Abstract
Euphorbia dentata (Euphorbiaceae), an invasive weed, is rarely eaten by herbivorous insects and could secrete a large amount of white latex, causing a serious threat to local natural vegetation, agricultural production and human health. In order to prevent this plant from causing more negative effects on humans, it is necessary to understand and utilize the chemical relationships between the latex of E. dentata and herbivorous insects. In this study, three new norsesquiterpenes (1–3), together with seven known analogues (4–10), were isolated and identified from the latex of E. dentata. All norsesquiterpenes (1–10) showed antifeedant and growth-inhibitory effects on H. armigera with varying levels, especially compounds 1 and 2. In addition, the action mechanisms of active compounds (1–3) were revealed by detoxifying enzyme (AchE, CarE, GST and MFO) activities and corresponding molecular docking analyses. Our findings provide a new idea for the development and utilization of the latex of E. dentata, as well as a potential application of norsesquiterpenes in botanical insecticides.
1. Introduction
Plants and herbivorous insects are important components of terrestrial ecosystems. Herbivorous insects need to adapt and depend on the host plants for survival, and these host plants affect the feeding of herbivorous insects through their chemicals. The two form many complex adaptive mechanisms in the process of long-term co-evolution [1]. Plant latex, usually stored in laticifers, is an opaque viscous liquid that can be secreted immediately when plants are subjected to herbivorous feeding insects or mechanical damage [2,3]. Plant latex contains a large number of secondary metabolites with diverse types and complex structures. Most of the metabolites have certain antifeedant and growth-inhibitory effects on herbivorous insects, acting as a chemical defense in resisting the feeding of herbivorous insects [4,5]. However, the secondary metabolites of most plant latexes against herbivorous insects have not been systematically studied, and their defense mechanisms against herbivorous insects are less researched. More research findings are needed to support it.
Euphorbia is the largest genus in the Euphorbiaceae family, with more than 2000 species worldwide. It is mainly distributed in tropical and subtropical regions of Africa and America [6]. In addition, Euphorbia is the largest genus of plants with latex. Some of the latexes of this genus have been used to kill fish or large animals [7]. The main characteristic of Euphorbia is that their latexes are rich in secondary metabolites, especially terpenoids with complex structural frameworks, including monoterpenes, sesquiterpenes, diterpenes, and triterpenes [6]. Among them, sesquiterpenes are naturally defensive compounds in plants, which have significant antifeedant and growth-inhibitory effects on herbivorous insects [8].
Euphorbia dentata Michx. is an annual plant of the genus Euphorbia in the family Euphorbia. It originated in North America and has spread to many countries in Europe, Australia, and Asia [9]. In recent years, the plant has grown rapidly in China and has a strong adaptability to new habitats. Some single-dominant communities have formed in Changsha of Hunan Province, Shijiazhuang and Baoding of Hebei Province, and Guanling of Guizhou Province, causing great harm to local natural vegetation, agricultural production, and human health. Field observations found that the whole plant of E. dentata contained white latex, which will flow out of the plant, especially when the plant is damaged by herbivorous insects. Therefore, we speculated that the strong invasive ability of the plant may be closely related to the successful chemical defense mechanisms of its latex against herbivorous insects. However, studies on secondary metabolites of E. dentata have not been conducted. Since E. dentata belongs to the genus Euphorbia, we hypothesized that E. dentata is also rich in sesquiterpenes.
Helicoverpa armigera (Lepidoptera: Noctuidae) is a herbivorous insect widely distributed in China and other areas of the world. The species is a euryphagous insect, and its host plants have more than thirty families [10]. In recent years, with the adjustment of agricultural planting structures, the occurrence and harm of H. armigera have become increasingly serious, causing great economic losses to local agriculture [11]. To effectively control H. armigera, extensive research has been carried out in chemical pesticides. Although some progress has been made, it has not stopped the large-scale trend of worldwide harm that H. armigera is causing. In addition, due to the long-term dependence on chemical pesticides, H. armigera has developed a strong resistance, and the ecological environment damage caused by chemical pesticides has become increasingly serious [12]. Therefore, it is vital to search for safe and effective defensive substances from chemical interactions between plant and herbivorous insects.
