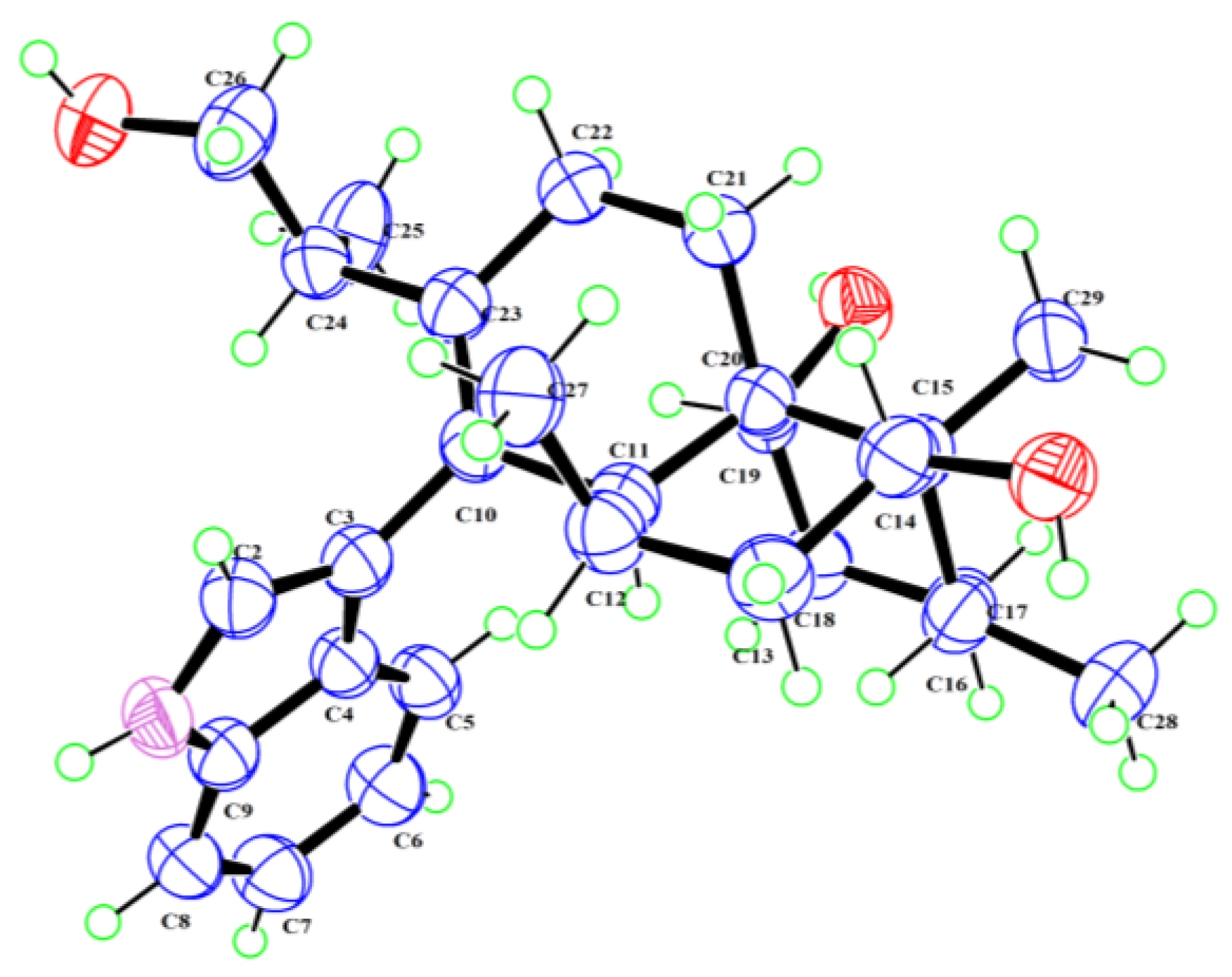
2.2. Structural Characterization of Isolated Compounds
2.3. Biological Activities
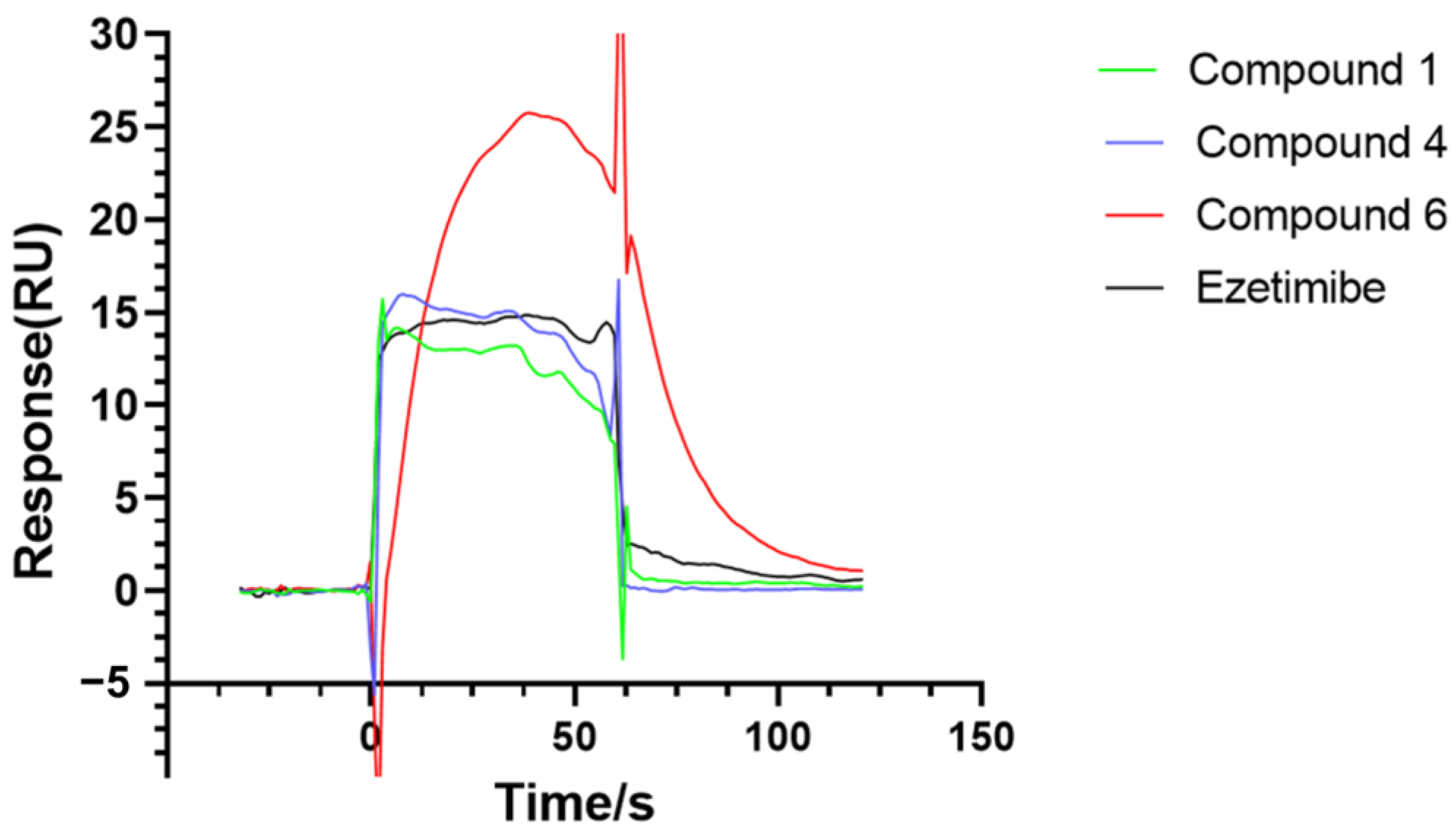
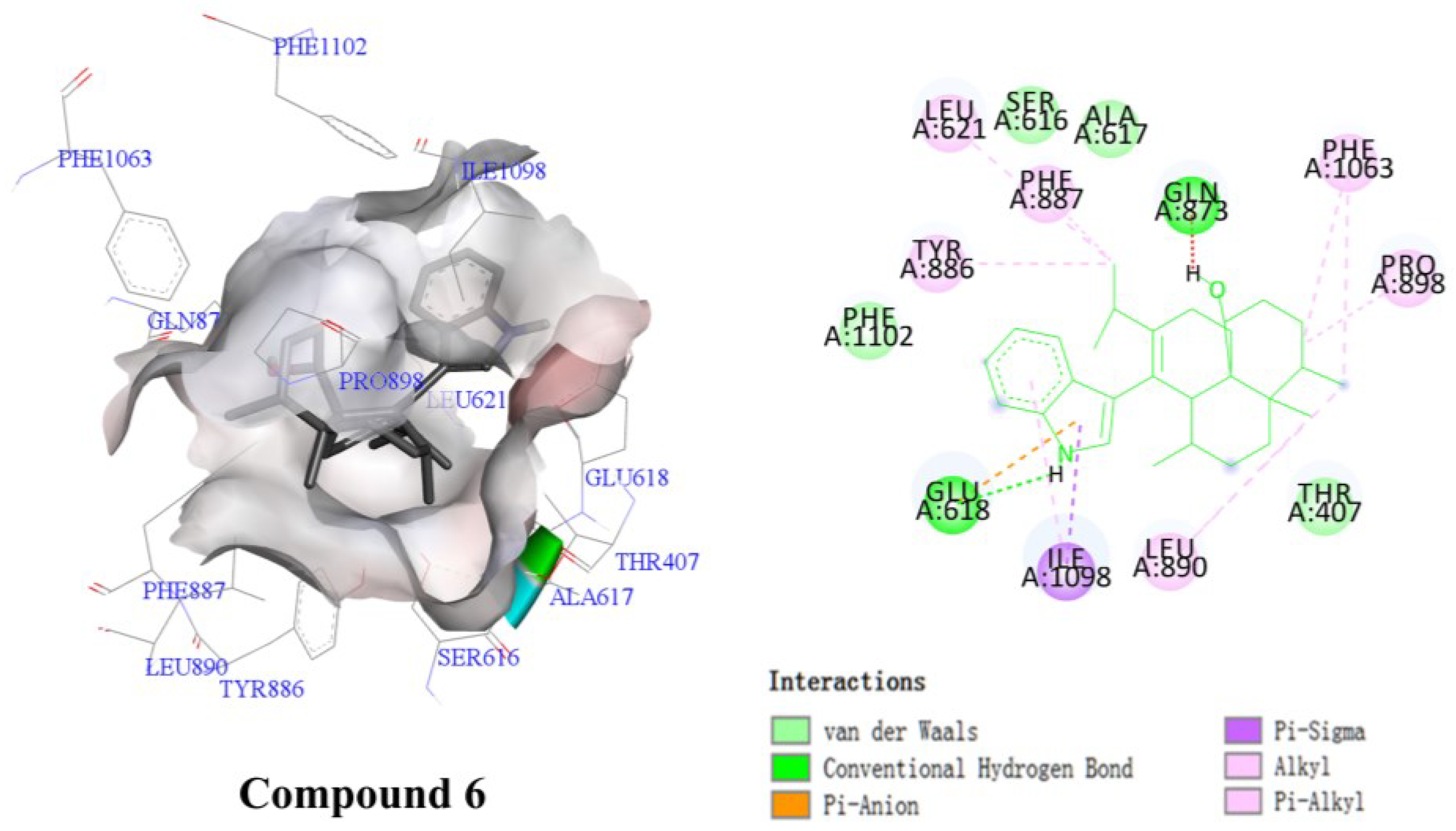
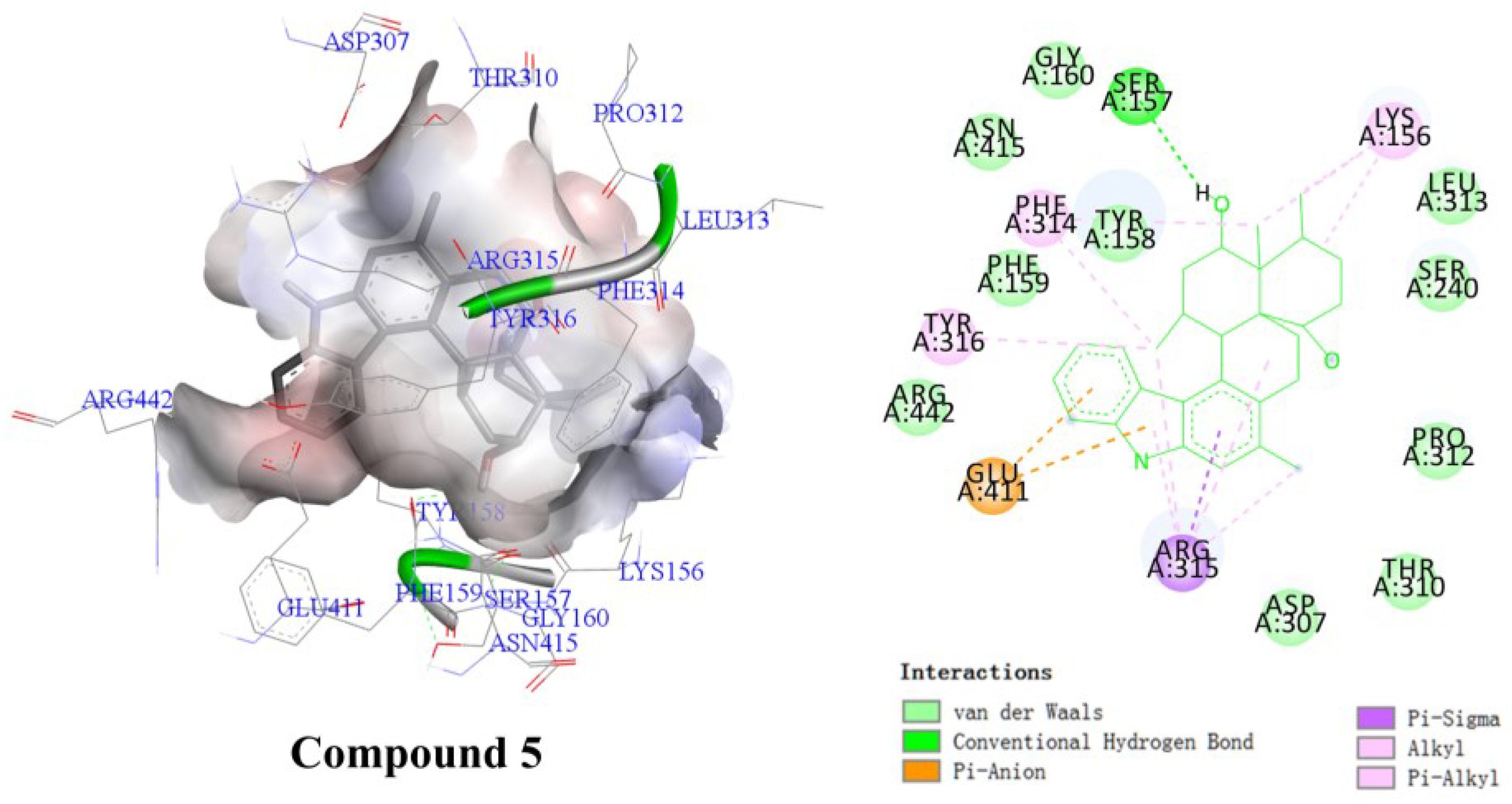
2.3.1. Results of Combining Ability with NPC1L1
2.3.2. α-Glucosidase Inhibitory Activity
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Strain Isolation and Fermentation
3.3. Extraction and Isolation
3.4. Physical Properties and Spectral Data of 1–6
3.5. Hydrolysis Reaction of Compound 1
3.6. Combining Ability Test with NPC1L1
3.7. α-Glucosidase Inhibitory Activity
3.8. Details of Docking Research
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gao, H.; Li, G.; Lou, H.-X. Structural Diversity and Biological Activities of Novel Secondary Metabolites from Endophytes. Molecules 2018, 23, 646. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.; Boyle, C.; Draeger, S.; Rommert, A.-K.; Krohn, K. Endophytic fungi: A source of novel biologically active secondary metabolites. Mycol. Res. 2002, 106, 996–1004. [Google Scholar] [CrossRef]
- Uzma, F.; Mohan, C.D.; Hashem, A.; Konappa, N.M.; Rangappa, S.; Kamath, P.V.; Singh, B.P.; Mudili, V.; Gupta, V.K.; Siddaiah, C.N.; et al. Endophytic Fungi-Alternative Sources of Cytotoxic Compounds: A Review. Front. Pharmacol. 2018, 9, 309. