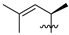
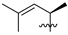
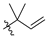
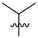
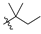
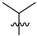
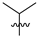
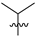
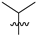
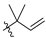
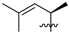
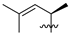
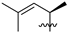
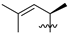
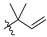
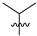
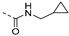
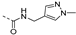
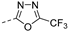
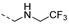Abstract
Peptide compounds play a significant role in medicinal chemistry as they can inhibit the activity of species that cause malaria. This literature review summarizes the isolation of antimalarial peptides, the synthesis method with the detailed structure and sequences of each peptide, and discusses the biological activity of the isolated and synthesized compounds. The synthetic routes and reactions for cyclic and linear antimalarial peptides are systematically highlighted in this review including preparing building blocks, protection and deprotection, coupling and cyclization reactions until the target compound is obtained. Based on the literature data and the results, this review’s aim is to provide information to discover and synthesize more antimalarial peptide for future research.
1. Introduction
Malaria is a common disease in Africa and certain Asian countries and is one of the leading causes of death worldwide. Indeed, the World Health Organization (WHO) reported that the number of deaths increased by 10% in 2020 compared with 2019, but declined to an estimated 619,000 deaths in 2021. Furthermore, nearly half of the world’s population is at risk of malaria [1].
Malaria is caused by a tiny protozoon belonging to the Plasmodium species that is spread to humans through the bite of an infected female Anopheles mosquito. Of the 172 Plasmodium species, five can infect humans, including Plasmodium malariae, P. falciparum, P. vivax, P. ovale, and P. knowlesi [2]. Many drugs have been produced to treat and prevent this disease. For example, several peptides have been identified as possessing antimalaria properties against P. falciparum. These antimalarial peptides were initially isolated from plants or animals and the low yields have stimulated great advances in the development of the synthetic methodologies and synthetic analogs. This review will discuss the relevant literature published between 2015 and 2023 regarding the isolation techniques, chemical synthesis, and biological activity of the antimalarial peptides.
2. Isolation of Antimalarial Peptide
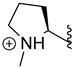
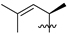
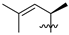
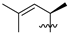
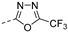
Nature has become a source of antimalarial peptides. Various types of peptides with antimalarial activity have been isolated with cyclic and linear structures (Table 1). Peptides with antimalarial activity are commonly found as cyclopeptides; for example, ribifolin is a cyclopeptide containing eight amino acid residues (ILGSIILG) that has antimalarial activity (Figure 1) and was first isolated by Pinto et al. in 2015 from the aerial parts of Jatropha ribifolia. The peptide was extracted using Strata C18-E solid-phase extraction catridge using H2O and CH3CN as mobile phases (0% 30%, 70%, and 100% CH3CN in H2O), then purified by semipreparative HPLC using a linear gradient of 40−80% of CH3CN, containing 0.036% TFA in H2O containing 0.045% TFA for 120 min [3].

Table 1.
Isolated antimalarial peptides.
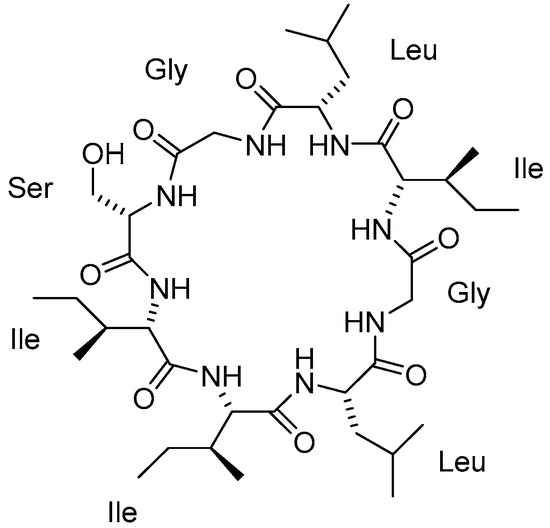
Figure 1.
Structure of ribifolin.
Antimalarial cyclopeptides can also be isolated from alkaloid-producing plants, such as the cyclopeptide alkaloids hymenocardine (2), hymenocardinol (3), hymenocardine N-oxide (4), and hymenocardine-H (5) (Figure 2) extracted from the root bark of Hymenocardia acida with 80% methanol by Tuenter et al. in 2016 [4]. The crude extract was fractioned by liquid–liquid partitioning followed by flash chromatography, and then the compounds were purified by semipreparative HPLC with DAD and ESIMS detection.
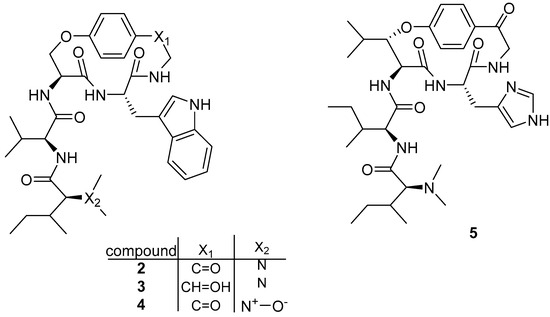
Figure 2.
Structure of cyclopeptide alkaloids isolated from Hymenocardia acida.
Tuenter et al. (2016) also isolated nine cyclopeptide alkaloids from two different batches of the roots of Ziziphus oxyphylla. The first batch was subjected to a general liquid−liquid partition and fractionation scheme, whereas a more alkaloid-specific fractionation procedure was used for the second batch to increase the number of alkaloids detected. The single compounds were purified by semipreparative HPLC with DAD and ESIMS detection with LC-DAD-SPE-NMR and identified as nummularine-R (6), O-desmethylnummularine-R (7), O-desmethylnummularine-R N-oxide (8), hemsine-A (9), hemsine-A N-oxide (10), ramosine-A (11), oxyphylline-C (12), oxyphylline-E (13), and oxyphylline-F (14) (Figure 3) [5].
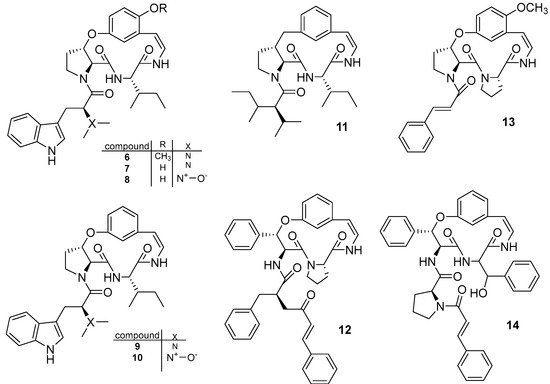
Figure 3.
Structure of cyclopeptide alkaloids isolated from Ziziphus oxyphylla.
Maluf et al. (2016) isolated a 42-amino acid polypeptide (YKCHKKGGHCFPKEKICLPPSSDFGKMDCRWRWKCCKKGSG, MW = 4889.81 Da) named crotamine (15) from rattlesnake venom [6]. Six hundred milligrams of crude dried venom was dissolved in 5 mL of 0.25 M ammonium formate buffer (pH 3.5) and the major component crotoxin was eliminated by slow-speed centrifugation before crotamine was recovered in a narrow protein peak by raising the NaCl concentration and acid hydrolysis. The purity of crotamine was further confirmed by analytical RP-HPLC with a C18-column, LCMS, and ESI-MS.
Two new cyclic octadepsipeptides octaminomycin A (16) and octaminomycin B (Figure 4) (17) discovered by Jang et al. (2017) exhibited activity against P. falciparum. The octadepsipeptides were isolated from a microbial metabolite fraction library of Streptomyces sp. RK85-270 based on Natural Products Plot (NPPlot) screening using liquid chromatography for extraction and purification [7].
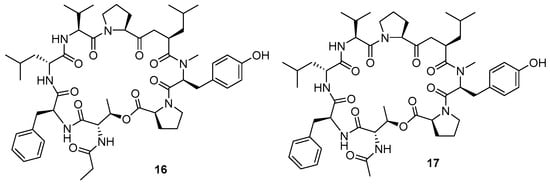
Figure 4.
Structure of cyclic octadepsipeptides isolated from Streptomyces sp. RK85-270.
Sweeney–Jones et al. (2020) isolated an antimalarial cyclopeptide named kakeromamide B (18) (Figure 5) from Fijian marine cynobacterium by VLC using 80 mL of silica gel in a glass vacuum funnel [8]. The fractions were collected using a stepped gradient of hexane, EtOAc, and MeOH (100% hexane, 10% EtOAc/90% hexane, 20% EtOAc/80% hexane, 40% EtOAc/60% hexane, 60% EtOAc/40% hexane, 80% EtOAc/20% hexane, 100% EtOAc, 25% MeOH/75% EtOAc, and 100% MeOH), then separated by reverse SPE phase (gradient decreasing polarity gradually starting with 15% MeOH in H2O to 100% MeOH). Fractions 3, 4, and 5 were separated again by HPLC using a C18-silica column to obtain kakeromamide B (tR = 17.5 min, 0.18 mg).
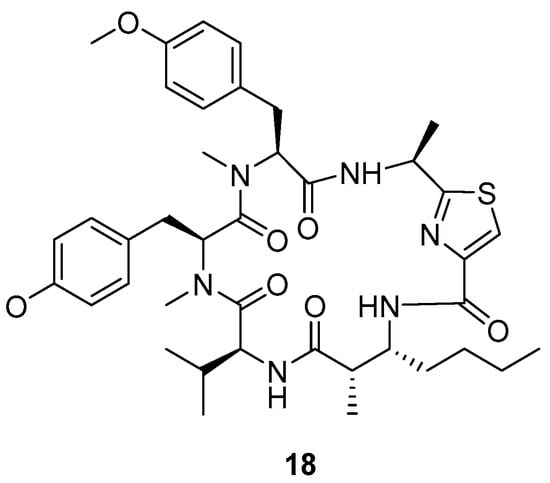
Figure 5.
Structure of kakeromide B.
Another cyclic peptide with antimalarial activity called pipecolisporin (19) (Figure 6) was isolated in 2021 by Fernández-Pastor et al. from the endophytic fungus Nigrospora oryzae CF-298113 [9]. A scaled-up culture of Nigrospora oryzae CF-298113 was extracted using methyl ethyl ketone (MEK) in BRFT medium and then purified using reversed-phase C18 medium pressure chromatography followed by semi-preparative reversed-phase HPLC on a phenyl column applying isocratic elution of H2O-CH3CN as the mobile phase (3.6 mL/min; 32% CH3CN for 40 min; UV detection at 210 nm). Pipecolisporin (19) was obtained as an amorphous white powder.
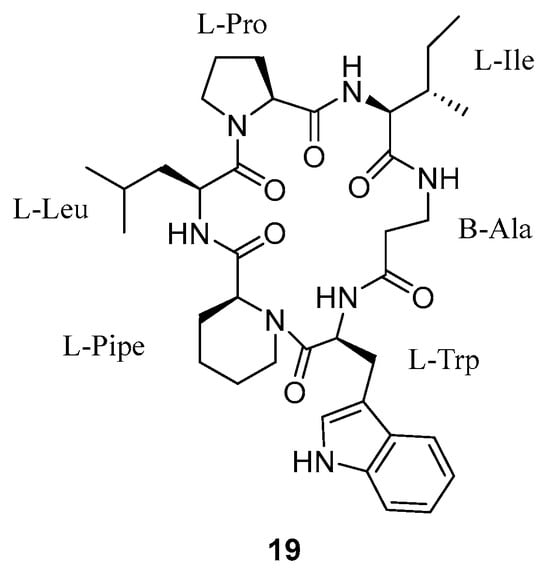
Figure 6.
Structure of pipecolisporin.
Subsequently, the antiplasmodial peptides kosidachin A (20) and kosidachin B (21) (Figure 7) were discovered by Watanabe et al. (2022) [10]. The compounds are cyclic tetrapeptides isolated from a culture broth of Pochonia binensis FKR-0564 by HPLC using Capcell Pak C18 column with an isocratic solvent system. Two new compounds were acquired from two fractions with retention times between 12.5–14.0 and 15.0–16.5 min.
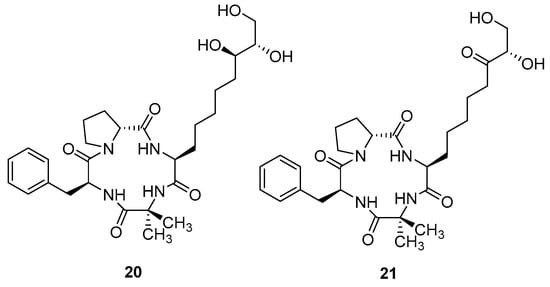
Figure 7.
Structure of koshidacin A and B.
Another antimalarial peptide georatusin (22) (Figure 8) was isolated by Shi et al. (2018) from Geomyces auratus, which is a mesophilic fungus [11]. The ethyl extract was fractionated using a flash purification system with a gradient elution of n-hexane:acetone and purified by semipreparative HPLC on a RP-HPLC with C-18 to obtain georatusin.
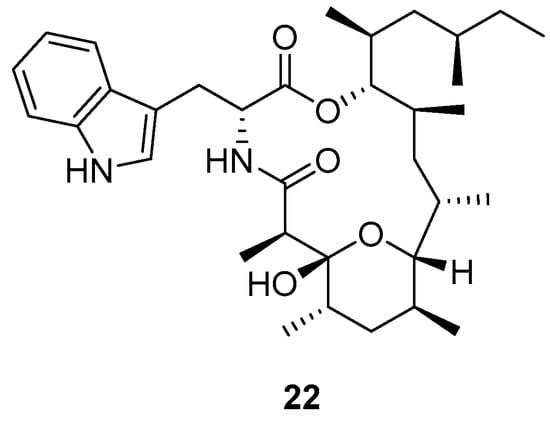
Figure 8.
Structure of georatusin.
Some linear peptides have also been reported to be active against malaria parasite. N-methylated linear peptides friomaramide B and shagamides A-F were purified from the marine sponge Inflatella coelosphaeroide by Bracegirdle et al. in 2022 (Figure 9) [12]. A collection of I. coelosphaeroides sponge samples was extracted, mounted onto silica gel and then partitioned using NP flash MPLC before fractionation by semipreparative C18 HPLC using a linear gradient. The individual fractions were then purified again by C18 HPLC to obtain friomaramide B (23), shagamide A (24), shagamide B (25), shagamide C (26), shagamide D (27), shagamide E (28), and shagamide F (29).
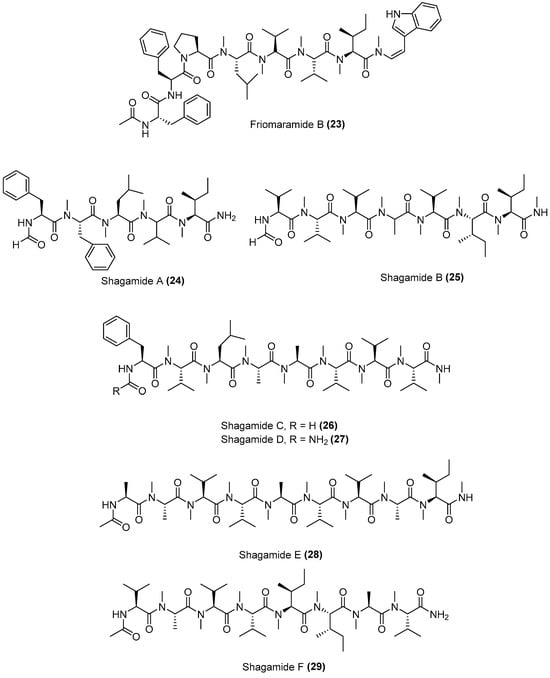
Figure 9.
Structure of linear antimalarial peptides isolated from Inflatella coelosphaeroide.
Recently, Cabrera-Muñoz et al. (2023) reported that the carboxypeptidase NpCI (30) inhibited P. falciparum growth (Figure 10) [13]. The compound was isolated from the mollusk Nerita versicolor and purified using a CPA-ehtyloxy-Sepharose CL-6B matrix. Figure 10 shows the sequence of NpCI and modelled folded of NpCI displaying a central compact region, a long N-tail, and a short C-tail.