In the process of adapting to external stress, insects could produce a class of detoxifying enzymes that could metabolize a large number of foreign substances. Acetylcholinesterase (AchE), carboxylesterase (CarE), glutathione-S-transferase (GST), and mixed-function-oxidase (MFO) are important detoxifying enzymes in insects, playing an important role in metabolizing toxic compounds and maintaining normal physiological activities [13,14]. In addition, molecular docking analysis has become an important technique for exploring the interactions between molecules, providing visual analyses to explore the binding sites and modes of action of enzymes and compounds [15]. Therefore, it is necessary to study the defense mechanisms of compounds by detoxifying enzyme activities and corresponding molecular docking analyses.
Field investigations found that H. armigera could feed on a small amount of E. dentata. At the same time, E. dentata secretes white latex from its damaged area. In addition, the latex of E. dentata and the methanol extract of latex have significant antifeedant effects on H. armigera (Figures S31 and S32), which also proved that chemical defense is the main form of defense against H. armigera. These phenomena suggest that the latex of E. dentata has a chemical defense mechanism against H. armigera. However, the secondary metabolites from the latex of E. dentata and their defense mechanisms against H. armigera are still unknown. In this study, we isolated and identified norsesquiterpenes (Figure 1) from the latex of E. dentata. Additionally, the antifeedant and growth-inhibitory effects of norsesquiterpenes on H. armigera were also investigated. In addition, the action mechanisms of active norsesquiterpenes were revealed by detoxifying enzyme activities and corresponding molecular docking analyses.
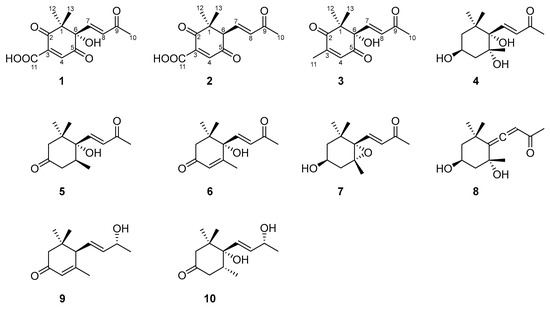
Figure 1.
Structures of compounds 1–10.
2. Results
2.1. Structural Elucidation
Norsescycldione A (1) was obtained as a yellowish oil with C13H14O6 (seven degrees of unsaturation) using its HR-ESIMS data (m/z 289.0628 [M + Na]+, calculated for C13H14O6Na, 289.0688). Its 1D-NMR spectra (Table 1) showed four carbonyl groups [δC 210.5 (C-2), 200.4 (C-9), 192.3 (C-5), 171.3 (C-11)], two trans-double bond groups [δH 7.82 (1H, s, H-4), 7.24 (1H, d, J = 15.8 Hz, H-7), 6.37 (1H, d, J = 15.8 Hz, H-8); δC 160.8 (C-4), 147.6 (C-7), 142.1 (C-3), 132.5 (C-8)], one tertiary alcohol group [δC 97.2 (C-6)], and three methyl groups [δH 2.34 (3H, s, H-10), 1.17 (3H, s, H-12), 0.96 (3H, s, H-13); δC 27.2 (C-10), 25.3 (C-12), 23.1 (C-13)]. In the HMBC spectrum (Figure 2), the key correlations between H-4/C-2, C-5, C-11; H-7/C-1, C-5, C-9; H-10/C-8, C-9; and H-12/C-2, C-13 established its planar structure. Compared with the literature, it was found that compound 1 possessed an ionone-type norsesquiterpene skeleton [16]. In addition, the computed ECD curve of (6S)-1a matched well with the experimental result of 1 (Figure 3), establishing its stereochemical structure. Thus, the structure of 1 was confirmed and named Norsescycldione A (Figure 1).

Table 1.
1H (600 MHz) and 13C NMR (150 MHz) data of compounds 1–3 in methanol-d4.
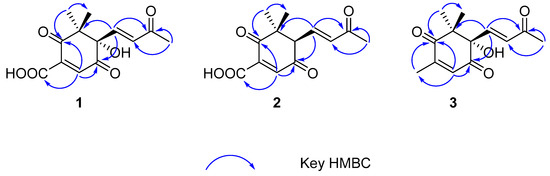
Figure 2.