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.S.; Silva, C.a.D.; Hamerski, L. Natural Products from Endophytic Fungi Associated with Rubiaceae Species. J. Fungi 2020, 6, 128. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, L.; Gong, Q.-H.; Zhu, G.; Yuan, C.; Zuo, M.; Rao, Q.; Zhu, W.; Hao, X.-J. Kojic Acid Derivatives and Sesquiterpenes from the Aspergillus flavus GZWMJZ-288, A Fungal Endophyte of Garcinia multiflora. Nat. Prod. Commun. 2018, 13, 1421–1424. [Google Scholar] [CrossRef]
- He, W.; Xu, Y.; Wu, D.; Wang, D.; Gao, H.; Wang, L.; Zhu, W. New alkaloids from the diversity-enhanced extracts of an endophytic fungus Aspergillus flavus GZWMJZ-288. Bioorg. Chem. 2021, 107, 104623. [Google Scholar] [CrossRef] [PubMed]
- Gloer, J.B.; Tepaske, M.R.; Sima, J.S.; Wicklow, D.T.; Dowd, P.F. Antiinsectan Aflavinine Derivatives from the Sclerotia of Aspergillus flavus. J. Org. Chem. 1988, 53, 5457–5460. [Google Scholar] [CrossRef]
- Zou, X.; Liu, S.; Zheng, Z.; Zhang, H.; Chen, X.; Liu, X.; Li, E. Two New Imidazolone-Containing Alkaloids and Further Metabolites from the Ascomycete Fungus sp. Chem. Biodivers. 2011, 8, 1914–1920. [Google Scholar] [CrossRef]
- Wicklow, D.T.; Dowd, P.F.; Tepaske, M.R.; Gloer, J.B. Sclerotial metabolites of Aspergillus flavus toxic to a detritivorous maize insect (carpophilus hemipterus, nitidulidae). Trans. Br. Mycol. Soc. 1988, 91, 433–438. [Google Scholar] [CrossRef]