Figure 10.
The derived putative 53-residue full protein sequence of NpCI and modelled folded of NpCI. The presence of six-cysteine (C) residues are highlighted in red.
3. Synthesis of Antimalarial Peptides
Cyclic and linear peptides have been synthesized by various methods as outlined in the following section.
3.1. Cyclopeptides
3.1.1. Mahafacyclin B
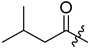
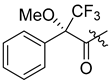
The antimalarial mahafacyclin B (32) is a cyclic heptapeptide that was synthesized by Fujita et al. (2013) using the soluble-tag assisted liquid–phase method [14]. This method allowed peptide head-to-tail cyclization leading to the total synthesis of mahafacyclin B assisted by hydrophobic benzyl alcohol. The synthesis was initiated by reductive amination of hydrophobic benzaldehydes using amino acid methyl esters to obtain hydrophobically tagged Gly-Ome from its reaction with coupling reagent Fmoc-Phe-OH, and a combination of HATU, HOAt, DIPEA, and THF. Basic deprotection of the hydrophobically tagged dipeptide produces diketopiperazine when the C-terminal residue is proline. The hydrophobically tagged tripeptide was deprotected to obtain the cyclized compound (Scheme 1). A less polar solvent was employed during cyclization to overcome the issue of the highly hydrophobic residues in the linear precursor. The amide nitrogen was used as the marking site allowing the rapid reaction checks and product isolation.
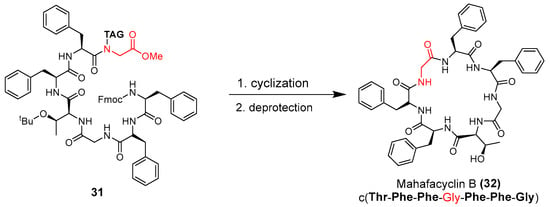
Scheme 1.
Synthesis of mahafacyclin B (32). Hydrophobically tagged Gly-OMe in compound 31 is highlighted in red. The untagged-glycine residue in compound 32 is highlighted in red.
3.1.2. Aerucylamide B
Aerucyclamide B (33) was synthesized by Peña et al. (2013) using two heterocycles and a dipeptide as building blocks (Scheme 2) [15]. Dipeptide 34 and thiazole 35 were synthesized in the first step and then HBTU was used as coupling reagent to produce the thiazole precursor 37 through cyclodehydration of β-hydroxythioamides and further oxidation methodology (Scheme 3).
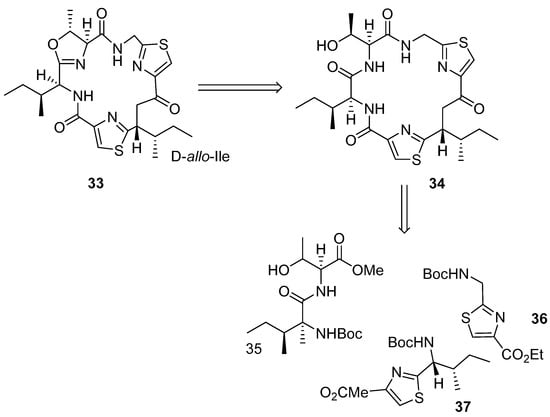
Scheme 2.
Retrosynthesis analysis of 33.
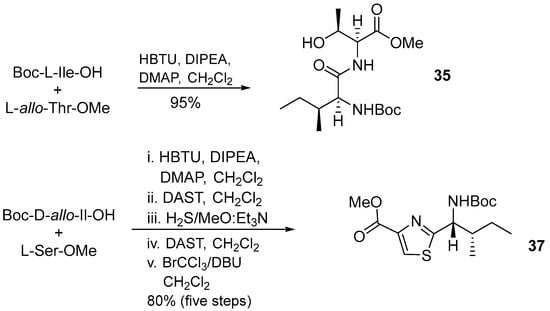
Scheme 3.
Synthesis of building blocks 35 and 37.
In the next step, two intermediate compounds 41 and 44 were prepared to obtain macrocycle 34. Compound 41 was synthesized via routes I and II (Scheme 4). Route I involved a coupling between the N-deprotected dipeptide 40 and intermediate 39, while route II involved coupling between compound 42 and 43.
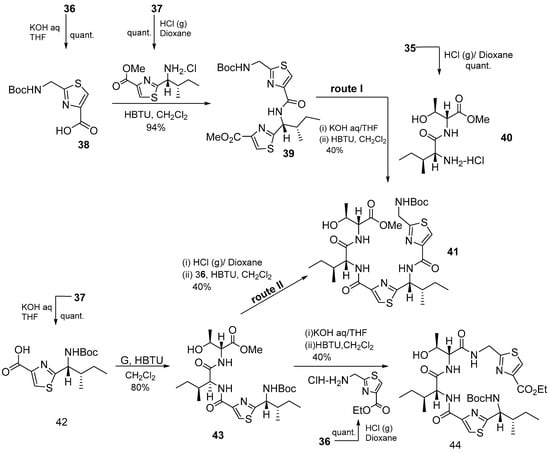
Scheme 4.
Synthesis of bis- and tris-heterocycles.
Macrocycle 34 was obtained from intermediates 41 and 44 using HBTU as a coupling agent (Scheme 5), and then aerucyclamide B was synthesized from 37 via intermediate 44, and from 36 via intermediate 41 [15].
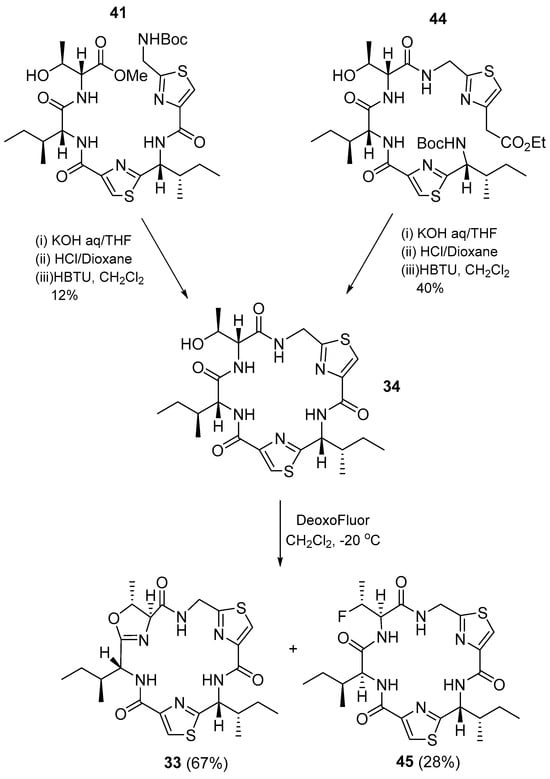
Scheme 5.
Synthesis of 34 and aerucyclamide B (33).
3.1.3. Cyclomarin
Cyclomarin A (46) (Figure 11) is a marine cycloheptapeptide with antimalarial activity that can be synthesized in an enantiomerically pure form [16]. Key steps in the synthesis of cyclomarin are asymmetric chelate−enolate Claisen rearrangement, asymmetric hydrogenation, and highly diastereoselective additions of organozinc and titanium reagents [16,17].
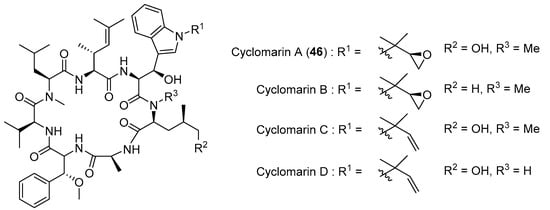
Figure 11.
Natural occuring cyclomarins.
The peptide synthesis developed to obtain the required building blocks is based on long-term experience in amino acid synthesis. Amino acid chelate enolate esters are used to obtain β-hydroxy through an aldol reaction, whereas γ,δ-unsaturated amino acids are obtained by transition metal-catalyzed allylic alkylation or Claisen enolate chelate rearrangement and syn-isomers are obtained by Claisen rearrangement. Thus, this approach was used to produce the desired N-protected amino acid (2S,3R)-50 (Scheme 6).
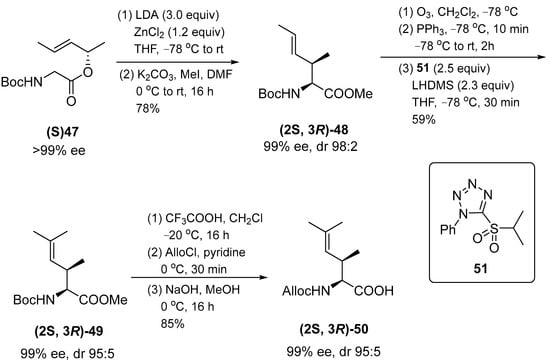
Scheme 6.
Synthesis of (2S,3R)–50.
The protected 5-hydroxyleucine 56 was synthesized with the commercially available Roche ester 52. The compound was reduced to the aldehyde 53, and used in an olefination using Schimdt’s phosphonoglycinate 57. The α,β-unsaturated amino acid derivative obtained was stereoselectively hydrogenated with (R)-monophos as a ligand. Following N-methylation and saponification, amino acid 56 was produced with good yield selectivity (Scheme 7).
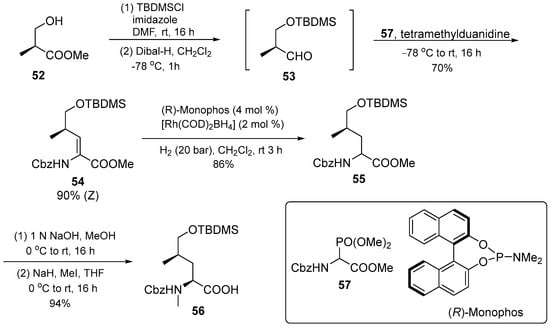
Scheme 7.
Synthesis of (2S,4R)–56.
β-methoxyphenylalanine 61 is the third unusual building block that was prepared through several attempts shown in Scheme 8. The first attempt using phenylmagnesium bromide (3 equiv.) and DIBAL, which gave an acceptable yield of the addition products (60%), but only moderate diastereoselective products (7:3 ratio). Consequently, the titanium reagent was added as the single diastereomer. Subsequent O-Methylation, cleavage of the silyl ether, and oxidation then gave rise to an excellent yield of the target acid 61. Another method was also reported for regioselectively prenylated from electron-withdrawing indole. The Pd-catalyzed protocol to 3-indolecarboxylate 62 was applied resulting in indole 63. The reaction temperature was maintained at 0 °C to suppress N-prenylation, the ester was then saponified directly before the peptide coupling to avoid decomposition of the labile acid.
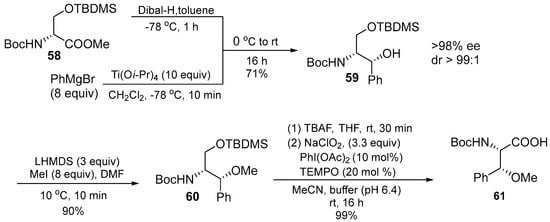
Scheme 8.
Synthesis of (2S,3R)–61.
The next building block was obtained by the ester saponification of compound 62. The prenylated indole 64 was converted to the 3-iodo derivative 65 and the terminal double bond was changed to epoxide 66. After several steps, niodosuccinimide (NIS) in MeOH produced methyl ester 72 (Scheme 9). The O-silylated cyclomarin A was obtained by removing the two protective silyl groups using a two-step protocol in which the primary OH group was deprotected first using NH4F.
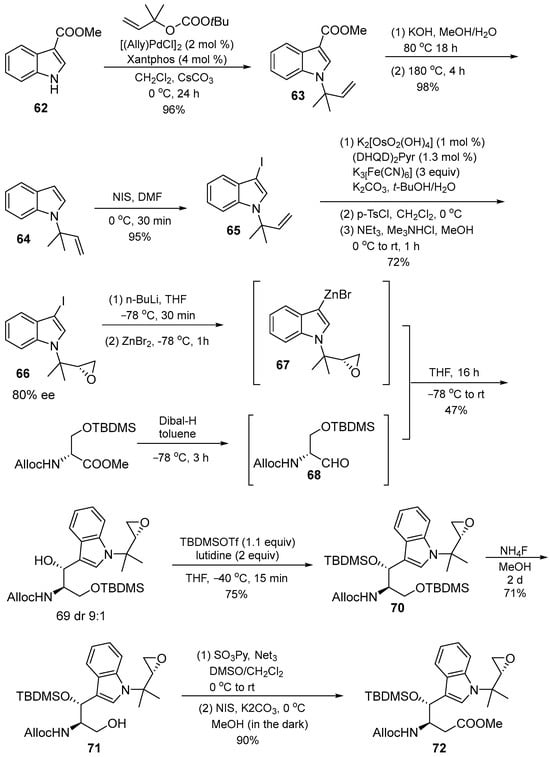
Scheme 9.
Synthesis of (2S,3R)–72.
After obtaining the necessary building blocks, cyclomarin A was then synthesized. Ring closure was planned between aminohexenoic acid and N-methylated leucine. BEP (2-bromo-1-ethylpyridinium tetrafluoroborate) 80 was used for coupling with protected valine. HCl was applied to suppress DKP formation during synthesis. The dipeptide salt was coupled to the next peptide with the activated compound 47. The same protocol was then applied in the next coupling step. The HOBT reagent was used to activate N-methyl-4-hydroxyleucine 56 to avoid epimerization during the coupling reaction. This step obtained enantiomerically pure pentapeptide 77. The methyl ester 72 was saponified and directly coupled using the BEP protocol to obtain compound 78. Under Pd catalysis, the alloc protecting group was removed and combined with amino acid 50, then the deprotected heptapetide was slowly added to an aqueous solution of PyBOP and base in CH2Cl2 to obtain O-sylilated cyclomarin A 46 (Scheme 10) [16].
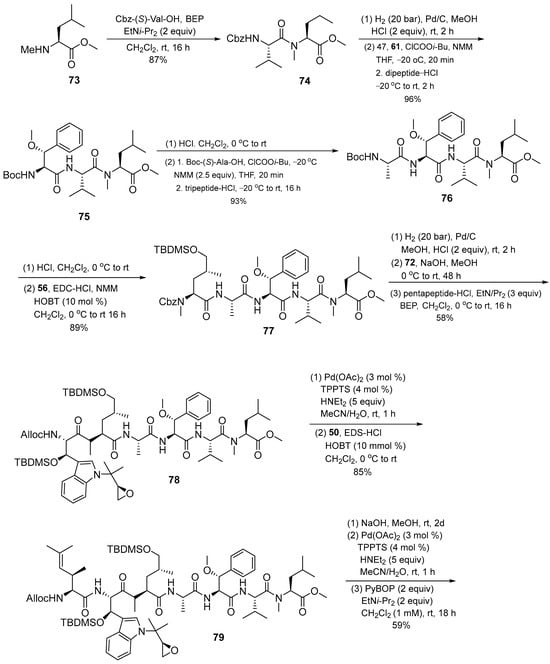
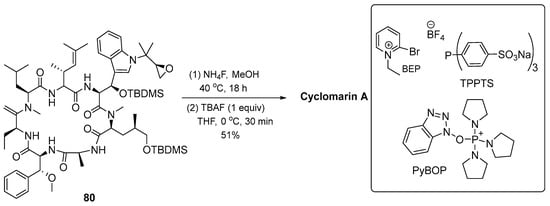
Scheme 10.
Synthesis of cyclomarin A (46).
The synthesis of another cyclomarin was also reported by Kiefer et al. (2019). Seventeen desoxycyclomarin-inspired derivatives (96–100, 106–110, 119–125) were synthesized using a straightforward solution-phase approach [17]. First, an analog library was prepared stepwise by varying the N′-side chain of the tryptophan moiety followed by simplifying the aminohexenoic acid unit. A Negishi reaction served as key step for the generation of N′-isopropyltryptophan, with final saponification yielding the N′-isopropyltryptophan derivative. d-hydroxyleucine units were replaced by d-hydroxynorvaline for additional structural changes in the peptide backbone. The synthesized fragment with protected glutamic acid (81) was activated as mixed anhydride and then reduced with NaBH4. The alcohol produced as TBS ether gives compound 82, which is saponified and N-methylated to obtain N-methyl-d-hydroxynorvaline 84. The first step of synthesis is focused on linear precursor assembly with varying tryptophan motifs and hexenoic amino acids (Scheme 11).
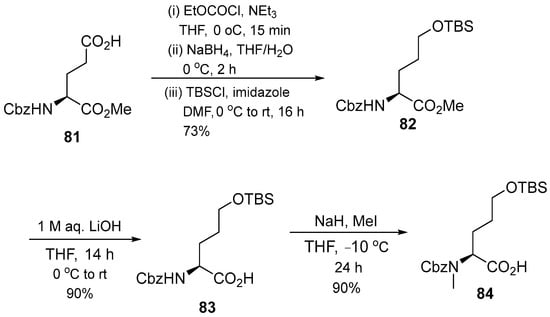
Scheme 11.
Synthesis of N-methylated d-hydroxy norvaline (82).
The intermediate compounds 86–90 were reacted with compounds 58/59 using the HOBt coupling reagent to produce compounds 91–95, which were then cyclized with PyBOP and DIPEA coupling reagents to produce compound 96–100 (Scheme 12). Then, compounds 106–110 were produced using the same synthesis steps. Intermediates 101–105 were cyclized with the same coupling reagent to produce the target compound. The second set of cyclomarin analogs was synthesized based on the key intermediate 106. Several stages of coupling and deprotection were conducted to produce linear peptide compounds 112–118. The linear compounds were then cyclized using PyBOP and DIPEA to produce compounds 119–125 (Scheme 13).
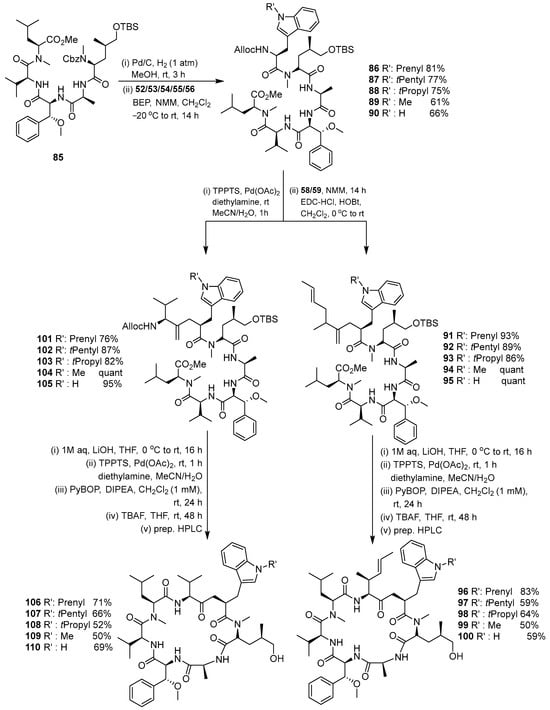
Scheme 12.
Coupling and macrolactamization of the first cyclomarin analogs.
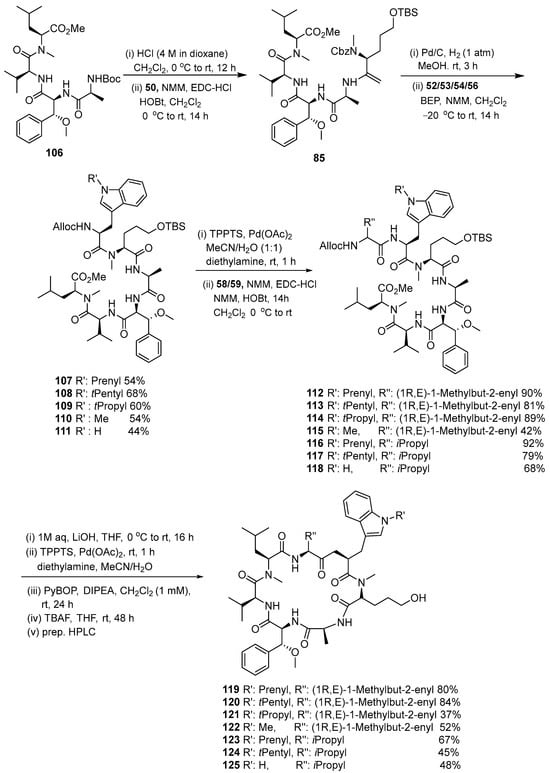
Scheme 13.
Synthesis of the second set of cyclomarin analogs based on key intermediate 106.
3.1.4. Chaiyaphumine
Gholap and Ugale (2017) reported the synthesis of chaiyaphumine-A (130) by convergent-based solution phase synthesis involving protection, deprotection, coupling and cyclization reactions [18]. This novel class of peptide will the subject of much research to develop new antimalarial drugs. Five amino acids, L-threonine, D-phenyl alanine, D-alanine, L-proline and L-tryptophan, were assembled by coupling reactions involving HATU. The macrocylization of the side chain (-OH) of L-threonine and C-terminus of carboxylic acid from L-tryptophan using T3P yields chaiyaphumine-A (Scheme 14).
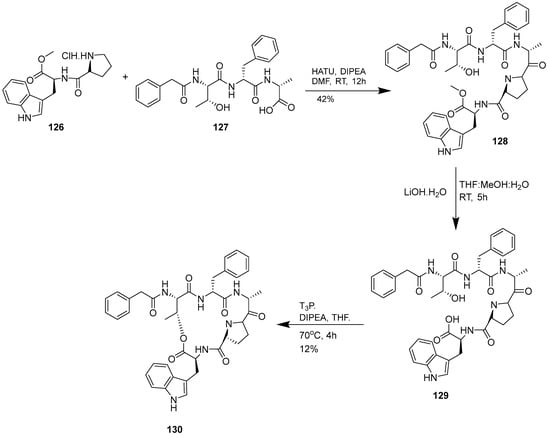
Scheme 14.
Synthesis of chaiyaphumine A (130).
The synthesis of chaiyapumines was also undertaken by Lu and Batey (2022) (Scheme 15). Fmoc-based solid-phase synthesis approaches were used for the synthesis of the requisite precursors. The solution-phase macrolactamization approach using EDC-HCl/HOBt was achieved without any detectable epimerization. This strategy was successfully applied for the total synthesis of the antimalarial compound chaiyaphumine A, as well as the natural products chaiyaphumine B, C, and D (136–139) using a late-stage acylation of the threonine amino group [19].
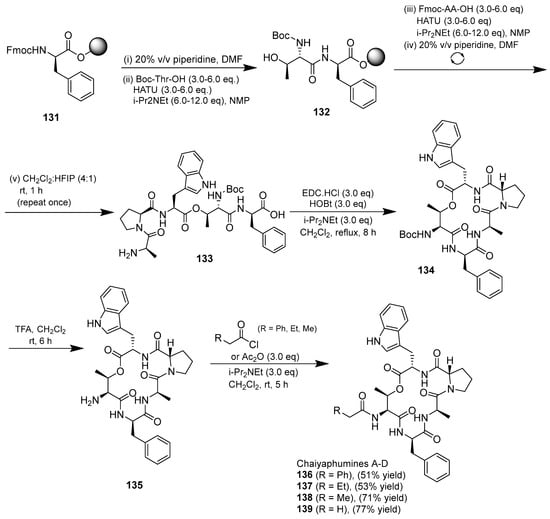
Scheme 15.
Synthesis of chaiyaphumines A–D.
Cyclized products were not produced by the macrolactonization method using MNBA/DMAP/iPr2Net, while the use of the additive Dy(OTf)3 resulted in epimerization macrocycles. The slow rate of macrolactonization is influenced by the weak nucleophilicity of oxygen and the steric hindrance of threonine in the seco acid precursor, which allows competitive epimerization/cyclization to occur. Successful macrocyclization can be achieved via a macrolactamization approach that allows the total synthesis of all chaiyaphumine compunds.
3.1.5. Cyclopeptides Anolgs Prepared by Macrocyclization
Fagundez et al. (2018) investigated the synthesis of a cyclopeptide using solid-phase peptide synthesis and on-resin macrocyclization, with various analytical techniques used to monitor reactions and purify compounds [20]. The investigation compared solution- and solid-phase ring-closing reactions following the synthesis of linear precursor on a 2-chrotrytil chloride resin (Scheme 16). Method A applied a combination of the solid-phase (linear precursor) and the solution-phase methods (macrocylization), whereas method B involved all reactions on-resin. A combination of DIC and Cl-HOBt as a coupling agent was used when the next amino acid added was Fmoc-L-Cys(Trt)-OH because it does not require the addition of a base and can decrease potent racemization. The solution-phase macrocyclization involved HBTU or HATU in the presence of DIPEA in DCM. The final products were obtained in moderate to good yields 26–80%.
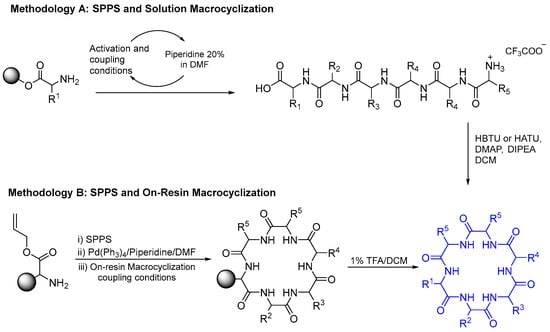
Scheme 16.
Methods A and B to synthesize cylopeptide.
Eleven cyclic peptides (140–150) (Figure 12) were synthesized (23 to 63% yield) using method A (Table 2). Method B constructed Fmoc-L-Glu-OH and all amino acids on 2-CTC resin and the capping step took advantage of DIPEA and MeOH (Table 3). Reagent [Pd(PPh3)4] in a solution of 10% piperidine in THF was used to remove the Fmoc-protected amino acid at the N-terminal group and produce a free carboxyl group at the C-terminal. This method yielded four antimalarial cyclopeptides (151–154) (Figure 13). A comparison of these methods is provided in Table 4.
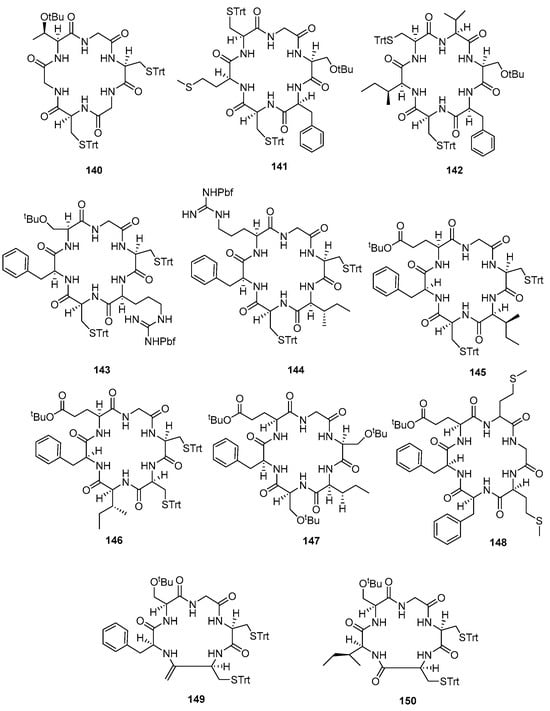
Figure 12.
Compounds 140–150.

