The Cell Adhesion Activity of the Joining Peptide of Proopiomelanocortin
Abstract
1. Introduction
2. Results and Discussion
2.1. Characterization of the JP Peptide of POMC
2.2. Preparation of the POMC
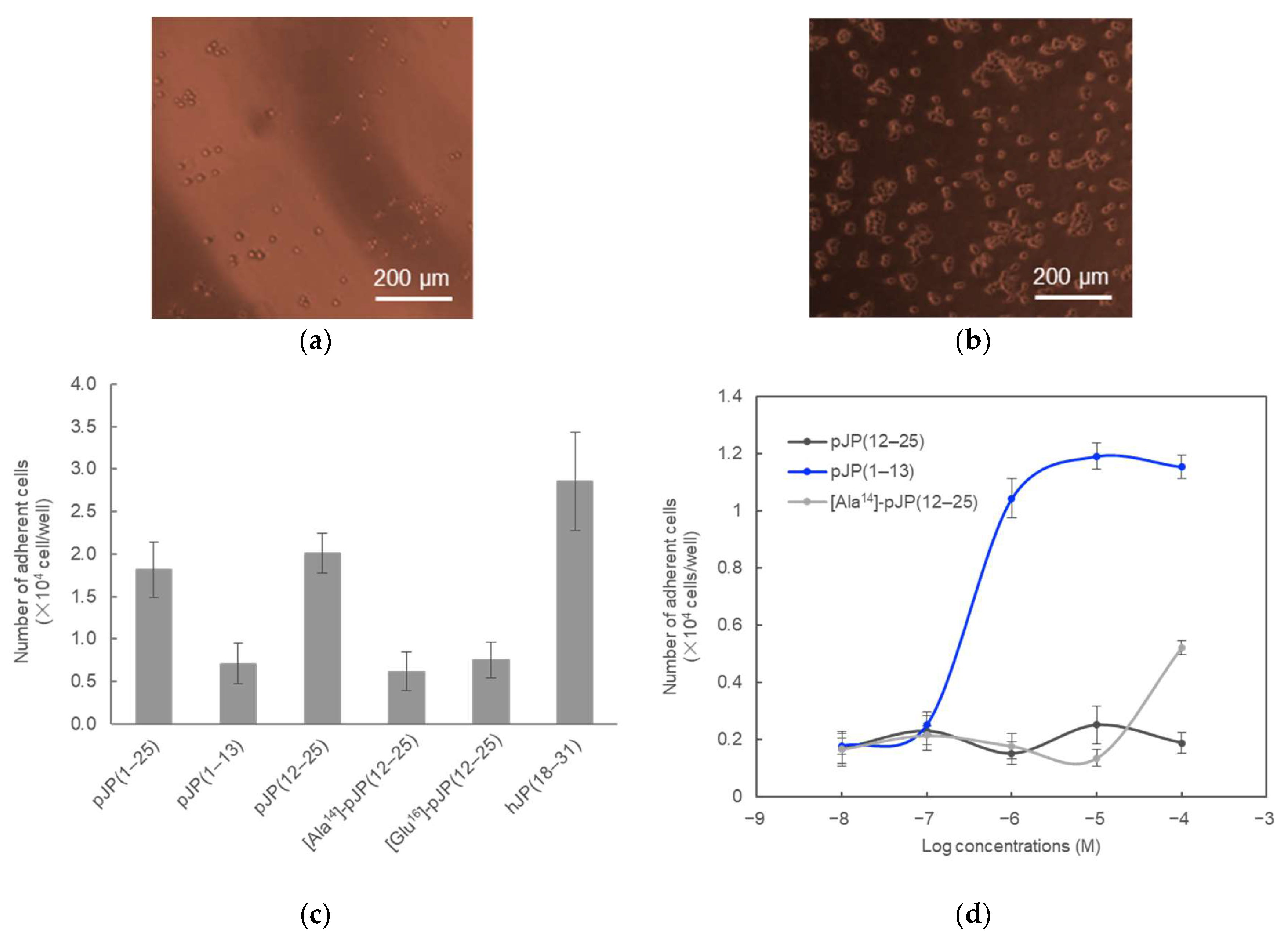
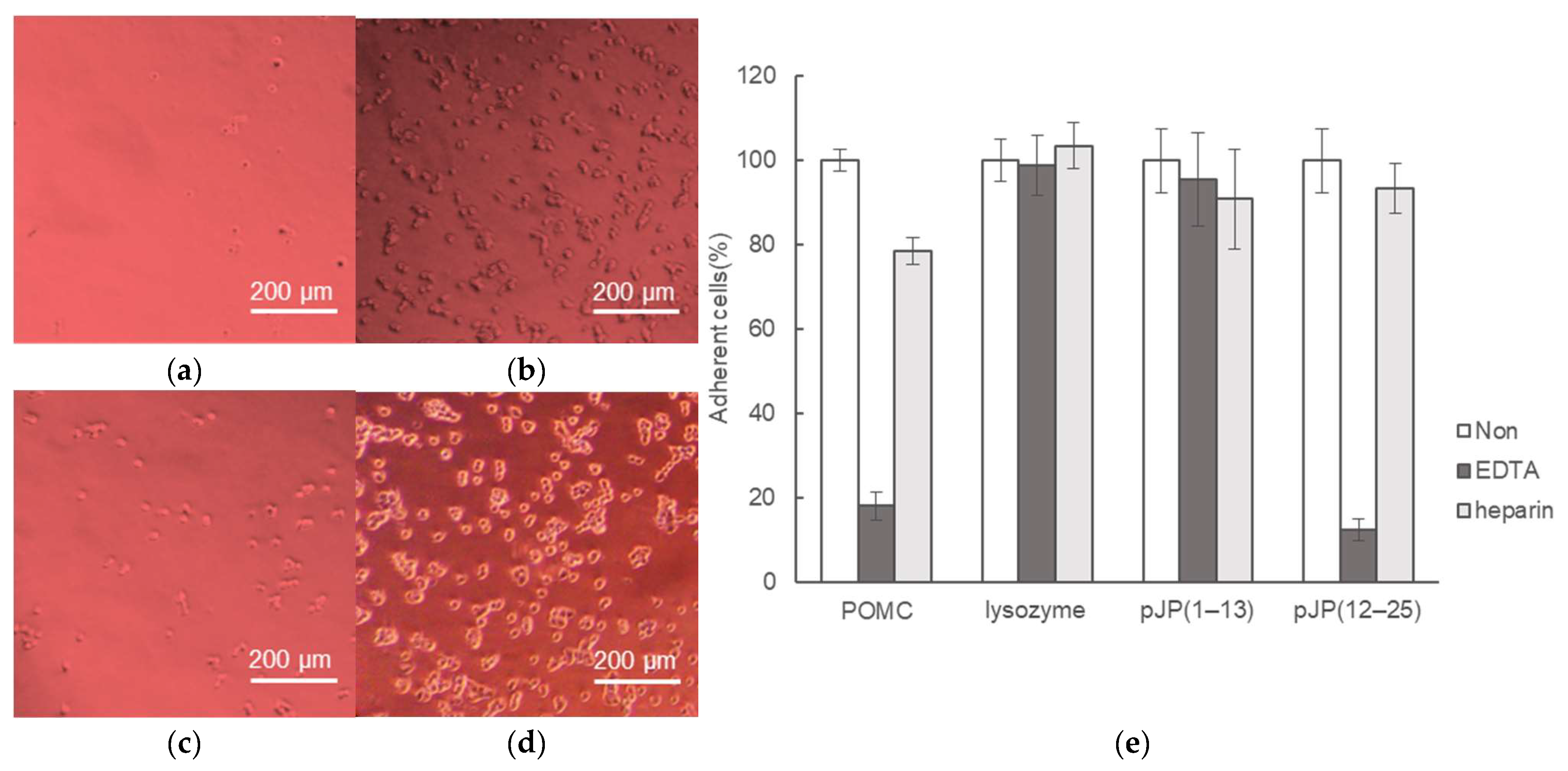
2.3. Cell-Binding Activity of POMCs
2.4. Conformational Analyses of the JP Peptides and the POMC Protein by CD Spectroscopy
3. Materials and Methods
3.1. Materials
3.2. Construction of Expression Vectors of Recombinant POMC in E. coli Cells
3.3. Protein Expression and Purification of Recombinant POMC
3.4. Peptide Synthesis
3.5. Reversed-Phase High-Performance Liquid Chromatography (RP-HPLC)
3.6. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF/MS)
3.7. CD Measurements
3.8. Cell Adhesion Assay
3.9. Competitive Binding Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Cawley, N.X.; Li, Z.; Loh, Y.P. 60 YEARS OF POMC: Biosynthesis, trafficking, and secretion of pro-opiomelanocortin-derived peptides. J. Mol. Endocrinol. 2016, 56, T77–T97. [Google Scholar] [CrossRef]
- Cool, D.R.; Loh, Y.P. Identification of a sorting signal for the regulated secretory pathway at the N-terminus of pro-opiomelanocortin. Biochimie 1994, 76, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Benjannet, S.; Rondeau, N.; Day, R.; Chretien, M.; Seidah, N.G. PC1 and PC2 are proprotein convertases capable of cleaving proopiomelanocortin at distinct pairs of basic residues. Proc. Natl. Acad. Sci. USA 1991, 88, 3564–3568. [Google Scholar] [CrossRef]
- Clark, A.J.; Lowry, P. 60 YEARS OF POMC: POMC: The consummate peptide hormone precursor. J. Mol. Endocrinol. 2016, 56, E1–E2. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bertagna, X.; Camus, F.; Lenne, F.; Girard, F.; Luton, J.P. Human joining peptide: A proopiomelanocortin product secreted as a homodimer. Mol. Endocrinol. 1988, 2, 1108–1114. [Google Scholar] [CrossRef] [PubMed]
- Toney, K.; Bateman, A.; Gagnon, C.; Bennett, H.P. Aspartimide formation in the joining peptide sequence of porcine and mouse pro-opiomelanocortin. J. Biol. Chem. 1993, 268, 1024–1031. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Hoess, R.H.; Bennett, J.S.; DeGrado, W.F. Use of phage display to probe the evolution of binding specificity and affinity in integrins. Protein Eng. 2003, 16, 65–72. [Google Scholar] [CrossRef]