- Li, H.; Chen, Q.; Lu, Z.; Li, A. Total Syntheses of Aflavazole and 14-Hydroxyaflavinine. J. Am. Chem. Soc. 2016, 138, 15555–15558. [Google Scholar] [CrossRef]
- Tepaskea, M.R.; Gloer, J.B.; Wicklowb, D.T.; Dowd, P.F. Three new Aflavinines from the Sclerotia of Aspergillus tubingensis. Tetrahedron 1989, 45, 4961–4968. [Google Scholar] [CrossRef]
- Gallagher, R.T.; Mccabe, T.; Hirotsu, K.; Clardy, J.; Nicholson, J.; Wilson, B.J. Aflavinine, a Novel Indole-mevalonate Metabolite from Tremorgen-producing Aspergillus flavus species. Tetrahedron Lett. 1980, 21, 243–246. [Google Scholar] [CrossRef]
- Cole, R.J.; Dorner, J.W.; Springer, J.P.; Cox, R.H. Indole Metabolites from a Strain of Aspergillus flavus. J. Agric. Food Chem. 1981, 29, 293–295. [Google Scholar] [CrossRef]
- Nozawa, K.; Sekita, S.; Harada, M.; Udagawa, S.; Kawai, K.-C. Isolation and Structures of Two new Indoloditerpenes related to aflavinine from a microsclerotium-producing strain of Aspergillus flavus. Chem. Pharm. Bull. 1989, 37, 626–630. [Google Scholar] [CrossRef][Green Version]
- Tang, M.-C.; Lin, H.-C.; Li, D.; Yi Zou, J.L.; Xu, W.; Cacho, R.A.; Hillenmeyer, M.E.; Garg, N.K.; Tang, Y. Discovery of unclustered fungal indole diterpene biosynthetic pathways through combinatorial pathway reassembly in engineered yeast. J. Am. Chem. Soc. 2015, 137, 13724–13727. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Bao, X.-F.; Wang, C.-X.; Xie, J.; Song, X.-J.; Dai, P.; Chen, G.