Table 2.
Synthesis of macrocyles by method A.

Table 3.
Synthesis of macrocycles by method B and DIC/Cl-HOBt.
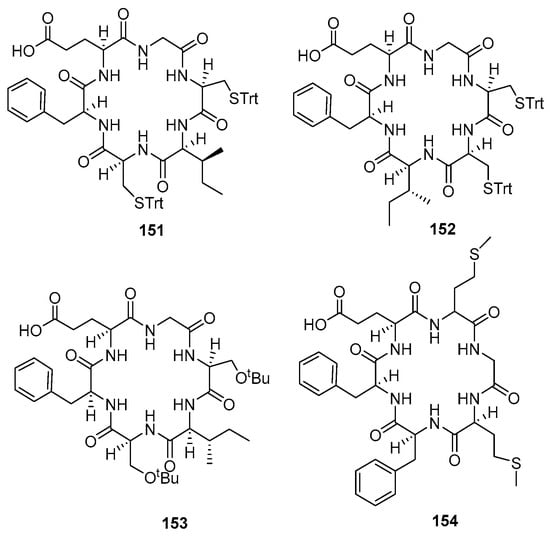
Figure 13.
Compounds 151–154.

Table 4.
Comparison of methods A and B.
A year later after synthesizing compounds 140–154 cyclopeptides, Fagundez et al. (2019) developed cyclopeptides containing N-methyl amino acids with promising antiplasmodial activity against both erythrocytic and liver stages of malaria in vitro. These compounds could potentially be used as new and safe drugs to combat malaria [21]. Compounds 156–161 were synthesized by employing solid-phase linear peptide synthesis and solution macrocyclization (Scheme 17). The 2-chlorotrityl resin (2-CTC) was used to decrease the diketopiperazine formation, and HBTU and DIPEA were employed as coupling reagents in most cases. HCTU and DIPEA reagents were used to combine N-Me-Gly with the next amino acids because they are more effective. DIC and Cl-HOBt were then used as reagents to activate the Fmoc-L-Cys(Trt)-OH and coupling to resin-attached peptides to minimize racemization of Cys residues.

Scheme 17.
Synthesis of cyclopeptides 156–161 by solution macrocyclization.
Compounds 162, 163, and 164 were synthesized employing on-resin macrocyclization (Scheme 18). First, Fmoc-L-Glu-O was anchored to 2-CTC resin with DIPEA, and then the Fmoc SPPS protocol was followed. After the removal of the allyl ester and Fmoc group with [Pd(PPh3)4] in a solution of 10% piperidine in THF, the ring closure was performed using DIC and Cl-HOBt. Figure 14 shows the structure of compounds 156–165.
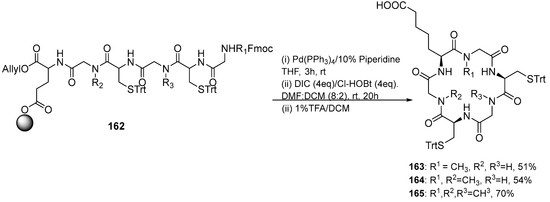
Scheme 18.
Synthesis of cyclopeptides 163–165 by on-resin cyclization.
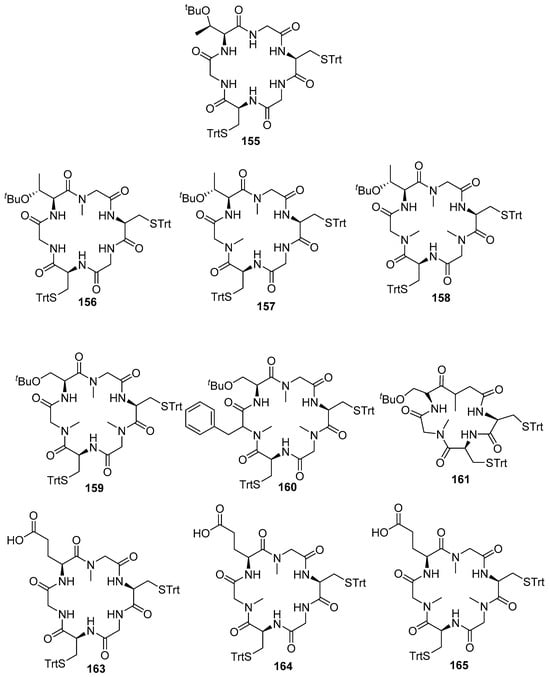
Figure 14.
Structure of cyclopeptides analogs (156–165) and lead compound 155.
3.1.6. Hirsutellide A and Its Analogues
Sahile et al. (2020) reported the synthesis and evaluation of hirsutellide A (Figure 15) and its analogs for their antimycobacterial and antiplasmodial activities [22]. Optimizing the structures of lipophilic antimicrobial cyclic peptides resulted in more membrane-permeable bioactive peptides, such as the lipophilic antimicrobial depsipeptide (hirsutellide A). The first report of the synthesis hirsutellide A was by Xu et al. in 2005; compound 167 had Ile residues instead of allo-Ile resulting in stereoisomers that do not have antimycobacterial potential [22,23].
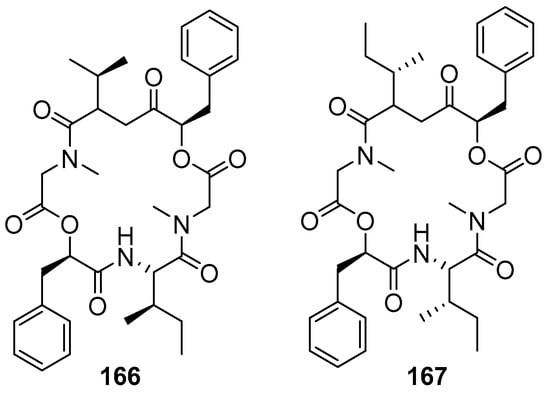
Figure 15.
Structure of hirsutellide A (166) and its synthetic stereoisomer (167).
The compound was synthesized by a solution-phase method and initiated by the preparation of protected tridepsipeptide (Scheme 19). D-Phe 168 was hydrolyzed into α-hydroxyl carboxyl acid 169. After several steps, dipepsipeptide 172 coupling with N-Boc-allo-Ile-OH using HATU/DIPEA to generate tridepsipeptide 173.
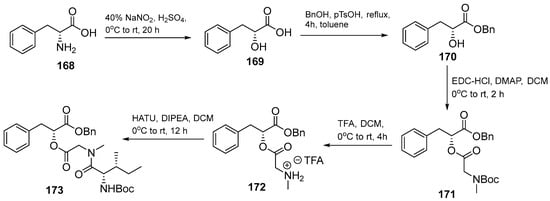
Scheme 19.
Synthesis of the protected tridepsipeptide, a precursor of hirsutellide A (173).
The desired cyclic depsipeptide 177 (Table 5) was obtained via macrocyclization of the precursor 176 using HATU/HOBt/DIPEA in DMF (Scheme 20). Furthermore, depsipeptide (178–181) analogs and peptide analogs (182–188) of hirsutellide A were designed and synthesized for preliminary SAR studies with a combination of solution and solid phase method (Table 5).