- Li, P.; Lu, Z.; Bao, H.; Li, D.; King, D.P.; Sun, P.; Bai, X.; Cao, W.; Gubbins, S.; Chen, Y.; et al. In-vitro and in-vivo phenotype of type Asia 1 foot-and-mouth disease viruses utilizing two non-RGD receptor recognition sites. BMC Microbiol. 2011, 11, 154. [Google Scholar] [CrossRef]
- Huettner, N.; Dargaville, T.R.; Forget, A. Discovering Cell-Adhesion Peptides in Tissue Engineering: Beyond RGD. Trends Biotechnol. 2018, 36, 372–383. [Google Scholar] [CrossRef]
- Barczyk, M.; Carracedo, S.; Gullberg, D. Integrins. Cell Tissue Res. 2010, 339, 269–280. [Google Scholar] [CrossRef]
- Schaffner, P.; Dard, M.M. Structure and function of RGD peptides involved in bone biology. Cell Mol. Life Sci. 2003, 60, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Boateng, S.Y.; Lateef, S.S.; Mosley, W.; Hartman, T.J.; Hanley, L.; Russell, B. RGD and YIGSR synthetic peptides facilitate cellular adhesion identical to that of laminin and fibronectin but alter the physiology of neonatal cardiac myocytes. Am. J. Physiol. Cell Physiol. 2005, 288, C30–C38. [Google Scholar] [CrossRef] [PubMed]
- Gullberg, E.; Keita, A.V.; Salim, S.Y.; Andersson, M.; Caldwell, K.D.; Soderholm, J.D.; Artursson, P. Identification of cell adhesion molecules in the human follicle-associated epithelium that improve nanoparticle uptake into the Peyer’s patches. J. Pharmacol. Exp. Ther. 2006, 319, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Davies, J.; Lu, D.; Xia, M.; Wattam, B.; Shang, D.; Sun, Y.; Scully, M.; Kakkar, V. The effect of the single substitution of arginine within the RGD tripeptide motif of a modified neurotoxin dendroaspin on its activity of platelet aggregation and cell adhesion. Cell Commun. Adhes. 2006, 13, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Bennett, H.P. Biosynthetic fate of the amino-terminal fragment of pro-opiomelanocortin within the intermediate lobe of the mouse pituitary. Peptides 1986, 7, 615–622. [Google Scholar] [CrossRef]