-D.; Hu, D.; Yao, X.-S.; Gao, H. Cladosporine A, a new indole diterpenoid alkaloid with antimicrobial activities from Cladosporium sp. Nat. Prod. Res. 2021, 35, 1115–1121. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Chen, T.; Tan, Q.; Zang, Z.; Chen, Y.; Ou, Y.; Li, G.; Hu, D.; Wang, B.; Yao, H.; et al. Plasmodium-Resistant Indole Diterpenoid Biosynthesis Gene Cluster Derived from Aspergillus oryzae Was Activated by Exogenous P450 Gene Ast B. J. Nat. Prod. 2023, 86, 1392–1401. [Google Scholar] [CrossRef]
- Huang, C.-S.; Yu, X.; Fordstrom, P.; Choi, K.; Chung, B.C.; Roh, S.-H.; Chiu, W.; Zhou, M.; Min, X.; Wang, Z. Cryo-EM structures of NPC1L1 reveal mechanisms of cholesterol transport and ezetimibe inhibition. Sci. Adv. 2020, 6, eabb1989. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, W.; Zeng, J.; Meng, J.; Jiang, H.; Wang, J.; Xing, D. Niemann-Pick C1-Like 1 inhibitors for reducing cholesterol absorption. Eur. J. Med. Chem. 2022, 230, 114111. [Google Scholar] [CrossRef]
- Zhang, R.; Song, Z.; Wang, X.; Xue, J.; Xing, D. One-step modification to identify dual-inhibitors targeting both pancreatic triglyceride lipase and Niemann-Pick C1-like 1. Eur. J. Med. Chem. 2021, 216, 113358. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, W.; Zeng, J.; Meng, J.; Shi, L.; Yang, S.; Chang, J.; Wang, C.; Xing, K.; Wen, J.; et al. Recent advances in the screening methods of NPC1L1 inhibitors. Biomed. Pharmacother. 2022, 155, 113732. [Google Scholar] [CrossRef] [PubMed]