Table 5.
List of synthesized depsipeptide and peptide analogs of hirsutellide A.
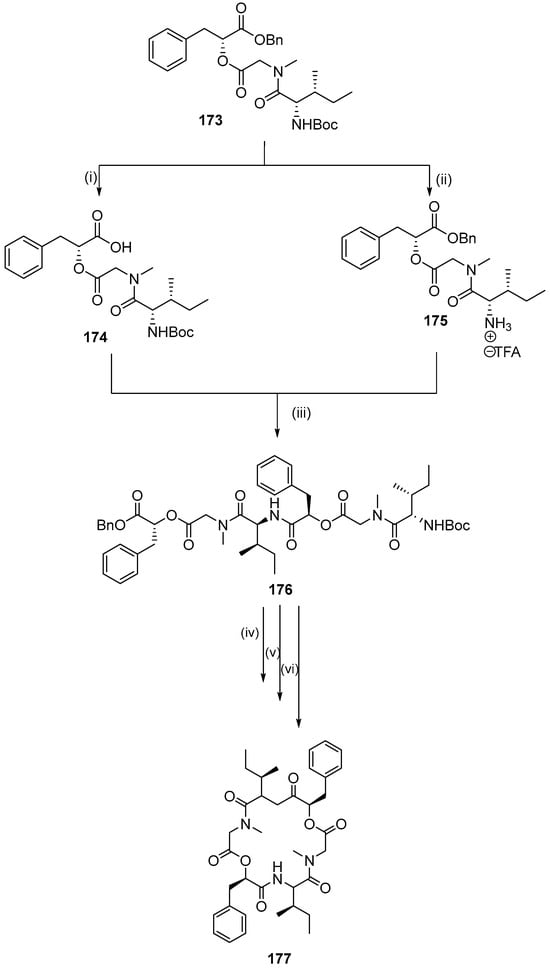
Scheme 20.
Synthesis of hexadepsipeptide precursor of hirsutellide A and macrolactamizations. Reagents and conditions: (i) 10% Pd/C, H2 (5 atm), EtOAc, rt, 12 h; (ii) TFA, DCM, 0 °C to rt, 4 h; (iii) BOP-Cl, DIPEA, 0 °C to rt, 18 h; (iv) 10% Pd/C, H2 (5 atm), EtOAc, rt, 12 h; (v) TFA, DCM, 0 °C to rt, 5 h; (vi) HATU, HOBt, DMF, 0 °C to rt, 3 days.
3.1.7. Macrocyclization Strategies
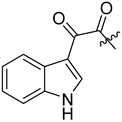
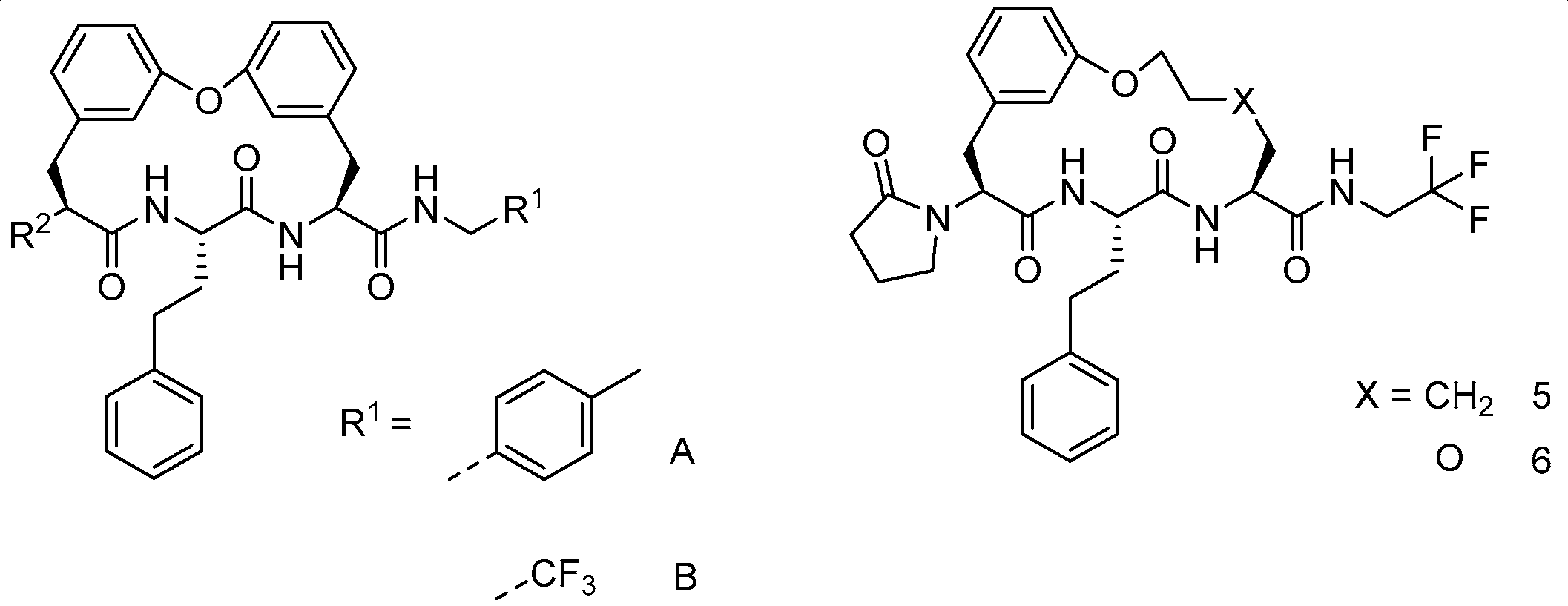
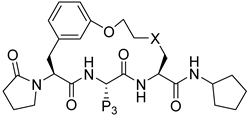
Recently, Zhang et al. (2022) synthesized macrocyclic peptides as species-selective antimalarial proteasome inhibitors [24]. The compounds were prepared through several macrocyclization strategies such as Suzuki coupling, intermolecular amidation, intermolecular alkylation, intramolecular alkylation, and ring-closing metathesis (RCM) reaction.
Synthesis of cyclic peptides 202, 203, and 204 took advantage of Suzuki coupling as a macrocyclization strategy (Scheme 21). Benzylation and borylation reactions of N-Boc-3-bromophenylalanine 189 produced boronate. Deprotection of Boc in compound 191 was followed by combining with Boc-L-homophenylalanine to produce dipeptide intermediate 193, which then underwent Boc deprotection and condensation with carboxylic acid 194 to produce compound 195. Oxidative hydrolysis of boronic ester to boronic acid in 195 by NaIO4 was followed by an intramolecular Chan–Lam coupling reaction to produce macrocycle 205. Removal of the benzyl protective group by catalytic hydrogenation, followed by a coupling reaction of urea, produces 198–199, which undergoes Boc deprotection to yield amines 209 and CP1. Furthermore, compounds 200 and CP1 undergo acetylation of free amines to produce macrocycles 202 and 203. Macrocycle 204 is produced from amine 210 through subsequent alkylation with 4-bromobutanoate, Boc-deprotection, and lactamization using EDCI and HOBt.
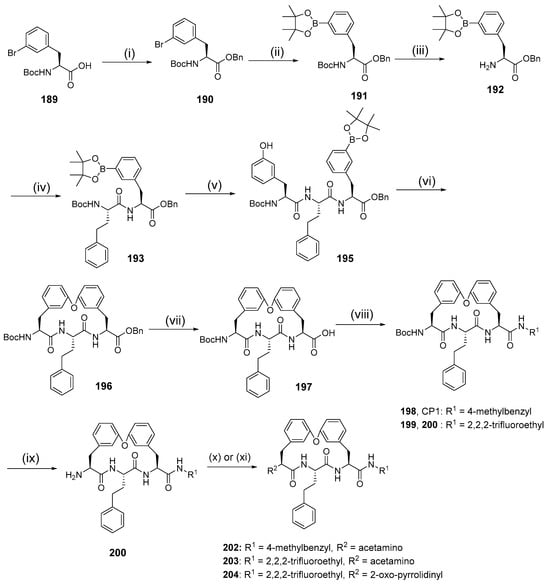
Scheme 21.
Synthesis of cyclic peptides 202, 203, 204 using the Suzuki coupling reaction as a macrocylization strategy. Reagents and conditions: (i) BnBr, K2CO3, acetone, r.t; (ii) Bis(pinacolato)diboron, KOAc, Pd(dppf)Cl2, dioxane, 80 °C; (iii) 4 N HCl/dioxane; (iv) Boc-L-homophenylalanine, HOBt, EDCI, DCM; (v) (1) 4 N HCl/dioxane, (2) carboxylic acid, HOBt, EDCIM DCM; (vi) (1) NaIO4, KOAc, acetone, H2O, (2) Cu(OAc)2, TEA, 4 Å molecular sieves, MeOH, DCM, r.t. 12 h; (vii) Pd/C, H2; (viii) R1NH2, HOBt, EDCI, DMF; (ix) TFA, DCM; (x) acetal chloride, TEA, DMF; (xi) (1) tert-butyl-4-bromobutanoate, K2CO3, DMF, 50 °C, (2) TFA, DCM, (3) EDCI, HOBt.
Intramolecular amidation as a macrocyclization strategy was used to synthesize compound 214 (Scheme 22) from compound 212 by hydrolysis of methyl ester and a coupling reaction using trifluoroethylamine. The synthesis began with deprotecting the tritile group in aziridine 205, then protecting it again with the Boc group present in Boc-aziridine 206. Compound 207 was then produced by an SN2-type ring opening reaction mediated by BF3·Et2O on aziridine 206 with 2-benzyloxyethanol. Boc-deprotection in compound 207 was followed by a coupling reaction with Boc-L-homophenylalanine to produce dipeptide 208, then benzyl deprotection and Mitsunobu reaction of fragment 209 yielded the macrocyclization precursor 210. Debenzylation and Boc deprotection of compound 210 was followed by intramolecular mediation to produce macrocycle 212.
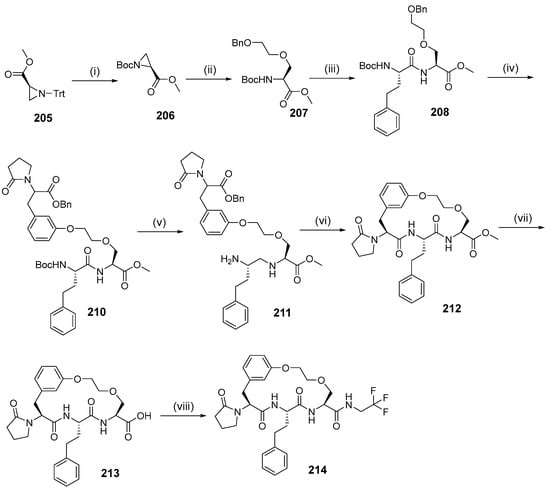
Scheme 22.
Synthesis of compound 214 using intramolecular amidation as the macrocyclization strategy. Reagen and conditions: (i) (1) TFA, DCM, MeOH, (2) Boc2O, TEA, MeCN; (ii) BF3.Et2O, DCM, 2-(benzyloxy)ethanol; (iii) (1) HCl, MeOH, (2) HBTU, DIOEA, DMF, Boc-L-homophenylalanine; (iv) (1) H2, Pd/C, MeOH, (2) DEAD, PPh3, THF, compound 16; (v) (1) Pd/C, H2, MeOH, (2) HCl/dioxane; (vi) EDCI, HOBt, DMF; (vii) NaOH, THF/H2O; (viii) 2,2,2-Trifluoroethylamine, EDCI, HOBt, pyridine.
The synthesis of compounds 229–237 were involved in intramolecular alkylation as a macrocyclization strategy (Scheme 23). Dipeptide 225 was prepared from amino acid 224 by methyl esterification and HATU-mediated coupling reaction with Boc-L-homophenylalanine. Boc deprotection of dipeptide 225 and a coupling reaction with fragment 226 gave tripeptide 227, which was then removed by two benzyl groups. The primary alcohol was chemoselectively protected with Tos groups, and Cs2CO3-mediated intramolecular. It underwent alkylation to give macrocycle 228. After treatment of the methyl ester with sodium hydroxide, coupling of the free acid with various primary amines yielded macrocycles 229, 230, 231, 232, 236, and 237. Reduction of the methyl ester 228 with lithium borohydride formed alcohol 238, and then Dess–Martin oxidation, followed by reductive amination with trifluoroethylamine, yielded macrocycle 233. Condensation of the methyl ester 228 with hydrazine, followed by coupling with methyl trifluoroacetimidate gave the hydrazide 239, which was thermally cyclized to give the 1,2,4-triazole 235. Subsequent hydrolysis and dehydration of hydrazide 239 in the presence of the Burgess reagent gave 1,3,4-oxadiazole 234.
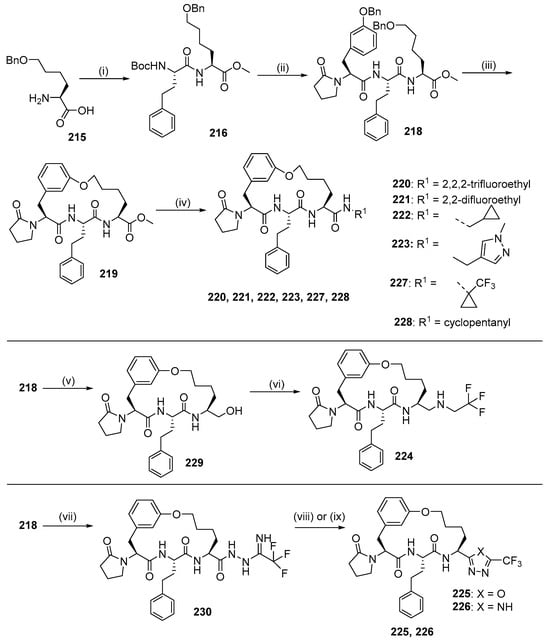
Scheme 23.
Synthesis of compounds 220–226 using intramolecular alkylation as a macrocyclization strategy. Reagents and conditions: (i) (1) HCl, MeOH, (2) Boc-L-homophenylalanine, HATU, DIEA, DMF; (ii) (1) HCl (4 M), dioxane, (2) compound 23, HATU, DIEA, DMF; (iii) (1) H2, Pd/C, Pd(OH)2, MeOH, (2) TosCl, pyridine, (3) Cs2CO3, DMF; (iv) (1) NaOH, THF, H2O, (2) HATU, DIEA, R1NH2; (v) LiBH4, THF; (vi) (1) Dess-Martin, THF, (2) 4 Å MS, NaBH(OAc)3, MeOH, 2,2,2-trifluoroethylamine; (vii) (1) N2H4.H2O, MeOH, (2) methyl trifluoroacetamide, DCM; (viii) 1,2-dichlorobenzene; (ix) (1) TFA, DCM, (2) Burgess reagent, THF.
Macrocyclization with the intramolecular alkylation strategy was used to synthesize compounds 247, 248, 249, and 254 (Scheme 24). Fragments 231–233 underwent an amide coupling reaction with fragments 234–235 and subsequent deprotection, yielding dipeptides 236–239, which were coupled to fragments 240–241 and 217 to yield tripeptides 242–246. The tripeptides 242–246 were sulfated with a hydroxyl group, debenzylation of aryl benzyl ether, and intramolecularly substituted between the phenol group and the sulfonate ester, giving macrocycles 250, 251, 247, 248 and 249. Removal of the tert-butyl from compounds 250–251 using TFA was followed by an amide coupling reaction with a primary amine, which produced macrocycles 254 and TDI8.
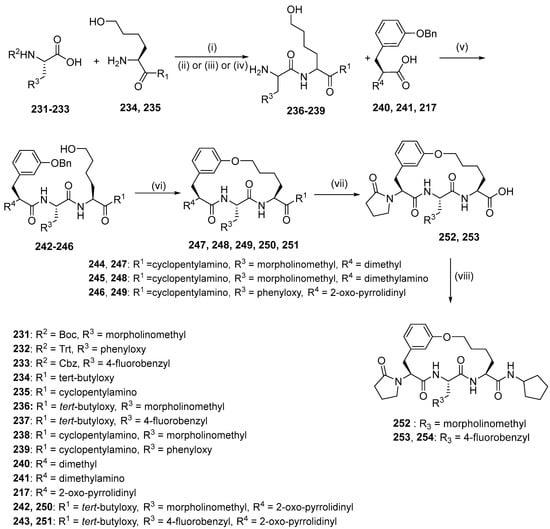
Scheme 24.
Synthesis of compounds 256, 257, 258, and 263 using intramolecular alkylation as macrocyclization strategy. Reagents and conditions: (i) T3P, DIEA, DMF 0–20 °C, 1 h; (ii) HCl (4 M, 4 eq.), dioxane, MeOH; (iii) HCl, dioxane, DCM, 20 °C, 3 h; (iv) H2, Pd/C, TFA, THF; (v) T3P, DIEA, DMF; (vi) (1) TosCl, pyridine, (2) H2, Pd(OH)2/C, MeOH, (3) Cs2CO3, DMF; (vii) TFA, DCM; (viii) cyclopentylamine, T3P, DIEA, DMF 0 °C, 1 h.
Synthesis of compounds 270, 271, 272, and 273 was achieved by the RCM reaction (Scheme 25). Fragments 231 and 255 underwent subsequent amide coupling reaction with fragment 256; Boc-deprotection and an amide coupling reaction with acids 261–264 afforded diolefins 265–268, which were subjected to RCM in the presence of Grubbs Catalyst second Generation and reducing of the C=C double bond, yielding macrocycles 270, 271, 272, and 269. Removal of the Boc protection of 269 provided macrocycle 273.
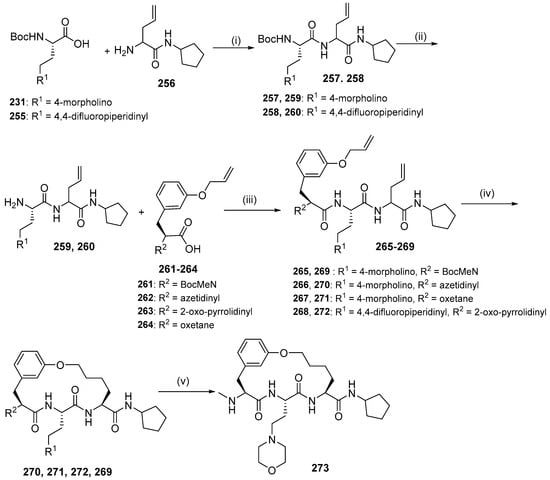
Scheme 25.
Synthesis of compounds 270, 271, 272, and 273 using RCM reaction as a macrocyclization strategy. Reagent and conditions: (i) T3P, DMF, DIEA; (ii) HCl/dioxane, MeOH; (iii) T3P, DIEA, DMF; (iv) (1) Grubbs 2nd, DCE, (2) H2, Pd/C, MeOH; (v) HCl, dioxane.
3.2. Linear Peptides
3.2.1. Falcitidin
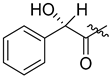
Somanadhan et al. (2013) succeeded in synthesizing linear antimalarial peptides. Falcitidin 284 is the first member of a new class of falcipain-2 inhibitors, which is a cysteine protease used by P. falciparum to degrade hemoglobin during the trophozoite stage of infection [25]. Falcitidin contains isovaleric acid-D-His-L-Ile-L-Val-L-Pro-NH2 and was synthesized by the solution-phase peptide synthesis method (Scheme 26). The synthesis begins with deprotecting the Boc group from N-Boc-proline carboxamide using TFA. The amino-proline TFA salt was coupled with freshly prepared L-Ile-L-Val using HATU/HOAt to give L-Ile-L-Val-L-Pro-NH2. The dipeptide was obtained by coupling L-Ile-NHBoc with the in situ prepared bis-trimethylsilane (TMS) ether of L-Val under mixed anhydride conditions. The Boc group of the tripeptide was then cleaved using TFA and coupled to Na-Fmoc-N(im)-trityl-D-histidine using HATU/HOAt to give the Fmoc–tetrapeptide. The Fmoc group of was removed using piperidine and HATU/HOAt. The final synthetic target falcitidin was obtained after trityl deprotection of Trt-falcitidin using TFA in the presence of triisopropylsilane.
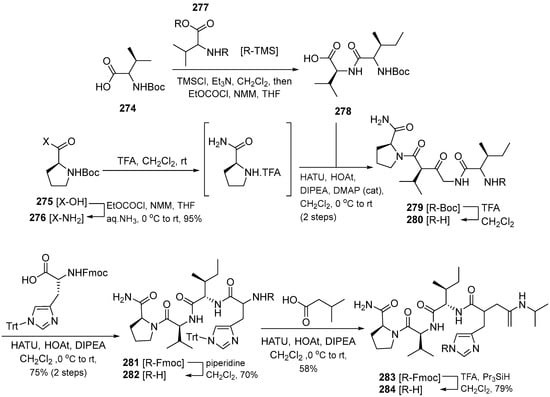
Scheme 26.
Synthesis of compound 284.
Kotturi et al. (2014) synthesized falcitidin and its analogs (296–303) using the same solution phase peptide synthesis (Scheme 27) [26]. First, Nim-trityl-D-histidine 288 was converted into bis-TMS ether with TMSCl/Et3N7 and coupled immediately with the mixed anhydride of isovaleric acid 287 using ethyl chloroformate/NMM to form the trityl protected N-acyl-D-histidine. In parallel, the N-Boc proline amide was deprotected with TFA, and the crude amine salt was coupled immediately with the known dipeptide N-Boc-L-Ile-L-Val by using HATU/HOAt to yield the key tripeptide intermediate N-Boc-L-Ile-L-Val-L-Pro-NH2. Lastly, the Boc group of tripeptide was cleaved using TFA and coupled to N-acyl-D-histidine using HATU/HOAt to give the Nim-trityl protected tetrapeptide 292 (Scheme 28). Falcitidin acylatide and its analogs produce eight analogs compounds (296–303) by diversifying the synthesis of the N-acyl tetrapeptide analog of falcitidin through the common tripeptide N-Boc-L-Ile-L-Val-L-Pro-NH2 (Scheme 29) (Table 6).
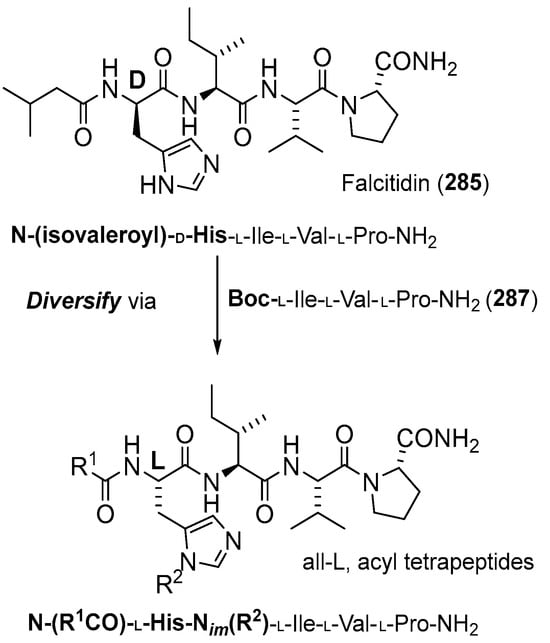
Scheme 27.
Falcitadin-derived 285 acylpeptide lead compound template.
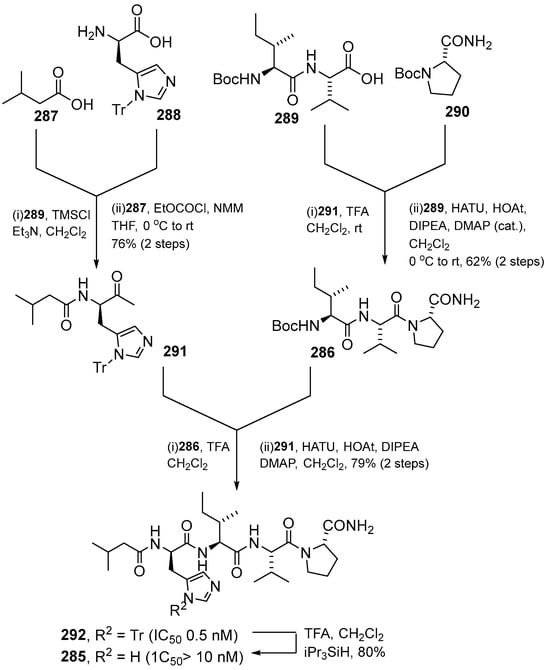
Scheme 28.
Convergent total synthesis of falcitidin 285 via the key tripeptide intermediate 286 and the whole-cell active N-trityl analog 292.
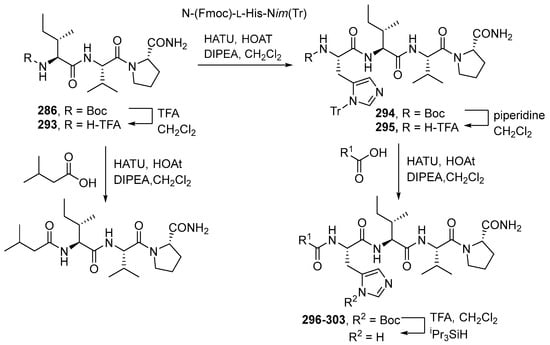
Scheme 29.
Diverted synthesis of all-L, N-acyl tetrapeptide analogs of falcitidin 285 via the common tripeptide N-Boc-L-Ile-L-Val-L-Pro-NH2.

Table 6.
Yield of falcitidin acylated analogs 296–303.
3.2.2. Gallinamide
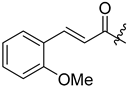
Gallinamide A is a linear peptide that has antimalarial activity. Conroy et al. (2014) designed and synthesized 18 gallinamide A analogs by solid-phase synthesis and substitution of amino acid residues [27]. Four gallinamide A analogs (309–312) were originally designed with varying levels of saturation, including compound 309, which is structurally identical to gallinamide A. Another analog was compound 310 with a reduced methoxy-enol moiety in the pyrolinone ring 309. Part of the olefinic component of the 4(S)-amino-2-(E)-pentenoic acid unit is reduced in compound 311 and 312 has reduced olefin groups. Analog synthesis was initiated by synthesizing the N-terminal fragment 304 by solid-phase peptide synthesis using the Fmoc strategy. The 2-CTC resin is filled with Fmoc-Leu-OH followed by the subsequent incorporation of the amino acid. Reductive amination of the resin, followed by cleavage of the resin using hexafluoroisopropanol (HFIP), produced an excellent yield of tripeptide 304 which was coupled with imide fragments 306 and 307 prepared using a similar protocol. The coupling reaction involved HATU at low temperature to avoid epimerization to produce good yields of analogs 309 and 310. At this stage, 309 and 310 underwent hydrogenation to give 311 and 312 (Scheme 30).
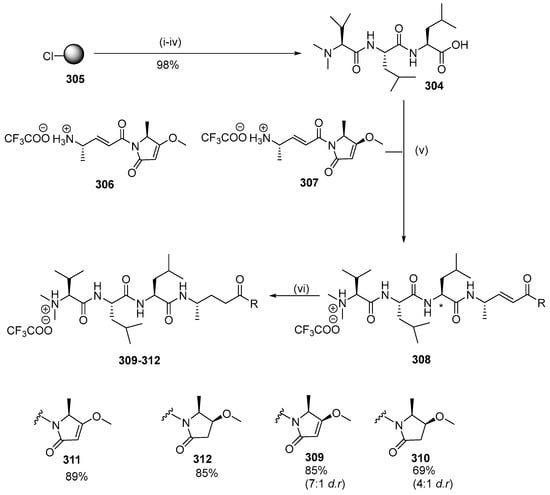
Scheme 30.
Synthesis of gallinamide A analogs 309–312. Reagents and conditions: (i) resin loading: Fmoc-Leu-OH, iPr2EtN, DMF/CH2Cl2; (ii) Fmoc-SPPS (deprotection, 10 vol % piperidine/DMF; coupling, 4 equiv Fmoc-AA-OH, 4 equiv PyBOP, 8 equiv NMM, DMF; capping, 10 vol % Ac2O/pyridine); (iii) HCHO, NaBH(OAc)3, AcOH, DMF; (iv) 30 vol % HFIP/CH2Cl2; (v) HATU, NMM, 0 °C for 20 min, rt for 2 h; (vi) H2, Pd/C, MeOH. * indicates the site of diastereoisomerism in 309 and 310.
Analogs 313–318 were synthesized from 2-CTC resin filled with Fmoc-Ala (Scheme 31; Table 7). The coupling of Fmoc-protected α,β-unsaturated amino acids was followed by elongation through the Fmoc solid-phase peptide synthesis strategy. With reductive methylation of the N-terminus using formaldehyde and sodium cyanoborohydride, the peptide was cleaved from the resin using HFIP to produce the C-terminal peptide acid. C-terminal functionality was achieved through benzylamine coupling using PyBOP at low temperatures, and compounds 313, 315, 317, and 318 were purified by RP-HPLC and demonstrated no significant epimerization. Compound 314 was produced by attaching 4-(R)-hydroxy L-proline methyl ester to the C-terminus of the two peptides with the addition of NMM as a hindered base. Aminothiazole was coupled to the C-terminus of one peptide acid using PyBOP at low temperature to yield compound 316 as a mixed diastereoisomer.
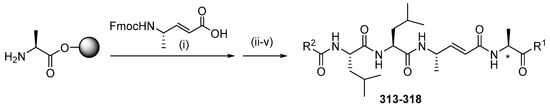
Scheme 31.
Synthesis of gallinamide A analogs 322–327. Reagents and conditions: (i) 4 equiv amino acid, 4 equiv PyBOP, 6 equiv NMM; (ii) Fmoc SPPS (deprotection, 10 vol % piperidine/DMF; coupling, 4 equiv Fmoc-AA-OH, 4 equiv PyBOP, 8 equiv NMM, DMF; capping, 10 vol % Ac2O/pyridine); (iii) HCHO, NaBH(OAc)3, AcOH, DMF; (iv) 30 vol % HFIP/DCM; (v) # R1-NH2, PyBOP, DMF, 0 °C. * indicates the site of diastereoisomerism in 313–318.