- Bennett, H.P. Isolation and characterization of the 1 to 49 amino-terminal sequence of pro-opiomelanocortin from bovine posterior pituitaries. Biochem. Biophys. Res. Commun. 1984, 125, 229–236. [Google Scholar] [CrossRef]
- Cool, D.R.; Fenger, M.; Snell, C.R.; Loh, Y.P. Identification of the sorting signal motif within pro-opiomelanocortin for the regulated secretory pathway. J. Biol. Chem. 1995, 270, 8723–8729. [Google Scholar] [CrossRef]
- Loh, Y.P.; Tam, W.W. Association of newly synthesized pro-opiomelanocortin with secretory granule membranes in pituitary pars intermedia cells. FEBS Lett. 1985, 184, 40–43. [Google Scholar] [CrossRef][Green Version]
- Matsunuma, M.; Kan, R.; Yamada, Y.; Hamada, K.; Kanagawa, M.; Nomizu, M.; Kikkawa, Y. Chain-specificity of laminin alpha1-5 LG45 modules in the recognition of carbohydrate-linked receptors and intramolecular binding. Sci. Rep. 2023, 13, 10430. [Google Scholar] [CrossRef]
- Gottschalk, K.E.; Kessler, H. The structures of integrins and integrin-ligand complexes: Implications for drug design and signal transduction. Angew. Chem. Int. Ed. Engl. 2002, 41, 3767–3774. [Google Scholar] [CrossRef]
- Leahy, D.J.; Aukhil, I.; Erickson, H.P. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell 1996, 84, 155–164. [Google Scholar] [CrossRef]
- Yamada, T.; Matsushima, M.; Inaka, K.; Ohkubo, T.; Uyeda, A.; Maeda, T.; Titani, K.; Sekiguchi, K.; Kikuchi, M. Structural and functional analyses of the Arg-Gly-Asp sequence introduced into human lysozyme. J. Biol. Chem. 1993, 268, 10588–10592. [Google Scholar] [CrossRef] [PubMed]
- Okumura, M.; Shimamoto, S.; Hidaka, Y. A chemical method for investigating disulfide-coupled peptide and protein folding. FEBS J. 2012, 279, 2283–2295. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T. Hydroxyapatite as a Liquid-Chromatographic Packing. J. Chromatogr. 1991, 544, 147–184. [Google Scholar] [CrossRef]
- Goto, M.; Yoshino, S.; Hiroshima, K.; Kawakami, T.; Murota, K.; Shimamoto, S.; Hidaka, Y. The Molecular Basis of Heat-Stable Enterotoxin for Vaccine Development and Cancer Cell Detection. Molecules 2023, 28, 1128. [Google Scholar] [CrossRef] [PubMed]
- Merrifield, R.B. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar] [CrossRef]