- Gür, F.; Gür, B.; Erkayman, B.; Halıcı, Z.; Karakoç, A. Investigation of serum and brain superoxide dismutase levels depending on atomoxetine used in attention-deficit/hyperactivity disorder treatment: A combination of and molecular docking studies. Bioorg. Chem. 2020, 105, 104435. [Google Scholar] [CrossRef] [PubMed]
- Yousuf, H.; Shamim, S.; Khan, K.M.; Chigurupati, S.; Kanwal; Hameed, S.; Khan, M.N.; Taha, M.; Arfeen, M. Dihydropyridines as potential α-amylase and α-glucosidase inhibitors: Synthesis, in vitro and in silico studies. Bioorg. Chem. 2020, 96, 103581. [Google Scholar] [CrossRef] [PubMed]
- Tepaske, M.R.; Gloer, J.B.; Wicklow, D.T.; Dowd, P.F. Aflavazole—A New Antiinsectan Carbazole Metabolite from the Sclerotia of Aspergillus-Flavus. J. Org. Chem. 1990, 55, 5299–5301. [Google Scholar] [CrossRef]
- Dean, M.; Moitra, K.; Allikmets, R. The human ATP-binding cassette (ABC) transporter superfamily. Hum. Mutat. 2022, 43, 1162–1182. [Google Scholar] [CrossRef] [PubMed]
- Menteşe, E.; Baltaş, N.; Emirik, M. Synthesis, α-glucosidase inhibition and in silico studies of some 4-(5-fluoro-2- substituted-1H-benzimidazol-6-yl) morpholine derivatives. Bioorg. Chem. 2020, 101, 104002. [Google Scholar] [CrossRef] [PubMed]