Table 7.
Yields of gallinamide A analogs 313–318.
A further six analogs (346–351) were designed, possessing the identical peptide backbones to 309 and 310 but with a variation in the side chain on the pyrolinone unit and the enol substitution of pyrolinone (Scheme 32). The synthesis of compounds 346–351 was initiated by preparation of the requisite pyrolinones 333–338 from Fmoc-protected amino acids 319–322. Then, Meldrum’s acid in ethyl acetate was coupled with amino acids 319–322 using EDC and DMAP followed by reflux of the Meldrum’s adduct in ethyl acetate. This step affected the condensate cyclization to give Fmoc-protected pyrolinones 323–326, which were then reacted with methanol under Mitsunobu conditions using DIAD and triphenylphosphine, respectively, to produce O-methylated pyrolinones 327–330. Compounds 323–324 were reacted with benzyl alcohol under the same conditions to produce 331–332. Compounds 333–338 were produced by treating piperidine in acetonitrile. After each pyrrolinone building block was obtained, amino acid 339 was activated as the corresponding pentafluorophenyl ester. Separately, pyrolinones 333–338 were deprotonated with n-butyllithium at low temperatures before the addition of pentafluorophenyl ester to yield 340–345 (36–59%). Due to the unfavorable results, acidolysis of the Boc groups from 340–345 was followed by coupling to the N-terminal tripeptide 304 using the coupling reagent HATU and NMM as the base to produce the desired gallinamide A analogs 346–351 with excellent yields.
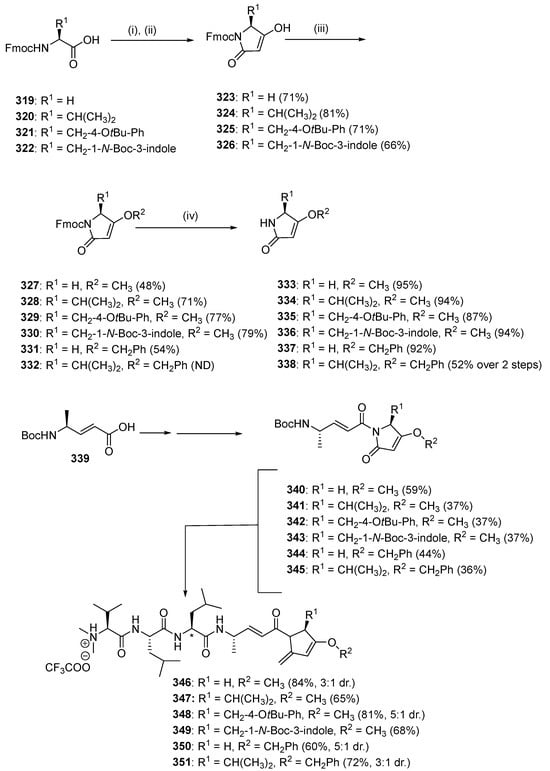
Scheme 32.
Synthesis of second-generation gallinamide A analogs 346–351. Reagents and conditions: (i) (1) Meldrum’s acid, EDC, DMAP, CH2Cl2, 0 °C to rt; (ii) EtOAc, 77 °C; (iii) R2-OH, DIAD, PPh3, CH2Cl2, 0 °C to rt; (iv) 1:4 v/v piperidine/MeCN, rt, 15 min; (v) CF3COOC6F5, pyridine, DMF, 0 °C; (vi) 333–337, n-BuLi, THF, −78 °C; (vii) 1:1 v/v TFA/CH2Cl2; (viii) tripeptide 333, HATU, NMM, DMF, 0 °C. ND = not determined due to co-contamination with DIAD by-product. * indicates the site of diastereoisomerism in 346, 348, 350, and 351.
The natural product gallinamide A (Figure 16) was also synthesized by Stoye et al. (2019) and possesses potent inhibitory activity against P. falciparum cysteine proteases, namely falcipain, and therefore shows promise as a potential malaria treatment to breakdown hemoglobin in the parasitic food vacuole [28]. Gallinamide A was synthesized using NMM in a solution of the imide fragments 353–354 (1.0 equiv, as the trifluoroacetate salt), tripeptide 334–336 (1.5 equiv, as the trifluoroacetate salt), HATU and HOAt in DMF/CH2Cl2 (1:1 v/v). After consumption of the starting material (as evidenced by LC-MS), the solvent was subsequently removed by an N2 stream and the residue was purified by preparative RP-HPLC (Scheme 33). This synthesis produced different yields of gallinamide A analogs 371–387 (Table 8 and Table 9).
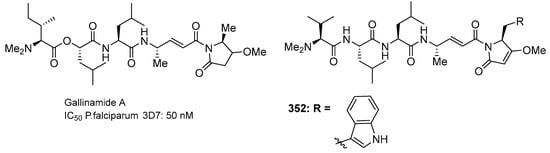
Figure 16.
Structure of gallinamide A and synthetic analog 352.
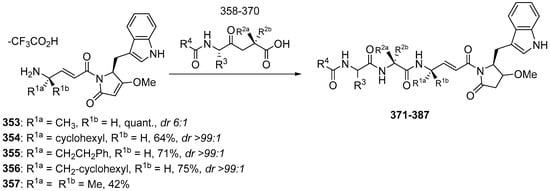
Scheme 33.
Synthesis of compounds 371–387.

Table 8.
List of tripeptides synthesized to generate compounds 371–387.

Table 9.
Overview of the yields of synthetic gallinamide A analogs [28].
3.2.3. PfSERA5 Analogs
PfSERA5 is an abundant asexual antigen that can inhibit parasitic growth in vitro and is a candidate malaria falciparum vaccine. Kanodia et al. (2014) reported the design and synthesis of peptides with similar sequences to the SERA5 protein, an inhibitor of malaria parasite development [29]. The solid-phase synthesis of all peptides was initiated on Rink amide resin. The appropriate Fmoc-amino acid was dissolved in a solution of TBTU/HOBT and DIPEA before being added to the resin. Fmoc was deprotected by piperidine in DMF and then acetylated by a mixture of acetic anhydride and DIPEA. The nine SE5 P1-P9 (388–396) were cleaved from the resin using TFA/H2O/TIS/Thioanisole/Phenol and their sequences are shown in Table 10.

Table 10.
Amino acid sequences of the peptides from PfSERA5.
3.2.4. Angiotensin
Silva et al. (2015) investigated angiotensin II, a peptide that has antiplasmodial activity, as an antimalarial drugs [30,31]. They synthesized 10 peptides (397–406) (Table 11) via manual solid-phase synthesis [30]. The Fmoc strategies were applied using Wang resins and deprotection was performed by treatment with 4-MePip in DMF. Couplings were conducted using DIC/HOBt in DCM/DMF (1:1, v/v) and were monitored using the Kaiser ninhydrin test. Dry-protected peptidyl resin was exposed to TFA/H2O/anisole (95:2.5:2.5, v/v/v) for 2 h at room temperature to produce the crude linear peptides. All crude peptides were precipitated with anhydrous diethyl ether, separated from the ether–soluble reaction components by filtration, extracted from the resin with 60% ACN in water, and lyophilized. They were purified by preparative RP-HPLC in 0.1% TFA/60% ACN in water on a Waters Associates system (Delta Prep 600). The peptides were loaded onto a Phenomenex C18 (21.2 × 250 mm, 15 µm particle size, 300 Å pore size) column at a flow rate of 10.0 mL/min and eluted using a linear gradient (slope 0.33% B/min) of TFA/ACN with detection at 220 nm. Selected fractions containing the purified peptides were pooled and lyophilized.

Table 11.
Purity of peptides determined by LC/MS [30].
In 2015, Silva et al. also synthesized nine octapeptides (407–415) of the renin-angiotensinsystem (RAS), which have been reported to have anti-plasmodium activity towards P. gallinaceum (88% sporozoite inactivation) [31]. The angiotensin II analogs (Figure 17) were synthesized by Fmoc/tert-butyloxycarbonyl (t-Boc) strategy on a solid phase, purified by liquid chromatography, and characterized by mass spectrometry. The amino acid Nα-terminal protecting group was removed with TFA in DCM in the presence of 2% anisole for 20 min. Coupling and deprotection reactions were carried out using the same reagent as before. Repeated couplings were performed for one hour using TBTU with DIPEA in DCM/NMP. Dry protected peptidylresin was exposed to 70% TFA/20% TFMSA in 10% anisole for 12 h. All crude peptides were precipitated with anhydrous diethyl ether, separated from the ether–soluble reaction components by filtration, and the fractions were lyophilized using 60% ACN (acetonitrile) in water.
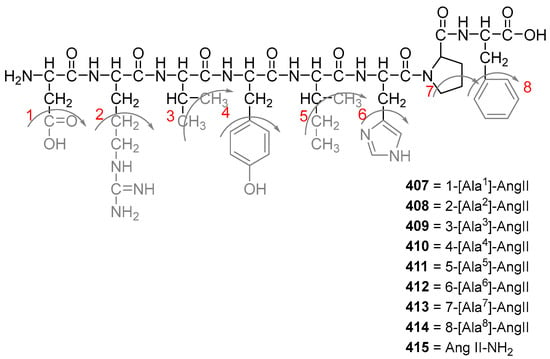
Figure 17.
Structure of compounds 407–415 [31] The additional amino acids on the native Ang II sequence are highlighted in red for each analogs.
Ten angiotensin analogs (416–425) were also synthesized by Torres et al. (2015) (Table 12) [32]. These lactams and sulfide bridge-containing peptides were synthesized manually by the solid-phase method using a t-Bu strategy with chloromethylated resin and a Fmoc strategy with Wang resin. Deprotection was conducted with TFA and DCM and the addition of 2% anisole. The resin was washed with anisol in isopropyl alcohol and TEA in DCM and MeOH, and the reaction was monitored with the Kaiser test. Recoupling and deprotecting the protective groups on each amino acid used the same reagents as before. Furthermore, the cyclization reaction was conducted with excess Castro reagent and DIPEA dissolved in DMSO/NMP, after which the resin was washed with TEA in DCM to obtain a dry peptidyl-resin.

Table 12.
Sequences of compounds 416–425 [32].
Deprotection of the sulfide bridge analogs was conducted by adding 4-MePip in DCM. The amino acid coupling reaction involved treating the protected amino acid acyl-resin with a molar excess of the Boc/Fmoc protected amino acid using the DIC/HOBt reagent. The disulfide bridges were formed by first dissolving the peptide in an acetic acid solution containing iodine. The peptides were then lyophilized after being extracted with water and diethyl ether and purified by RP-HPLC.
Silva et al. (2017) synthesized eight angiotensin II hormones (426–433) (Table 13) [33]. They optimized the synthesis of these peptides for their antiplasmodial activity and to reduce their vasoconstriction and rapid degradation characteristic. The peptides were synthesized by manual solid-phase synthesis and characterized by circular dichroism spectroscopy. The synthesis was similar to previously reported and involved manual phase peptide synthesis using the Fmoc strategy and Wang resin. The amino acid residues were coupled using the DIC/HOBt reagent in DCM. Each step was followed by alternating washing with DMF, methanol, and DCM to change the degree of resin swelling and promote the removal of excess reagents. Dry peptidyl resin was added to TFA/water/anisole to obtain unprotected peptides. The S-S bonds were formed by first dissolving the crude peptide in an iodine-containing acetic acid solution. The peptides were extracted with water and diethyl ether, evaporated, and then lyophilized.

Table 13.
Design of angiotensin II-derived synthetic peptides [33].
3.2.5. Decoralin
Torres et al. (2018) reported the re-engineering of a wasp venom peptide, decoralin, into a synthetic anti-malaria agent through modifications that removed its hemolytic activity [34]. Dec-NH2 and eight analogs (434–441) were synthesized and designed to preserve specific physicochemical structures. Certain amino acid substitutions significantly improved the antiplasmodial activity, giving new sequence principles for creating potent anti-malaria drugs such as replacing the original sequence by Arg, Phe and Trp. The synthesis involved a solid-phase method and purification by chromatography and characterization using MS. The antiplasmodial activities were assessed by fluorescence microscopy. All residue substitutions resulted in increased anti-Plasmodium activity (Table 14), with [Arg]1-Dec-NH2, [Pro]4-Dec-NH2, and [Phe]2-Dec- NH2 being the most active peptides tested.

Table 14.
Characterization and theoretical physicochemical properties of decoralin and synthetic analogs.
3.2.6. Carmabin and Dragomabin
Ye et al. (2018) synthesized carmabin A and dragomabin, which demonstrated antimalarial activity [35]. Carmabin A and dragomabin synthesis was initiated by retrosyntethic analysis (Scheme 34). The C-terminal amide of carmabin A (442) and dragomabin (443) was prepared via amidation of the C-terminal methyl esters of compounds 444 and 445. Compounds 448 were further disconnected into two parts: the Mdya/Moya fragment and protected tetrapeptide 454 (Scheme 35 and Scheme 36).
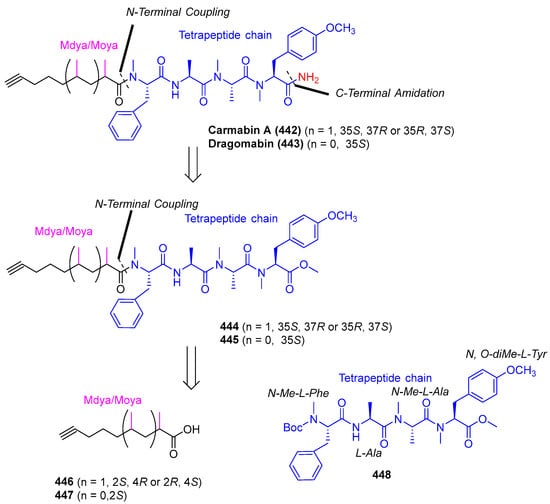
Scheme 34.
Retrosynthetic analysis of carmabin A (442) and dragomabin (443).
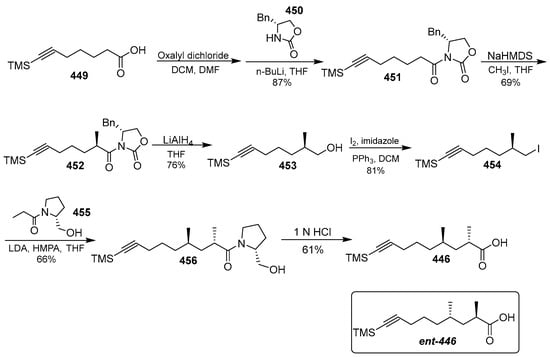
Scheme 35.
Synthesis of Mdya 446.
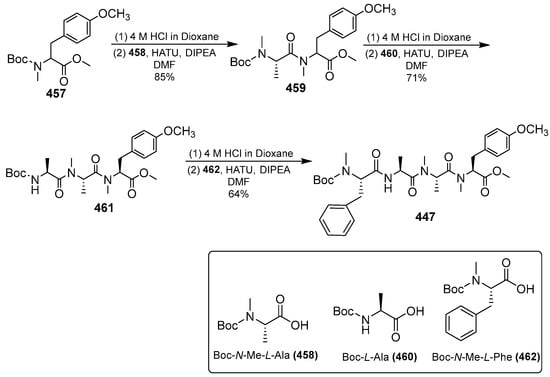
Scheme 36.
Synthesis of tetrapeptide 447.
Tetrapeptide 448 was prepared by repeated condensation of amino acids (Scheme 35). The methyl group of Mdya/Moya 446/447 could be stereoselective. Carboxylic acid 454 was converted to the corresponding alcohol 453 in four steps involving acyl chlorination, amidation with benzyl-2-oxazolidinone, diastereoselective-methylation, and reduction. The addition of carboxylic 454 with N HCl led to the hydrolysis of the TMS group and the amide bond to yield compound 446.
Scheme 36 shows the construction of tetrapeptide 447, the treatment of compound 457, and TFA resulting in Boc deprotection. The condensation of compound 458 with the coupling reagent HATU/DIPEA produced compound 459 and repeated condensation under the same conditions yielded tetrapeptide 447.
Carmabin A was synthesized from the building blocks 446, ent-446, and 447 (Scheme 37 and Scheme 38). The coupling reaction between Boc-deprotected 447 and 446 and ent-446 was followed by the addition of ammonia to produce compounds 442 and 442a. The configuration of carmabin A was further investigated using NMR, showing that only the 442 NMR data matched that of natural carmabin A.
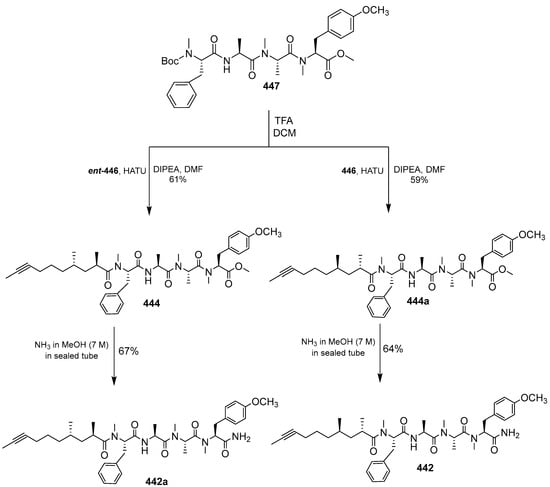
Scheme 37.
Total synthesis of carmabin A (442).
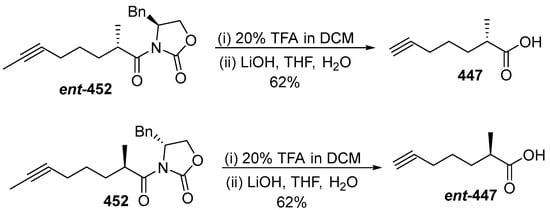
Scheme 38.
Synthesis of Moya 447 and ent-447.
Dragomabin was synthesized using the building blocks Moya 447 and ent-447. First, Moya 447 is generated from ent-452 by deprotecting with TMS and TFA, then the chiral auxiliary is removed (Schema 38). Dragomabin is obtained by deprotecting 448 with TMF under DCM to provide 445, which is amidated with NH3 to form compound 443. The correct structure for dragomabin was revised as shown in 443a (Scheme 39). As dragomabin and dragonamide differ in the stereochemistry on the Moya fragment, the stereochemistry of the alkyne fragment in these lipopeptides is varied, and correlation with other natural products is not reliable. The absolute stereochemistry at C35 and C37 of carmabin A was assigned as 35R, 37S and the absolute stereochemistry at C35 of dragomabin has been revised as 35R.
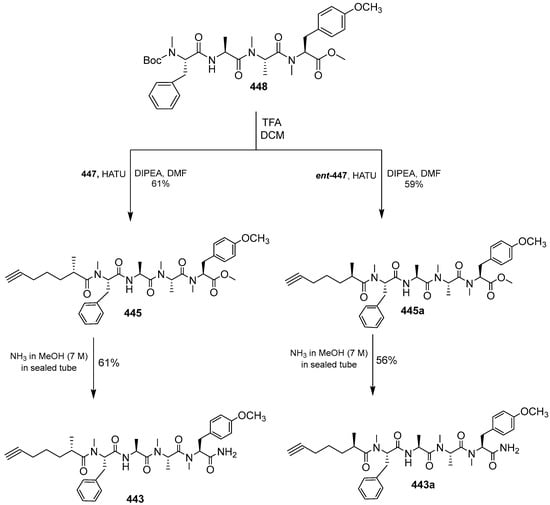
Scheme 39.
Total synthesis of dragomabin (443).
3.3. Biology Activity of Isolated and Synthesized Compounds
Ribifolin (1) is moderately effective against the Plasmodium falciparum 3D7 strain with an IC50 of 42 µM (Table 15), while its linear analogue 1a had an IC50 of 519 µM, demonstrating the importance of cyclization to enhance the biological activity [3]. None of the tested compounds showed cytotoxic potential against human cells (HEK293aT), although normal growth was observed in the concentration range of 0.001–100 μM.