- Sakata, N.; Murakami, Y.; Miyazawa, M.; Shimamoto, S.; Hidaka, Y. A Novel Peptide Reagent for Investigating Disulfide-Coupled Folding Intermediates of Mid-Size Proteins. Molecules 2023, 28, 3494. [Google Scholar] [CrossRef] [PubMed]
- Sakata, N.; Ogata, A.; Takegawa, M.; Murakami, Y.; Nishimura, M.; Miyazawa, M.; Hagiwara, T.; Shimamoto, S.; Hidaka, Y. Degradation-Suppressed Cocoonase for Investigating the Propeptide-Mediated Activation Mechanism. Molecules 2022, 27, 8063. [Google Scholar] [CrossRef]
- Nomizu, M.; Kim, W.H.; Yamamura, K.; Utani, A.; Song, S.Y.; Otaka, A.; Roller, P.P.; Kleinman, H.K.; Yamada, Y. Identification of cell binding sites in the laminin alpha 1 chain carboxyl-terminal globular domain by systematic screening of synthetic peptides. J. Biol. Chem. 1995, 270, 20583–20590. [Google Scholar] [CrossRef]




| Peptide Names | Amino Acid Sequences | [M + H]+calc. | [M + H]+obs. |
|---|---|---|---|
| pJP | 1EEEEVAAGEGPGPRGDGVAPGPRQD25 * | 2475.1 | 2476.5 |
| pJP(1–13) | 1EEEEVAAGEGPGP13 | 1291.6 ** | 1291.3 |
| pJP(12–25) | 12GPRGDGVAPGPRQD25 | 1377.7 | 1377.1 |
| [Ala14]-pJP(12–25) | 12GPAGDGVAPGPRQD25 | 1292.6 | 1295.4 |
| [Glu16]-pJP(12–25) | 12GPRGEGVAPGPRQD25 | 1391.7 | 1392.0 |
| hJP(18–31) | 18EPRSDGAKPGPREG31 | 1451.7 | 1451.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hiroshima, K.; Sakata, N.; Konogami, T.; Shimamoto, S.; Hidaka, Y. The Cell Adhesion Activity of the Joining Peptide of Proopiomelanocortin. Molecules 2023, 28, 7754. https://doi.org/10.3390/molecules28237754
Hiroshima K, Sakata N, Konogami T, Shimamoto S, Hidaka Y. The Cell Adhesion Activity of the Joining Peptide of Proopiomelanocortin. Molecules. 2023; 28(23):7754. https://doi.org/10.3390/molecules28237754
Chicago/Turabian StyleHiroshima, Kyona, Nana Sakata, Tadafumi Konogami, Shigeru Shimamoto, and Yuji Hidaka. 2023. "The Cell Adhesion Activity of the Joining Peptide of Proopiomelanocortin" Molecules 28, no. 23: 7754. https://doi.org/10.3390/molecules28237754
APA StyleHiroshima, K., Sakata, N., Konogami, T., Shimamoto, S., & Hidaka, Y. (2023). The Cell Adhesion Activity of the Joining Peptide of Proopiomelanocortin. Molecules, 28(23), 7754. https://doi.org/10.3390/molecules28237754