- Aispuro-Pérez, A.; López-Ávalos, J.; García-Páez, F.; Montes-Avila, J.; Picos-Corrales, L.A.; Ochoa-Terán, A.; Bastidas, P.; Montaño, S.; Calderón-Zamora, L.; Osuna-Martínez, U.; et al. Synthesis and molecular docking studies of imines as a-glucosidase and a-amylase inhibitors. Bioorg. Chem. 2020, 94, 103491. [Google Scholar] [CrossRef]


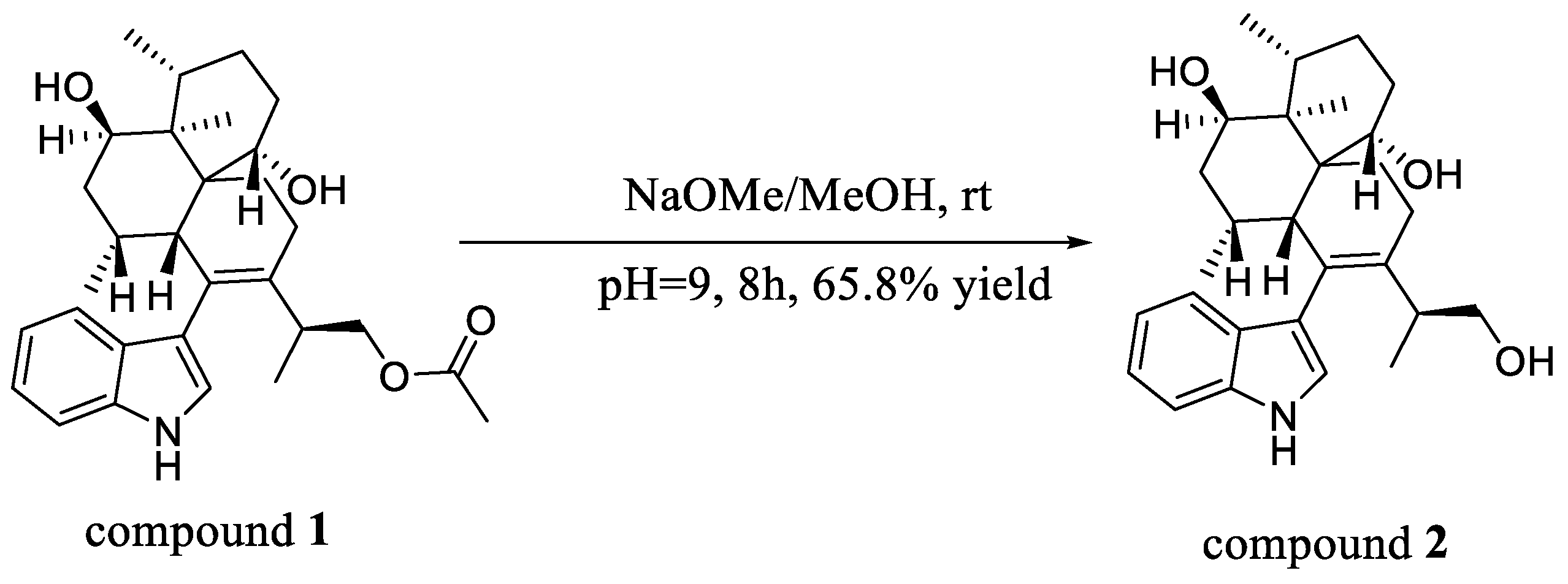

| Position | 1 | 2 | Hydrolysate of 1 | ||
|---|---|---|---|---|---|
| δC | δH, (J in Hz) | δC | δH, (J in Hz) | δH, (J in Hz) | |
| 2 | 122.3, CH | 6.93, d (2.0) | 122.5, CH | 6.98, d (2.0) | 6.97, d (2.1) |
| 3 | 116.1, C | 118.4, C | |||
| 4 | 126.8, C | 127.4, C | |||
| 5 | 118.4, CH | 7.29, d (7.9) | 118.7, CH | 7.35, d (8.1) | 7.34, d (7.8) |
| 6 | 118.5, CH | 6.95, t (7.9) | 116.4, CH | 6.94, t (8.1) | 6.93, t (7.8) |
| 7 | 120.8, CH | 7.05, t (7.6) | 120.7, CH | 7.04, t (7.7) | 7.04, t (7.5) |
| 8 | 111.6, CH | 7.35, d (7.6) | 111.5, CH | 7.34, d (7.7) | 7.32, d (7.5) |
| 9 | 135.8, C | 135.9, C | |||
| 10 | 128.9, C | 126.9, C | |||
| 11 | 43.2, CH | 2.34, d (5.6) | 43.2, CH | 2.35, d (6.3) | 2.33, d (5.9) |
| 12 | 29.5, CH | 1.96–2.00, overlap | 29.6, CH | 1.92–2.02, overlap | 1.92–2.01, overlap |
| 13 | 35.3, CH2 | 1.36, d (12.3) 1.76–1.81, m | 35.4, CH2 | 1.35, d (12.2) 1.79, ddd (12.2, 12.2, 5.9) | 1.34, d (12.5) 1.78, ddd (12.5, 12.5, 5.7) |
| 14 | 69.5, CH | 3.80–3.83, overlap | 68.1, CH | 3.83, ddd (11.9, 4.3, 3.7) | 3.81, ddd (12.5, 4.5 3.8) |
| 15 | 42.9, C | 43.9, C | |||