Table 15.
Orbitides isolated from Jatropha species and their potential antiplasmodial activities.
The isolated cyclopeptide alkaloids (2–5) from the root bark of Hymenocardia acida demonstrated antiplasmodic activity against P. falciparum K1, with IC50 values ranging from 12.2 to 27.9 μM. Only hymenocardine (2) exhibited cytotoxic properties against MRC-5 cells with an IC50 of 51.1 ± 17 μM (Table 16) [4].

Table 16.
Antiplasmodial activity against against P. falciparum strain KI and cytotoxicity against MRC5-Cells (IC50 μM) of compounds 2 (Hymenocardine), 3 (Hymenocardinol), 4 (Hymenocardine N-Oxide), and 5 (Hymenocardine-H) from the root bark of Hymenocardia acida.
The antiplasmodic activity of nummularine-R (6), O-desmethylnummularine-R (7), hemsine-A (9), ramosine-A (11), and oxyphylline-F (14) was evaluated against the P. falciparum strain KI, while cytotoxicity was tested on MRC-5 cells (human fetal lung fibroblast cells). Based on the structural features and IC50 values for compounds 6, 7, 9, and 11, it can be assumed that the tryptophan moiety in the side chain is important for antiplasmodic activity. Compound 14 does not contain a tryptophan unit and also showed antiplasmodic activity, indicating that tryptophan could mediate, but is not vital for the antiplasmodic activity. The most promising compound was O-desmethylnummularine-R (7), which exhibited an IC50 of >64.0 µM against MRC-5 cells (Table 17) [5].

Table 17.
Antiplasmodial activity against against P. falciparum strain K1 and cytotoxicity against MRC5-Cells (IC50 μM) for compounds 6, 7, 9, 11, and 14 from the roots of Ziziphus oxyphylla.
Maluf et al. (2016) reported that crotamine (15) selectively enters infected erythrocytes (Figure 18A) and has potent anti-plasmodial activity with an IC50 value of 1.87 µM (Figure 18B) [6].
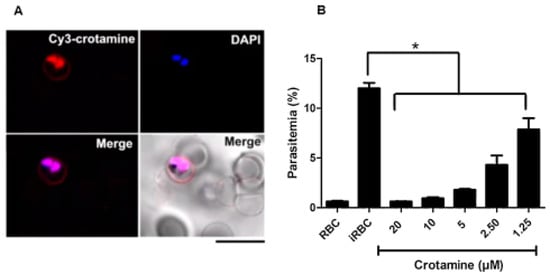
Figure 18.
Uptake of crotamine (15) by parasite-infected RBCs and effect on P. falciparum survival in vitro. (A) P. falciparum culture was incubated (10 µM) for 1 h, at room temperature. (B) Crotamine was added to 20, 10, 5, 2.50, and 1.25 µM P. falciparum culture. * p < 0.05.
The instability of H+ homeostasis by cr otamine was also confirmed (Figure 19B,C). The authors suggest that crotamine alters the internal pH of the vesicle due to its abundant Lys and Arg residues and the resulting high net charge (8+). Internal pH regulation is important for parasite survival, as it regulates the activity of certain intracellular enzymes required for parasite growth. Thus, this polypeptide is a promising lead molecule for the development of potential new peptidomimetics that have selectivity for infected erythrocytes and the ability to inhibit malaria infection by their natural affinity for acid vesicles [6].
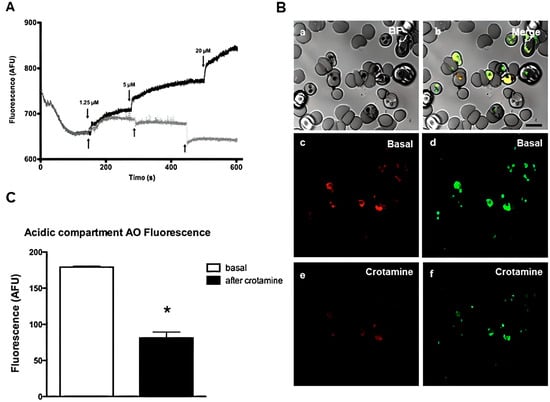
Figure 19.
Effect of chloroquine and crotamine (15) on intracellular acridine orange (AO) mobilization from acidic compartments of isolated P. falciparum parasites. (A) The lysosomotropic fluorochrome AO (5 M) were added in isolated parasites solution. Different concentrations of crotamine (1.25, 5, and 20 µM) were added during AO fluorescence acquisition in spectrofluorometer cuvette. Fluorescence intensities (arbitrary fluorescence units—AFU) represent at least three different cell preparations. Crotamine (gray line) and chloroquine (black line). (B) The alterations of acridine orange (AO) fluorescence was performed in confocal microscopy. iRBC was loaded with 5 µM of AO during 30 min. After addition of 20 M of crotamine, the effects on fluorescence of AO were monitored. (a–f) Confocal images: (a) Brightfield (BF); (b) Merged; (c) basal AO red channel; (d) basal AO green channel; (e,f) AO red and green channels after crotamine addition; (C) Histogram of AO fluorescence (red channel) with mean ± SD (N = 3). * p < 0.05.
The cyclic octadepsipeptides, octaminomycins A (16) and B (17) showed activity against P. falciparum (Table 18). According to Jang et al. (2017), compounds 16 and 17 were not significantly cytotoxic at a concentration of 30 μM against human cervical cancer cells (HeLa), human promyelocytic leukemia cells (HL-60), mouse temperature-sensitive cdc2 mutant cells (tsFT210), and rat kidney cells, which were infected with ts25 (srcts-NRK). They were also evaluated for antimicrobial activity against Staphylococcus aureus 209, Escherichia coli HO141, Aspergillus fumigatus Af293, Pyricularia oryzae kita-1, and Candida albicans JCM1542, as well as antimalarial activity against the P. falciparum 3D7, Dd2, and K1 strains. Chloroquine was less effective against strains resistant to Dd2 and K1, whereas compounds 16 and 17 showed the same in vitro antimalarial activities against chloroquine-sensitive 3D7 and chloroquine-resistant Dd2 and K1 strains, with no antimicrobial activity up to 30 μM [7].

Table 18.
Antimalarial activities for octaminomycins A (16) and B (17) with chloroquine as positive control against chloroquine-sensitive (3D7) and resistant (Dd2 and K1) strains of P. falciparum (IC50 in μM).
The antimalarial activity of the isolated kakeromamide B peptide (18) was evaluated against asexual blood-stage and liver-stage P. falciparum (Table 19). Compound 18 exhibited moderate activity against the blood stage of P. falciparum with an EC50 value of 8.9 µM as well as moderate liver-stage antimalarial activity against P. berghei liver schizonts with EC50 values of 11 µM. Although 18 displayed only moderate antimalarial activity, its ability to inhibit both the blood and liver life stages of Plasmodium, coupled with its low cytotoxicity in human cell lines, make it a promising lead compound for drug discovery [8].

Table 19.
Antimalarial activity and cytotoxicity of kakeromamide B (18).
The novel cyclic antimalarial and antitrypanosomal hexapeptide, pipecolisporin (19), was isolated from cultures of Nigrospora oryzae CF-298113 and exhibited interesting activity against P. falciparum and Trypanosoma cruzi Tulahuen C4 parasites [9]. The activity against the T. cruzi Tulahuen C4 parasites was the most remarkable, with an IC50 of 8.46 µM, comparable to that of the standard drug benznidazole, currently used in the treatment of Chagas disease (IC50 in the same assay of 2.21 µM) (Figure 20 and Figure 21). The activity against P. falciparum was also in the micromolar range, with an IC50 of 3.21 µM (Figure 22). It was not cytotoxic to the human cancer cell lines A549 (lung carcinoma), A2058 (metastatic melanoma), MCF7 (breast adenocarcinoma), MIA PaCa-2 (pancreatic carcinoma), and HepG2 (hepatocyte carcinoma) at the highest concentration tested of 50 µM (Figure 23).
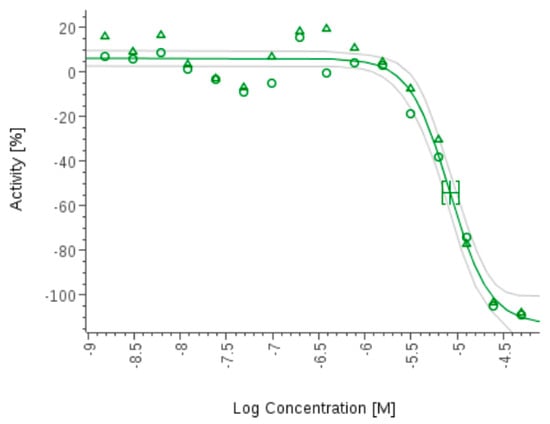
Figure 20.
Pipecolisporin (19) inhibits the growth of Tripanosoma cruzi.
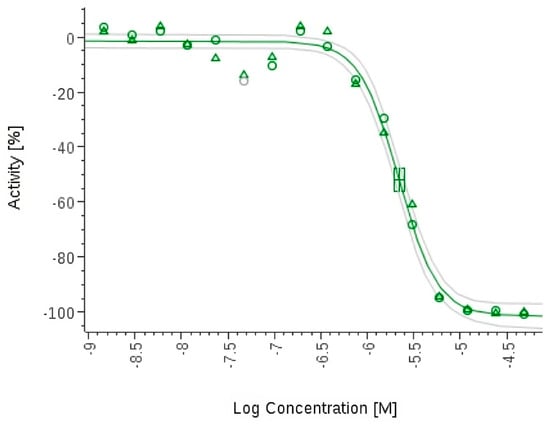
Figure 21.
Benznidazole inhibits the growth of Tripanosoma cruzi.
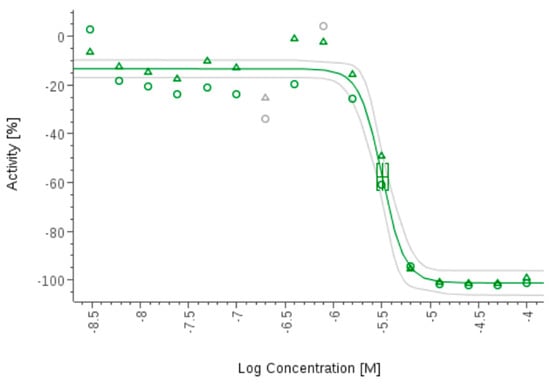
Figure 22.
Pipecolisporin (19) inhibits the growth of P. falciparum 3D7.
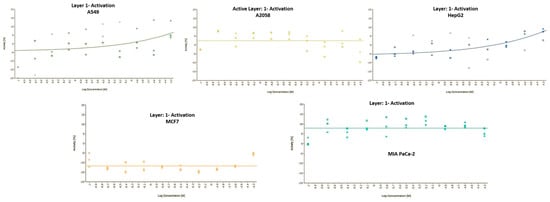
Figure 23.
Growth inhibition curves of pipescoliporin (19) against human cell lines.
Koshidacins A (20) and B (21) demonstrated in vitro antiplasmodial activity and cytotoxicity against human MRC-5 cells [10]. Compounds 20 and 21 demonstrated antiplasmodial activity against a chloroquine-sensitive P. falciparum FCR3 strain with IC50 values of 17.1 and 0.89 μM, respectively. Similarly, compounds 20 and 21 also exhibited antiplasmodial activity against chloroquine-resistant P. falciparum K1 strain with IC50 values of 12.5 and 0.83 μM, respectively. Compounds 20 and 21 showed cytotoxicity against human MRC-5 cells, with IC50 values of 6.8 and 14.7 μM, respectively, suggesting selectivity indices ranging from 0.4 to 18.4 (Table 20). In addition, when given intraperitoneally at a dose of 30 mg/kg/day for 4 days, compound 21 inhibited 41% of malaria parasites in vivo (Table 21).

Table 20.
In vitro antiplasmodial activity, cytotoxicity, and selectivity index of koshidacins A (20) and B (21) and artemisinin.

Table 21.
In vivo antiplasmodial activity in the Peters’ 4-day suppressive test of koshidacins A (20) and B (21) and artesunate (30 mg/kg/day × 4 days, i.p.) in a mouse model.
Shi et al. (2018) evaluated georatusin (22) produced by a soil fungus Geomyces auratus [11]. It had no obvious cytotoxicity, but displayed antiparasitic activities against Leishmania donovani (IC50 = 9.1 μM) and P. falciparum (IC50 = 1.6 μM) (Table 22). This discovery offers new insight into the metabolic potential and ecological importance of Geomyces and may encourage further exploration of this genus.

Table 22.
IC50 (µM) of compounds 23–29 against blood-stage P. falciparum strains.
New highly N-methylated linear peptides, friomamaride B (23) and shagamides A-F (24–29), exhibited activity against three strains of blood-stage P. falciparum [12]. All compounds were tested for their effectiveness against three blood-stage of P. falciparum using the blood-stage antiplasmodial and cytotoxicity assay. Friomamaride B (23), shagamides C (26) and D (27) with values less than 10 µg/mL all possess potential activity. The N-terminal of phenylalanine residue is essential for this activity. None of the isolated compounds demonstrated cytotoxic activity.
The carboxypeptidase inhibitor NpCI peptide (30), which is related to the model enzymes of bovine carboxypeptidase A (bCPA) and porcine carboxypeptidase B (pCPB), was discovered by Cabrera-Muñoz et al. (2023) [13]. The kinetic characterization of NpCI revealed that it was a slow inhibitor of bCPA and pCPB. While pCPB inhibition was not significant, the evaluation of NpCI inhibition was also performed by comparing the decrease in bCPA inhibitory activity caused by the substrate with the increase in substrate concentration. The Dd2 strain was used for in vitro antiplasmodial activity against P. falciparum, showing that the cycle was significantly delayed with an IC50 of 5.5 µmol/L. Parasite growth can be slowed down by increasing inhibitor concentration. The growth inhibition by NpCI occurs during parasite development. The 3D7 strain displayed comparable inhibition with a delayed maturation mechanism. NpCI was not cytotoxic to human cells (IC50 < 25%).
Kiefer et al. (2019) synthesized 17 cyclomarin analogues (96–100, 106–110, 119–125) and evaluated their biological activities against chloroquine-sensitive Pfalcp strain 3D7 and multi-resistant strain Dd2, as well as against Mtb wild-type strain Erdma (Table 23) [17]. The antitubercular activity was also evaluated, and the resazurin reduction microtiter assay (REMA) was used to measure the growth inhibition of Mtb.

Table 23.
The biological activity of the new cyclomarin derivatives against Pfalcp (chloroquine sensitive 3D7, multi-resistant Dd2 strains, and Mtb (Erdman wt strain)), as well as the overall synthesis processes.
Three compounds (96, 97, 99) demonstrated excellent parasitic growth suppression (Table 23). Compound 99 consists of a simplified γ,δ-unsaturated side chain, and the N’-methyl tryptophan unit. Desoxycyclomarin C, which was inspired by a natural product compound, has a similar range of bioactivity, but it is significantly shortened. The anti-mycobacterial activity was demonstrated by the five derivatives (96, 98, 99, 108 and 110) when applied to Erdman wild-type strain. Due to a remarkable simplification by replacing two of the four non-canonical amino acids by L-valine and L-tryptophan, compound 110 represents a highly appealing natural product-derived lead structure for battling Mtb.
Fagundez et al. (2018) revealed that most of the synthesized peptides have the potential to be antimalarials [20]. The corresponding cyclopeptide effectiveness was evaluated against the chloroquine-resistant K1 strain of P. falciparum. Cyclo-Cys(Trt)-Gly-Thr(tBu)-Gly-Cys(Trt)-Gly (compound 140) showed potent in vitro and selective activity against this parasite with an EC50 = 28 µM. The inclusion of a carboxylic group from Glu (151, 152) can increase solubilization, and the substitution of hydrophobic amino acids can boost biological activity (Table 24) [20].

Table 24.
Antiplasmodium activities of macrocycles.
Fagundez et al. (2018) also developed cyclopeptides containing N-methyl amino acids that demonstrated promising antiplasmodial activity, compounds 156–161 and 163–165 (Table 25) [21]. In addition, a new class of antimalarial cyclopeptides that contain N-methyl Gly has been developed that exhibits enhanced antiplasmodial activity. The in vitro evaluation of the compounds against P. falciparum revealed that N-Me-Gly is required to maintain the activity in the presence of a Glu with a free carboxyl group. Moreover, none of the active compounds are toxic against HepG2 cells.

Table 25.
Parasite growth inhibition and selectivity indexa.
Compounds 158 and 160 were assessed for their antimalarial activity in comparison to other antimalarial drugs (Figure 24). These results point to an antimalarial mode of action that does not immediately affect parasite viability. The prophylactic potential of compounds 158 and 160 is demonstrated by their low and submicromolar EC50 values (0.018 and 0.355 µM) in the liver stage of the parasite.
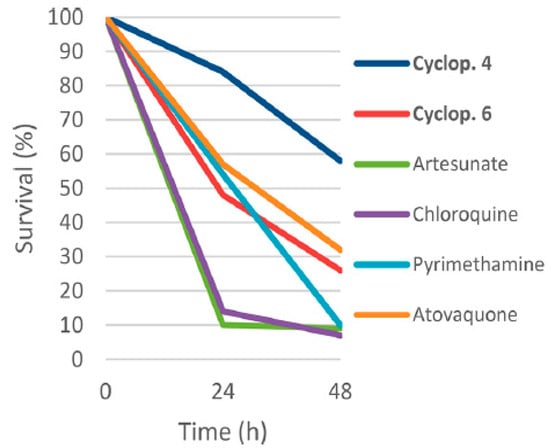
Figure 24.
Killing profile against P. falciparum 3D7A strain of compounds 158 and 160, artesunate, chloroquine, atovaquone, and pyrimethamine tested at 10 times the EC50 (number of replicates n = 2). cyclop. 4 = 158; cyclop. 6 = 160.
Compounds 157 and 165 are effective against P. berghei parasites, reducing parasitemia by 71 and 66% on day 5. To evaluate the oral bioavailability, the plasma pharmacokinetic of compound 157 in male Swiss Albino mice, following a single oral dosage, was examined, and shows a considerable half-life of 4.93 h.
The hirsutellide A analog, compound 177, exhibited moderate antiplasmodial activity (IC50 = 2.3 μM) similar to that reported for hirsutellide A (IC50 = 4.2 μM). Compound 177 is not cytotoxic (IC50 > 100 μM) to Hep2G cells. ADME profiling for compound 177 displayed moderate stability in humans, but low stability in mouse microsomes (Table 26) [22].

Table 26.
Biological activities and ADME profile of compound 177.
Additionally, the analogs have little to no activity against Mtb H37Rv. Peptide analogs generally have higher antiplasmodial activity (IC50 = 1.8−7.7 μM) than the depsipeptide analogs (IC50 = 7.5−20.1 μM), exhibiting a higher aqueous solubility, a high plasma stability and mouse plasma stabilities. Thus, the ester-to-amide substitution and the membrane permeability of hirsutellide A analogs appear to be dependent on the nature of the amino acid substituents. This study provides insight into the structural features relevant to the cyclic peptide-related drugs. Some compounds exhibited promising antimycobacterial activity and low cytotoxicity, making them potential candidates for further research and development as anti-TB agents.
The developed macrocyclic peptides as proteasome inhibitors has potential for antimalarial drugs (Zhang et al., 2022) [24]. Cyclic peptide 201 is a noncovalent inhibitor with strong antimalarial activity and high species selectivity, but has poor pharmacokinetics, therefore a docking model was developed. Compound 201 was acetylated using the N-Terminal amino group to boost its antiparasitic and inhibition activities. Five compounds were created, and P1, P3, P5, and P2–P4 linkers were examined for their contribution to the potency and drug-like properties of macrocyclic peptides (Table 27, Table 28, Table 29 and Table 30) [24].

Table 27.
Optimization of the N-Terminal Cap and Linker.

Table 28.
Optimization of the P1 Group.

Table 29.
Optimization of the P3 Group.

Table 30.
Optimization of the P5 Group.
Compound 220 demonstrated good potency against parasite and pharmacokinetics and made optimal use of P1 amide moiety modification (Table 28). Compound 223 showed moderate inhibition against hu-β2c (IC50 = 97.4 μM). Compounds 272 and 270 exhibited moderate inhibition of β1c and β1i. All other compounds showed <50% inhibition against all the four subunits, even at 100 μM.
Similar to the observation of the simultaneous inhibition of β5 and β2 in tumors, co-inhibition of β5 and β2 is synergistic and the inhibition of Pf20s β5 is sufficient to kill Plasmodium. The compounds were optimized for aqueous solubility, passive membrane permeability, metabolic stability, and a clean off-target profile against CYP450s and the hERG channel. The proteasome function appears to be critical for parasites to survive.
Falcitidin and its analogs (296–303) were synthesized by Kotturi et al. (2014) and showed moderate activity against P. falciparum 3D7, but only when N-tritylated on its histidine residue [25]. The IC50 activity of the new compounds, which ranged from 1 to 5 µM, was typically modest in whole cells. Compound 302 exhibited the highest activity (IC50 0.14 µM). These new compounds represent an important new peptide chemotype that may be elaborated into improved antimalarial leads (Table 31).

Table 31.
Falcitidin acylated analogs and their IC50 values against P. falciparum 3D7 in a whole-cell assay (standardized with commercial chloroquine; IC50 0.020 ± 0.002 µM).
Conroy et al. (2014) developed a new class of antimalarial drugs based on the natural product gallinamide A, which inhibits the falcipain cysteine proteases essential for malaria parasite survival [27]. These gallinamide A analogs were as effective as chloroquine against the P. falciparum parasite. Gallinamide A analog 309 has strong inhibitory activity against FP-2, FP-3, and P. falciparum in vitro (Table 32). Reducing the enol moiety in the acyl-pyrrolinone unit in compound 310 resulted in a two-fold reduction in antiplasmodial activity (IC50 = 210 nM), but a slight improvement in activity against FP-2 and FP-3. Analog 311 showed no measurable inhibition of FP-3 and a 3 orders of magnitude drop in inhibitory effectiveness against FP-2 (IC50 = 3710 nM), as well as significantly decreased activity against P.falciparum. Analog 312 lost the measurable inhibitory activity against the FPS and the parasite after losing both of its olefinic moieties.

Table 32.
Inhibiton of FP-2, FP-3, and P. falciparum by gallinamide A analogs 309–312 (n = 3).
The activities of analogs 313, 315, and 317 were similar: they inhibited FP-2 at low micromolar concentrations (IC50 = 3.4–11.5 µM), did not significantly inhibit FP-3 at 25 µM, and inhibited P. falciparum at nanomolar concentrations (IC50 = 320–5400 nM) (Table 33). Interestingly, peptide 318 demonstrated less potent antiparasitic activity (IC50 = 6.6 µM) but inhibited FP-2 and FP-3 by adding an N-methylproline functionality at the N-terminus of the peptide while keeping the C terminal benzylamide. The lack of activity was especially noticeable for 315 because it has the same structure as analog 309. The addition of a more flexible and highly functionalized hydroxyproline methyl ester to the C-terminus in 314 produced inhibitory activity against FP-2 and P. falciparum, which was comparable to the benzylamide-derived compounds. Low micromolar antiparasitic activity (IC50 = 1.1 µM) and moderate inhibitory activity against FP-2 and FP-3 were achieved by C-terminal functionalization as a thiazole amide in compound 316 [27].

Table 33.
Inhibition of FP-2, FP-3, and the 3D7 Strain of P. falciparum by gallinamide A analogs 313–318 a.
Like analog 309, the C-terminal N-acyl-pyrrolinone in 346–351 restored strong inhibitory activity against the FPs and P. falciparum (Table 34). Compound 346 displayed similar inhibitory activity to 309 against FP-2, FP-3, and P. falciparum despite having no modification on the pyrrolinone molecule. The pyrrolinone ring of 347 was modified to include a more hydrophobic substituent, which improved its inhibitory efficacy against FP-3 and P. falciparum. As with compound 347, the addition of aromatic side chains to the pyrrolinone ring did not modify the inhibitory activity against FP-2 but significantly increased activity against FP-3 and P. falciparum. The most effective inhibitor of FP-3 and P. falciparum was compound 349, which had an indole side chain attached to the pyrrolinone ring [27].

Table 34.
Inhibition of FP-2, FP-3, and the 3D7 strain of P. falciparum by gallinamide A analogs 309 and 346–351 a.
The compounds were screened against the P. falciparum, namely AP M1, AP M17, and AP M18 aminopeptidase (AP) enzymes. The effective N-acyl pyrrolinone analogs 309 and 346–349, as well as the C-terminal amide derivatives 309, 315, and 317, had IC50 values of less than 600 nM against the 3D7 strain of P. falciparum (Table 35). The CQ-resistant, Dd2 strain of P. falciparum was powerfully inhibited by every tested compounds (IC50 = 29.0–421 nM). The compounds were selective inhibitors of P. falciparum over HEK298 cells, with 309, 315, and 317 showing no measurable inhibition of this cell line at a concentration of 50 µM, but having strong inhibitory activity against human cathepsin [27].

Table 35.
Inhibition of the CQ-Resistant Dd2 Strain of P. falciparum, HEK298 Cells, and P. falciparum aminopeptidase M1, M17, and M18 (AP M1, M17, and M18) by gallinamide A analogs a.
The gallinamide A analogs, synthesized by Stoye et al. (2019), were also tested for activity against FP2, FP3 also CQ-sensitive 3D7 and CQ-resistant W2 strains of P. falciparum in vitro (Table 36) [28]. Most substitutions in the compound structure were well tolerated, except for analog 387 which had a more potent activity parasite (IC50 3D7 = 1 nM; W2 = 4 nM) and substituents on R1 which markedly enhanced the stability in plasma and blood. Five gallinamide A analogs were assessed in vitro for their stability in mouse blood and plasma (Table 37). The half-life in plasma and blood was significantly prolonged due to the substituents at R1 and R3. Additional cytotoxicity assays in the HEK293 cell line of the compounds displayed no measurable cytotoxicity at 25 µm. The last three analogs 372, 373 and 387 were assessed in vivo in a mouse model of cerebral malaria (CM), P. berghei ANKA (PbA) infection. Severe signs of disease as well as the rise in parasitemia were both significantly delayed by analog 387.

Table 36.
Inhibition of FP2, FP3 and 3D7 and W2 Strains of P. falciparum a.

Table 37.
In Vitro degradation half-life in mouse blood and plasma.
The peptides derived from PfSERA5 by Kanodia et al. (2014) inhibited the enzymatic activity of PfSERA5P50 protein, which in turn blocked the development of the parasites in an in vitro culture [29]. Evaluation of PfSERA5 derivative against P. falciparum development and growth revealed that SE5 P1 (388) and SE5 P2 (389) have the highest parasite growth inhibition and invasion values of 60–70% (Figure 25).
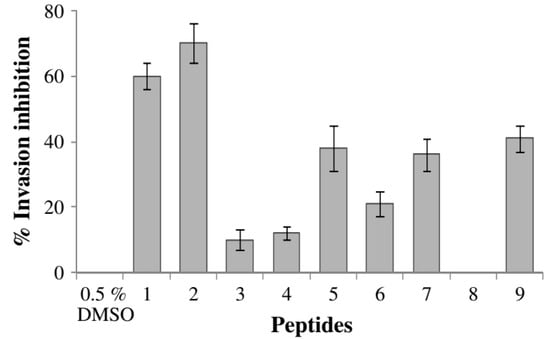
Figure 25.
The ability of SE5 P1 (388) and SE5 P2 (389) to specifically inhibit PfSERA550 activity.
Analysis of the synthetic substrate Suc-LLVY-AMC revealed that PfSERA550′s proteolytic activity was greatly reduced by the two C-terminal peptides SE P1 (388) and SE5 P2 (389). Kanodia et al. also describe how they used computational modeling to examine molecular docking studies with the known crystal structure of PfSERA5 to explore the effects of SE5 P1 (388) and SE5 P2 (389) peptides. SE5 P1 (388) and SE5 P2 (389) occupied 50.1 and 57.5% larger than the substrate when the Suc-LLVY-AMC was applied to see the bond (Table 38), suggesting that the peptides must interact with Glu638 and Ser640/Ser641 to be inhibitory [29].

Table 38.
The shared percentage volume of each peptide and Suc-ILVY-AMC substrate according to the poses and conformations selected in the docking studies.
P. falciparum-infected RBCs uptake of labeled SE5 P1 and SE5 P2 peptides when the localization of biotinylated peptides therein inhibited the protease activity similarly to the non-biotinylated peptides. This indicates that the peptides have access to the intracellular parasites and are co-localized with the PfSERA5 protein. PfSERA5 plays an important role in parasite development and the final proteolytic cleavage, which can be produced as a new drug design and offers information on the potential use of these peptides as antimalarial therapeutics [29].
Angiotensin II analogs synthesized by Silva et al. (2015) and tested in vitro to identify a short bioactive peptide as well as to verify the hydrophobic cluster’s influence on parasite-membrane interaction on both P. gallinaceum and P. falciparum [30]. Fluoresence microscopy was utilized to examine the effects of the peptides on P. gallinaceum sporozoites produced by the salivary glands and the therapeutic index (MHC/MIC ratio). Higher values in the therapeutic index indicate more antimicrobial specificity. It is a parameter that measures the specificity of an antimicrobial agent and is calculated by the ratio MHC and MIC. The MHC/MIC ratio, which was higher in peptide 401, indicates that the peptides have varied specificities (Table 39). New peptides related to Ang II were designed, including the most hydrophobic amino acid residues (Val, Ile, Pro, and Phe), aromatic residues (Tyr, His, Pro, and Phe) and residues from the Ang II hydrophobic cluster (Tyr, Ile and His) in an attempt to verify the peptide–parasite interactions.