| 16 | 31.0, CH | 1.96–2.00, overlap | 31.0, CH | 1.92–2.02, overlap | 1.92–2.01, overlap |
| 17 | 27.3, CH2 | 1.01, d (11.9) 1.66–1.71, m | 27.3, CH2 | 1.02, d (11.8) 1.65–1.73, m | 1.01, d (11.5) 1.65–1.69, m |
| 18 | 30.2, CH2 | 1.57–1.60, m 1.82–1.87, m | 30.3, CH2 | 1.56–1.60, m 1.84–1.91, m | 1.55–1.59, m 1.84–1.88, m |
| 19 | 68.0, CH | 4.26, brs | 69.6, CH | 4.27, brs | 4.25, brs |
| 20 | 43.9, C | 43.0, C | |||
| 21 | 21.5, CH2 | 1.61–1.65, overlap 2.16, dd (12.4, 7.7) | 21.7, CH2 | 1.61–1.64, m 2.14, dd (12.3, 7.4) | 1.59–1.62, m 2.12, dd (12.5, 7.3) |
| 22 | 20.5, CH2 | 1.91–1.96, m 2.26–2.32, m | 21.3, CH2 | 1.92–2.02, overlap 2.25–2.32, m | 1.92–2.01, overlap 2.26–2.30, m |
| 23 | 135.6, C | 137.1, C | |||
| 24 | 35.3, CH | 2.67–2.72, m | 38.7, CH | 2.50–2.53, m | 2.51–2.54, m |
| 25 | 15.3, CH3 | 0.77, d (7.2) | 15.4, CH3 | 0.75, d (7.0) | 0.74, d (7.1) |
| 26 | 66.6, CH2 | 3.84, dd (10.3, 7.1) 3.93, dd (10.3, 9.9) | 65.1, CH2 | 3.20–3.27, m 4.30, t (4.6) | 3.20–3.23, m 4.27, t (5.0) |
| 27 | 19.1, CH3 | 1.11, d (7.5) | 19.3, CH3 | 1.10, d (7.2) | 1.09, d (7.2) |
| 28 | 19.3, CH3 | 0.93, d (6.5) | 19.4, CH3 | 0.94, d (7.1) | 0.93, d (6.8) |
| 29 | 13.3, CH3 | 1.14, s | 13.3, CH3 | 1.15, s | 1.13, s |
| 30 | 170.2, C | ||||
| 31 | 20.8, CH3 | 1.98, s | |||
| 14-OH | 4.08, d (4.5) | 4.06, d (4.3) | 4.04, d (4.5) | ||
| 19-OH | 4.27, d, (4.2) | 4.22, d (4.6) | 4.21, d (4.2) | ||
| NH | 10.93, d (1.8) | 10.89, d (1.5) | 10.89, d (1.4) | ||
| Compounds | 1 | 2 | 3 | 4 | 5 | 6 | Acarbose |
|---|---|---|---|---|---|---|---|
| IC50 ± SD (μM) | >300 | >300 | >300 | >300 | 29.22 ± 0.83 | >300 | 387.27 ± 19.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Zhuang, X.; Yin, Y.; Wu, D.; He, W.; Zhu, W.; Xu, Y.; Zuo, M.; Wang, L. Indole Diterpene Derivatives from the Aspergillus flavus GZWMJZ-288, an Endophytic Fungus from Garcinia multiflora. Molecules 2023, 28, 7931. https://doi.org/10.3390/molecules28237931
Wang D, Zhuang X, Yin Y, Wu D, He W, Zhu W, Xu Y, Zuo M, Wang L. Indole Diterpene Derivatives from the Aspergillus flavus GZWMJZ-288, an Endophytic Fungus from Garcinia multiflora. Molecules. 2023; 28(23):7931. https://doi.org/10.3390/molecules28237931
Chicago/Turabian StyleWang, Dongyang, Xiaohong Zhuang, Ying Yin, Dan Wu, Wenwen He, Weiming Zhu, Yanchao Xu, Mingxing Zuo, and Liping Wang. 2023. "Indole Diterpene Derivatives from the Aspergillus flavus GZWMJZ-288, an Endophytic Fungus from Garcinia multiflora" Molecules 28, no. 23: 7931. https://doi.org/10.3390/molecules28237931
APA StyleWang, D., Zhuang, X., Yin, Y., Wu, D., He, W., Zhu, W., Xu, Y., Zuo, M., & Wang, L. (2023). Indole Diterpene Derivatives from the Aspergillus flavus GZWMJZ-288, an Endophytic Fungus from Garcinia multiflora. Molecules, 28(23), 7931. https://doi.org/10.3390/molecules28237931