Table 39.
Therapeutic index determination (MHC/MIC ratio).
Due to the influence of hydrophobic clusters, side chain aromatic rings, and hydrophobic residues, these peptides showed antiplasmodial activity in P. gallinaceum sporozoite (64–94%) and activity between 89 and 94% (Figure 26). The three peptides with the highest antiplasmodial activity were 1 (397), 5 (401), and 6 (402) with 94, 89, and 94%, respectively [30].
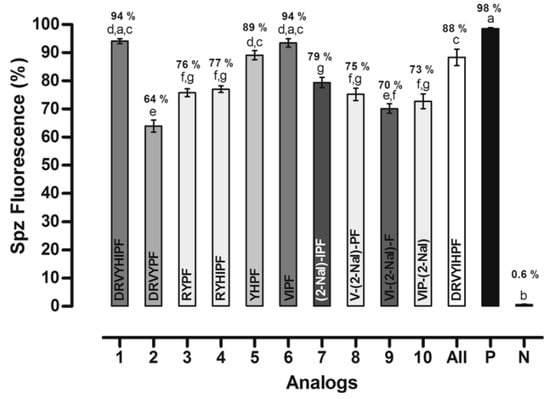
Figure 26.
Effects of peptides on membrane permeability expressed as the percent of fluorescent mature sporozoites (mean ± standard deviation, n = 9). Letters indicate those results not significantly different from each other at the p < 0.05 level. Positive control group (+): digitonin/PBS; negative control group (−) Positive control group (+): digitonin/PBS: negative control group (−): PBS.
The effect of the peptides in the P. falciparum erythrocytic cycle was assessed in vitro. All peptides reduced new ring formation at 10−8 mol L−1, which was studied by Saraiva et al. as the ideal concentration for these inhibition assays. Four analogs reduced the ring formed in the blood stage, but only analogs 5 (401) and 6 (402) showed inhibition that was higher by 50% than control (Figure 27). Figure 27 showed that after 24 h of incubation with 2–3% schizont-infected erythrocyte cultures in the absence (control) or presence of 10−8 mol L−1 peptides, the percentage age of rings was evaluated (* p < 0.05 compared to the control, *** p < 0.001). The result is statistically significant compared to the (mean ± standard deviation, n = 2), as indicated by the dark grey shading [30].
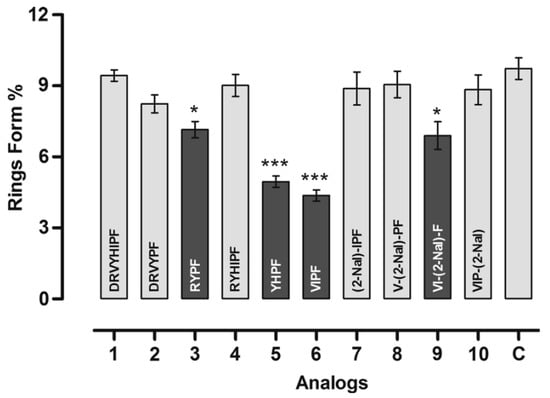
Figure 27.
Effect of peptides in the new ring formation. * Statistically significant compared with control value p < 0.05. *** Statistically significant compared with control value p < 0.001. Analog 1 = 397; 2 = 398; 3 = 399; 4 = 400; 5 = 401; 6 = 402; 7 = 403; 8 = 404; 9 = 405; 10 = 406.
Another technique for determining the hemolytic effect of peptides demonstrated that peptides 401 and 402 had an effect of the Ang II to define the hemolysis. These peptides did not exhibit hemolytic effects (Figure 28).
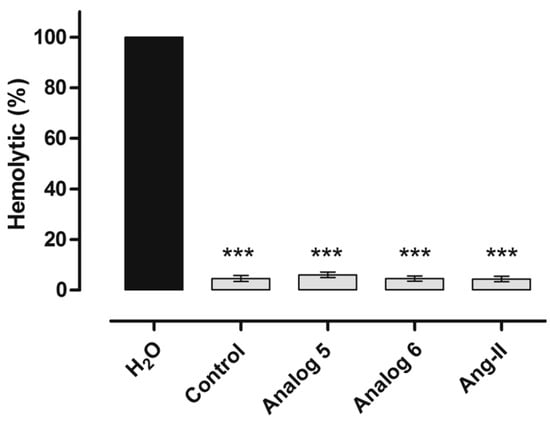
Figure 28.
Haemolytic assay of human red blood cells. *** Statistically significant compared with distilled water value p < 0.05. Analog 5 = 401; 6 = 402.
The hydrophobic portion and the Arg, Tyr, Pro, and Phe residues increased the antiplasmodial activity when they were present in the primary sequence. Furthermore, these peptides did not display hemolysis or contractile response activities, as shown in Figure 29 (*** p < 0.05 compared to control, n = 2) [30].
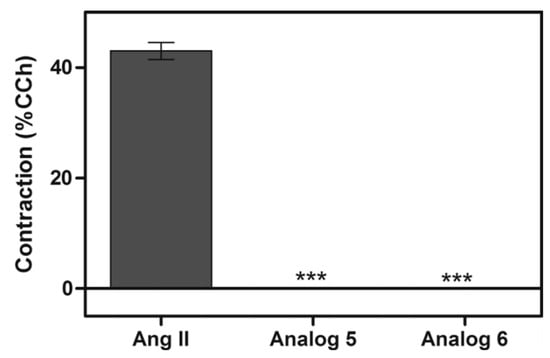
Figure 29.
Effect of Ang II, peptide 5 (401) and 6 (402) on the contractile responses of muscle tissue compared to carbachol (CCh) activity. *** Statistically significant compared with control value p < 0.05 (mean ± standard deviation, n = 2)
The IC50 values are more promising than the Ang II results, which were demonstrated by evaluating seven concentrations giving 7–65% inhibition (Figure 30). The results imply that intramolecular interactions cause conformational tendencies that are crucial for antiplasmodial activity. When present on the peptide primary sequence, the hydrophobic portion and the residues of Arg, Tyr, Pro, and Phe increased the antiplasmodial activity. Following in vivo model tests of this class of peptides, this type of research aids the development of new chemotherapeutics, which can be explored as antimalarial drugs [30].
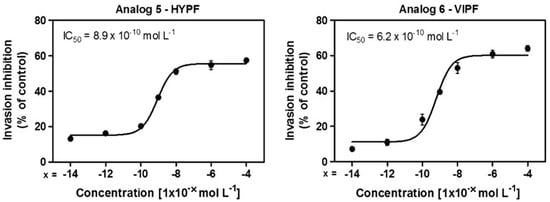
Figure 30.
IC50 value for each assay. Peptide 5 (401) and 6 (402) were diluted in seven concentration [(10−4, 10−6, 10−8, 10−10, 10−12, 10−14) mol L−1] giving 7–65% inhibition.
The synthesized RAS octapeptides were tested for their effectiveness against P. falciparum and P. gallinaceum, revealing that only [Ala5]-Ang II showed equipotent Ang II activity with 45% of biological activity [31]. At 10−8M concentration, P. gallinaceum erythrocytic cycle [Ala6]-Ang II decreased parasite invasion by 49%. The efficiency of anti-plasmodial activity was significantly impacted by the presence or absence of amino acid substitutions. The anti-plasmodial activity of the Ang II molecule depends on specific amino acid side chains. The biological activities and binding affinities are affected when the biologically active peptide is replaced by an alanine residue. From Figure 31, the percentage of rings was determined after 24 h of incubation of erythrocyte culture infected with 2–3% schizonts in the absence (control) or in the presence of 10−8 M analogs. Dark grey shading denotes that the result is statistically significant compared to the control.
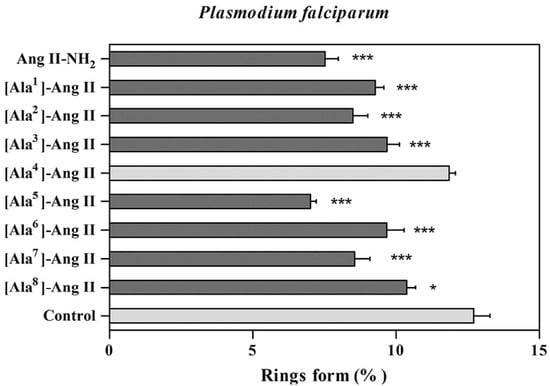
Figure 31.
Effect of analogues in the new ring formation. * Statistically significant compared with control value p < 0.05. *** Statistically significant compared with control value p < 0.01. Dark grey shading indicates that the result is statistically significant compared with control.
Regarding the contractile response, [Ala5]-Ang II and [Ala6]-Ang II did not promote contractile activity compared to Ang II, and carbachol [Ala5]-Ang II and [Ala6]-Ang II reduced parasite invasion in red blood cells in the erythrocyte hemolysis assays, and presented no effect on cells integrity at 10−8 M concentration. This model directs new possibilities for peptide design that can act more effectively in preventing the erythrocytic cycle of the parasite and other phases of the human–malaria cycle. Figure 32 demonstrates that after erythrocyte cultures were infected for 24 h with 3% parasitemia, the percentage of total parasites was determined in the absence (control) or presence of 10−8 M analogs. Dark grey shading denotes that the result is not statistically significant compared with control [31].
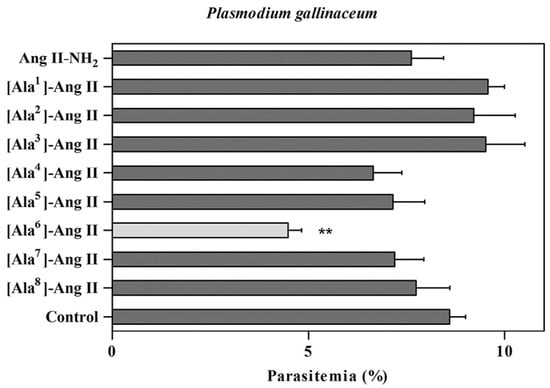
Figure 32.
Effect of analogs in the erythrocytic cycle of P. gallinaceum. ** Statistically significant compared with control value p < 0.05. Dark grey shading indicates that the result is not statistically significant compared with control
Other synthesized angiotensin analogs presented high antiplasmodial activity in the P. gallinaceum test [32]. They have similar hydrophobic interactions between the isolated guanidyl (Arg2) and hydroxyl (Tyr4) groups and the side chains of alkyl and aromatic amino acid. The guanidyl (Arg2) and carboxyl (Asp1) groups seem to be less significant in this case. Lactam bridges have an important role in constricting the conformation of bioactive peptides. When evaluated against P. falciparum, all lactam bridge analogs lacked significant activity against P. gallinaceum and perform better against P. falciparum because of the rigidity of the peptide bridge and possibe increased resistance to degradation under this kind of restriction.
Analog 416 and 418 presented significant antiplasmodial activity, however analog 425, a disulfide bridge analog, was the most active peptide against P. gallinaceum (Figure 33). It was also active against P. falciparum because it has the same restriction size and implications when considering hydrophobic and hydrophilic interactions between side chains. All analogs frequently adopt a β-turn conformation, which is consistent with that of the analog that is believed to be most active against P. gallinaceum [32].
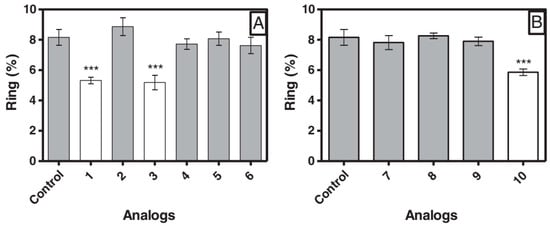
Figure 33.
Effect of synthetic peptides in erythrocyte invasion by Plasmodium falciparum. (A) Analog 1–6; (B) Analog 7–10. *** Statistically significant compared with control value p < 0.05 Analog 1 = 416; 2 = 417; 3 = 418; 4 = 419; 5 = 420; 6 = 421; 7 = 422; 8 = 423; 9 = 424; 10 = 425.
Angiotensin II hormones were also synthesized to investigate erythrocytic cycle invasion [33]. Peptides 426 and 427 exhibited >80% activity on P. gallinaceum and >40% activity on P. falciparum. The hemolytic effect, contractile response, and stability in human serum were determined (Figure 34), demonstrating that the peptides did not present hemolytic effects as they were inactive when compared to positive controls and were resistant to degradation in human serum after a prolonged exposure (6 h), especially peptides 426 and 427, which displayed excellent stability.
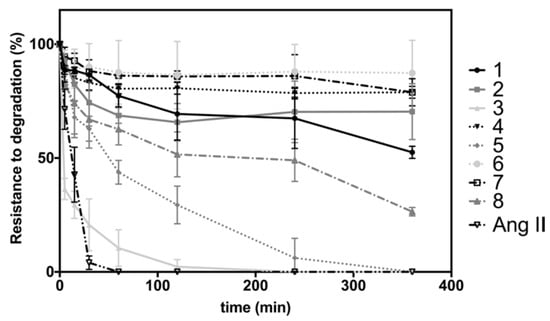
Figure 34.
Stability of Ang II derivatives in human serum. Analog 1 = 426; 2 = 427; 3 = 428; 4 = 429; 5 = 430; 6 = 431; 7 = 432; 8 = 433.
The study of synthesized Dec-NH2 and eight analog peptides reported that certain amino acid substitutions significantly improved the peptide’s antiplasmodial activity, providing new sequence principles for creating potent anti-malaria drugs, such as replacing the original sequence by Arg, Phe, and Trp [34]. The most active peptides examined were [Arg]1-Dec-NH2 (438), [Pro]4-Dec-NH2 (437), and [Phe]2-Dec-NH2 (439); all compounds substitutions improved the effectiveness, with the gihest antiplasmodial activities achieved with mutations to the N-terminus of Dec-NH2. A higher antiplasmodial activity was achieved by the introduction of a positive charge, increased hydrophobicity, and introduction of a restrictor residue (Figure 35).
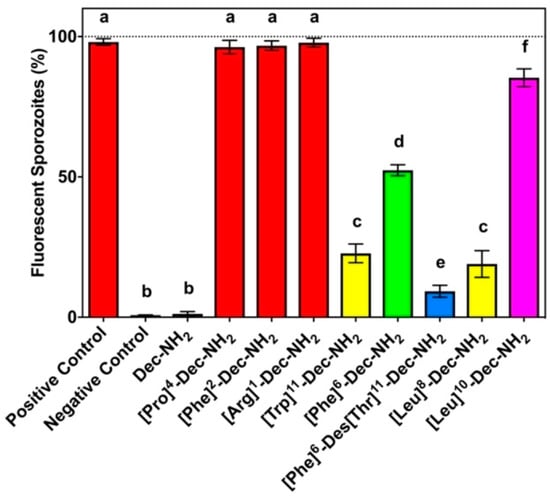
Figure 35.
Effects of Dec-NH2 and analogs on membrane permeability. Positive control group (+): digitonin/PBS; negative control group (–): PBS. Peptide concentration = 60 mmol L−1. Letters indicate those results that are not significantly different from each other at p < 0.05 by Turkey’s test.
4. Conclusions
This review on antimalarial peptides discussed the natural sources, chemical access and antimalarial properties of the peptides, showing that nature (higher plant, microbes, venom) is a source of these antimalarial peptides. Alternatively, they can be chemically synthesized using a variety of techniques, such as solution phase, solid phase and a combination of solid and solution phase synthesis. The selection of coupling reagent, cyclization site, and protection groups are important factors to consider for the successful synthesis. Most isolated and synthesized peptides are active against P. falciparum with some exhibiting inhibition of P. gallinaceum and P. berghei. Moreover, several peptides were also cytotoxic against MRC5-cells, HEK2931, and HepG2. The information presented in this review may be useful in the design and synthesis of new potent antimalarial agents.
Author Contributions
Conceptualisation, N.K. and R.M.; writing—review and editing, N.K., R.M., A.T.H. and U.S.; funding acquisition, N.K. and R.M. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by UNIVERSITAS ISLAM BANDUNG (Grant number 1549/UN6.3.1/PT.00/2023)
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not Applicable.
Acknowledgments
The authors would like to thank UNIVERSITAS PADJADJARAN for the facilities provided in the process of writing the literature review.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization. Available online: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2021 (accessed on 11 September 2023).
- Talapko, J.; Škrlec, I.; Alebić, T.; Jukić, M.; Včev, A. Malaria: The past and the present. Microorganisms 2019, 7, 179. [Google Scholar] [CrossRef]
- Pinto, M.E.F.; Batista, J.M.; Koehbach, J.; Gaur, P.; Sharma, A.; Nakabashi, M.; Cilli, E.M.; Giesel, G.M.; Verli, H.; Gruber, C.W.; et al. Ribifolin, an orbitide from jatropha ribifolia, and its potential antimalarial activity. J. Nat. Prod. 2015, 78, 374–380. [Google Scholar] [CrossRef]
- Tuenter, E.; Exarchou, V.; Baldé, A.; Cos, P.; Maes, L.; Apers, S.; Pieters, L. Cyclopeptide Alkaloids from Hymenocardia acida. J. Nat. Prod. 2016, 79, 1746–1751. [Google Scholar] [CrossRef]
- Tuenter, E.; Ahmad, R.; Foubert, K.; Amin, A.; Orfanoudaki, M.; Cos, P.; Maes, L.; Apers, S.; Pieters, L.; Exarchou, V. Isolation and Structure Elucidation by LC-DAD-MS and LC-DAD-SPE-NMR of Cyclopeptide Alkaloids from the Roots of Ziziphus oxyphylla and Evaluation of Their Antiplasmodial Activity. J. Nat. Prod. 2016, 79, 2865–2872. [Google Scholar] [CrossRef]
- Maluf, E.; Mas, C.; Oliveira, E.B.; Melo, P.M.; Carmona, A.K.; Gazarini, M.L.; Hayashi, M.A.F. Inhibition of malaria parasite Plasmodium falciparum development by crotamine, a cell penetrating peptide from the snake venom. Peptides 2016, 78, 11–16. [Google Scholar] [CrossRef]
- Jang, J.P.; Nogawa, T.; Futamura, Y.; Shimizu, T.; Hashizume, D.; Takahashi, S.; Jang, J.H.; Ahn, J.S.; Osada, H. Octaminomycins A and B, cyclic octadepsipeptides active against Plasmodium falciparum. J. Nat. Prod. 2017, 80, 134–140. [Google Scholar] [CrossRef]
- Sweeney-Jones, A.M.; Gagaring, K.; Antonova-Koch, J.; Zhou, H.; Mojib, N.; Soapi, K.; Skolnick, J.; McNamara, C.W.; Kubanek, J. Antimalarial peptide and polyketide natural products from the Fijian marine cyanobacterium moorea producens. Mar. Drugs 2020, 18, 167. [Google Scholar] [CrossRef]
- Fernández-Pastor, I.; González-Menéndez, V.; Annang, F.; Toro, C.; Mackenzie, T.A.; Bosch-Navarrete, C.; Genilloud, O.; Reyes, F. Pipecolisporin, a novel cyclic peptide with antimalarial and antitrypanosome activities from a wheat endophytic nigrospora oryzae. Pharmaceuticals 2021, 14, 268. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Hachiya, K.; Ikeda, A.; Nonaka, K.; Higo, M.; Muramatsu, R.; Noguchi, C.; Honsho, M.; Asami, Y.; Inahashi, Y.; et al. Koshidacins A and B, Antiplasmodial Cyclic Tetrapeptides from the Okinawan Fungus Pochonia boninensis FKR-0564. J. Nat. Prod. 2022, 85, 2641–2649. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.M.; Richter, C.; Challinor, V.L.; Grün, P.; Girela Del Rio, A.; Kaiser, M.; Schüffler, A.; Piepenbring, M.; Schwalbe, H.; Bode, H.B. Georatusin, a Specific Antiparasitic Polyketide-Peptide Hybrid from the Fungus Geomyces auratus. Org. Lett. 2018, 20, 1563–1567. [Google Scholar] [CrossRef] [PubMed]
- Bracegirdle, J.; Casandra, D.; Rocca, J.R.; Adams, J.H.; Baker, B.J. Highly N-Methylated Peptides from the Antarctic Sponge Inflatella coelosphaeroides Are Active against Plasmodium falciparum. J. Nat. Prod. 2022, 85, 2454–2460. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Muñoz, A.; Sierra-Gómez, Y.; Covaleda-Cortés, G.; Reytor, M.L.; González-González, Y.; Bautista, J.M.; Avilés, F.X.; Alonso-del-Rivero, M. Isolation and Characterization of NpCI, a New Metallocarboxypeptidase Inhibitor from the Marine Snail Nerita peloronta with Anti-Plasmodium falciparum Activity. Mar. Drugs 2023, 21, 94. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Fujita, S.; Okada, Y.; Chiba, K. Soluble tag-assisted peptide head-to-tail cyclization: Total synthesis of mahafacyclin B. Org. Lett. 2013, 15, 1155–1157. [Google Scholar] [CrossRef] [PubMed]
- Peña, S.; Scarone, L.; Manta, E.; Serra, G. First total synthesis of aerucyclamide B. Tetrahedron Lett. 2013, 54, 2806–2808. [Google Scholar] [CrossRef]
- Barbie, P.; Kazmaier, U. Total Synthesis of Cyclomarin A, a Marine Cycloheptapeptide with Anti-Tuberculosis and Anti-Malaria Activity. Org. Lett. 2016, 18, 204–207. [Google Scholar] [CrossRef]
- Kiefer, A.; Bader, C.D.; Held, J.; Esser, A.; Rybniker, J.; Empting, M.; Müller, R.; Kazmaier, U. Synthesis of New Cyclomarin Derivatives and Their Biological Evaluation towards Mycobacterium Tuberculosis and Plasmodium Falciparum. Chem.-A Eur. J. 2019, 25, 8894–8902. [Google Scholar] [CrossRef]
- Gholap, S.S.; Ugale, S.R. A Total Synthesis of the Cyclic Depsipeptide Chaiyaphumine-A. ChemistrySelect 2017, 2, 7445–7449. [Google Scholar] [CrossRef]
- Lu, H.; Batey, R.A. Total synthesis of chaiyaphumines A-D: A case study comparing macrolactonization and macrolactamization approaches. Tetrahedron Lett. 2022, 108, 154138. [Google Scholar] [CrossRef]
- Fagundez, C.; Sellanes, D.; Serra, G. Synthesis of cyclic peptides as potential anti-malarials. ACS Comb. Sci. 2018, 20, 212–219. [Google Scholar] [CrossRef]
- Fagundez, C.; Sellanes, D.; Peña, S.; Scarone, L.; Aguiar, A.C.C.; De Souza, J.O.; Guido, R.V.C.; Stewart, L.; Yardley, V.; Ottilie, S.; et al. Synthesis, Profiling, and in Vivo Evaluation of Cyclopeptides Containing N-Methyl Amino Acids as Antiplasmodial Agents. ACS Med. Chem. Lett. 2019, 10, 137–141. [Google Scholar] [CrossRef]
- Sahile, H.A.; Sahile, H.A.; Martínez-Martínez, M.S.; Dillenberger, M.; Becker, K.; Imming, P. Synthesis and Evaluation of Antimycobacterial and Antiplasmodial Activities of Hirsutellide A and Its Analogues. ACS Omega 2020, 5, 14451–14460. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chen, L.; Duan, X.; Meng, Y.; Jiang, L.; Li, M.; Zhao, G.; Li, Y. Total synthesis of hirsutellide A. Tetrahedron Lett. 2005, 46, 4377–4379. [Google Scholar] [CrossRef]
- Zhang, H.; Ginn, J.; Zhan, W.; Liu, Y.J.; Leung, A.; Toita, A.; Okamoto, R.; Wong, T.T.; Imaeda, T.; Hara, R.; et al. Design, Synthesis, and Optimization of Macrocyclic Peptides as Species-Selective Antimalaria Proteasome Inhibitors. J. Med. Chem. 2022, 65, 9350–9375. [Google Scholar] [CrossRef] [PubMed]
- Somanadhan, B.; Kotturi, S.R.; Yan Leong, C.; Glover, R.P.; Huang, Y.; Flotow, H.; Buss, A.D.; Lear, M.J.; Butler, M.S. Isolation and synthesis of falcitidin, a novel myxobacterial-derived acyltetrapeptide with activity against the malaria target falcipain-2. J. Antibiot. 2013, 66, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Kotturi, S.R.; Somanadhan, B.; Ch’Ng, J.H.; Tan, K.S.W.; Butler, M.S.; Lear, M.J. Diverted total synthesis of falcitidin acyl tetrapeptides as new antimalarial leads. Tetrahedron Lett. 2014, 55, 1949–1951. [Google Scholar] [CrossRef]
- Conroy, T.; Guo, J.T.; Elias, N.; Cergol, K.M.; Gut, J.; Legac, J.; Khatoon, L.; Liu, Y.; McGowan, S.; Rosenthal, P.J.; et al. Synthesis of gallinamide A analogues as potent falcipain inhibitors and antimalarials. J. Med. Chem. 2014, 57, 10557–10563. [Google Scholar] [CrossRef] [PubMed]
- Stoye, A.; Juillard, A.; Tang, A.H.; Legac, J.; Gut, J.; White, K.L.; Charman, S.A.; Rosenthal, P.J.; Grau, G.E.R.; Hunt, N.H.; et al. Falcipain Inhibitors Based on the Natural Product Gallinamide A Are Potent in Vitro and in Vivo Antimalarials. J. Med. Chem. 2019, 62, 5562–5578. [Google Scholar] [CrossRef]
- Kanodia, S.; Kumar, G.; Rizzi, L.; Pedretti, A.; Hodder, A.N.; Romeo, S.; Malhotra, P. Synthetic peptides derived from the C-terminal 6 kDa region of Plasmodium falciparum SERA5 inhibit the enzyme activity and malaria parasite development. Biochim. Et Biophys. Acta-Gen. Subj. 2014, 1840, 2765–2775. [Google Scholar] [CrossRef]
- Silva, A.F.; Torres, M.D.T.; Silva, L.D.S.; Alves, F.L.; Pinheiro, A.A.D.S.; Miranda, A.; Capurro, M.L.; Oliveira, V.X. New linear antiplasmodial peptides related to angiotensin II. Malar. J. 2015, 14, 433. [Google Scholar] [CrossRef]
- Silva, A.F.; Silva, L.D.S.; Alves, F.L.; TorossianTorres, M.D.; de SáPinheiro, A.A.; Miranda, A.; LaraCapurro, M.; Oliveira, V.X. Effects of the angiotensin II Ala-scan analogs in erythrocytic cycle of Plasmodium falciparum (in vitro) and Plasmodium gallinaceum (ex vivo). Exp. Parasitol. 2015, 153, 1–7. [Google Scholar] [CrossRef]
- Torres, M.D.T.; Silva, A.F.; Silva, L.D.S.; de Sá Pinheiro, A.A.; Oliveira, V.X. Angiotensin II restricted analogs with biological activity in the erythrocytic cycle of Plasmodium falciparum. J. Pept. Sci. 2015, 21, 24–28. [Google Scholar] [CrossRef]
- Silva, A.F.; Torres, M.D.T.; Silva, L.S.; Alves, F.L.; de Sá Pinheiro, A.A.; Miranda, A.; Capurro, M.L.; De La Fuente-Nunez, C.; Oliveira, V.X. Angiotensin II-derived constrained peptides with antiplasmodial activity and suppressed vasoconstriction. Sci. Rep. 2017, 7, 14326. [Google Scholar] [CrossRef]
- Torres, M.D.T.; Silva, A.F.; Pedron, C.N.; Capurro, M.L.; de la Fuente-Nunez, C.; Junior, V.X.O. Peptide Design Enables Reengineering of an Inactive Wasp Venom Peptide into Synthetic Antiplasmodial Agents. ChemistrySelect 2018, 3, 5859–5863. [Google Scholar] [CrossRef]
- Ye, B.; Jiang, P.; Zhang, T.; Sun, Y.; Hao, X.; Cui, Y.; Wang, L.; Chen, Y. Total synthesis of the highly N-methylated peptides carmabin a and dragomabin. Mar. Drugs 2018, 16, 338. [Google Scholar] [CrossRef] [PubMed]
- Auvin, C.; Baraguey, C.; Blond, A.; Lezenven, F.; Pousset, J.L.; Bodo, B. Curcacycline B, a cyclic nonapeptide from Jatropha curcas enhancing rotamase activity of cyclophilin. Tetrahedron Lett. 1997, 38, 2845–2848. [Google Scholar] [CrossRef]
- Baraguey, C.; Auvin-Guette, C.; Blond, A.; Cavelier, F.; Lezenven, F.; Pousset, J.L.; Bodo, B. Isolation, structure and synthesis of chevalierins A, B and C, cyclic peptides from the latex of Jatropha chevalieri. J. Chem. Soc.-Perkin Trans. 1998, 18, 3033. [Google Scholar] [CrossRef]
- Baraguey, C.; Blond, A.; Correia, I.; Pousset, J.L.; Bodo, B.; Auvin-Guette, C. Mahafacyclin A, a cyclic heptapeptide from Jatropha mahafalensis exhibiting β-bulge conformation. Tetrahedron Lett. 2000, 41, 325–329. [Google Scholar] [CrossRef]
- Baraguey, C.; Blond, A.; Cavelier, F.; Pousset, J.L.; Bodo, B.; Auvin-Guette, C. Isolation, structure and synthesis of mahafacyclin B, a cyclic heptapeptide from the latex of Jatropha mahafalensis. J. Chem. Soc. Perkin Trans. 2001, 17, 2098–2103. [Google Scholar] [CrossRef]
- Auvin-Guette, C.; Baraguey, C.; Blond, A.; Xavier, H.S.; Pousset, J.L.; Bodo, B. Pohlianins A, B and C, cyclic peptides from the latex of Jatropha pohliana ssp. molissima. Tetrahedron 1999, 55, 11495–11510. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).