Key HMBC correlations of compounds 1–3.
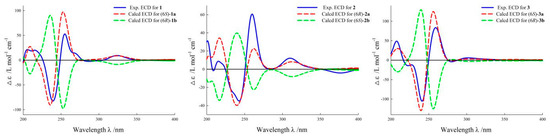
Figure 3.
ECD curves of compound 1–3.
Norsescycldione B (2) was obtained as a yellowish oil with C13H14O5 (seven degrees of unsaturation) using its HR-ESIMS data (m/z 273.0736 [M + Na]+, calculated for C13H14O5Na, 273.0739). Its 1D-NMR spectra (Table 1) showed four carbonyl groups [δC 212.3 (C-2), 204.5 (C-9), 194.0 (C-5), 172.6 (C-11)], two trans-double bond groups [δH 7.75 (1H, s, H-4), 7.28 (1H, dd, J = 15.3 Hz, H-7), 6.28 (1H, d, J = 15.3 Hz, H-8); δC 161.4 (C-4), 144.2 (C-7), 140.5 (C-3), 136.0 (C-8)], and three methyl groups [δH 2.37 (3H, s, H-10), 1.19 (3H, s, H-12), 0.98 (3H, s, H-13); δC 28.2 (C-12), 27.3 (C-10), 26.1 (C-13)]. A comparison of its 1D NMR data with Norsescycldione A indicated that they were very similar except for one less hydroxyl group, which showed that compound 2 also possessed a norsesquiterpene skeleton similar to Norsescycldione A. In the HMBC spectrum (Figure 2), the key correlations of H-4/C-2, C-5, C-11; H-7/C-1, C-5, C-9; H-10/C-8, C-9; H-12/C-2, C-13 established its planar structure, which also proved that the missing hydroxyl group was at the position of C-6. In addition, the computed ECD curve of (6R)-2a matched well with the experimental result of 2 (Figure 3), establishing its stereochemical structure. Thus, the structure of 2 was confirmed and named Norsescycldione B (Figure 1).
Norsescycldione C (3) was obtained as a yellowish oil with C13H16O4 (six degrees of unsaturation) using its HR-ESIMS data (m/z 259.0949 [M + Na]+, calculated for C13H16O4Na, 259.0946). Its 1D-NMR spectra (Table 1) showed three carbonyl groups [δC 207.1 (C-2), 202.6 (C-9), 195.6 (C-5)], two trans-double bond groups [δH 7.22 (1H, d, J = 16.1 Hz, H-7), 6.77 (1H, s, H-4), 6.41 (1H, d, J = 16.1 Hz, H-8); δC 146.2 (C-7), 144.2 (C-3), 136.0 (C-4), 134.1 (C-8)], one tertiary alcohol group [δC 99.3 (C-6)], and four methyl groups [δH 2.42 (3H, s, H-10), 2.26 (3H, s, H-11), 1.18 (3H, s, H-12), 0.95 (3H, s, H-13); δC 27.5 (C-10), 25.7 (C-12), 23.9 (C-13), 14.7 (C-11)]. A comparison of its 1D NMR data with Norsescycldione A indicated that they were very similar except for one less carboxyl group and one more methyl group, showing that compound 2 also possessed a norsesquiterpene skeleton similar to Norsescycldione A. In the HMBC spectrum (Figure 2), the key correlations of H-4/C-2, C-5, C-11; H-7/C-1, C-5, C-9; H-10/C-8, C-9; H-12/C-2, C-13; H-11/C-2 established its planar structure, which also proved that the missing carboxyl group was at the position of C-3 and the extra methyl group was attached to the position of C-3. In addition, the computed ECD curve of (6S)-3a matched well with the experimental result of 3 (Figure 3), establishing its stereochemical structure. Thus, the structure of 3 was confirmed and named Norsescycldione C (Figure 1).
The other compounds 4–10 were known and identified as (3S, 5R, 6S, 7E)-3,5,6-Trihydroxy-7-megastigmen-9-one (4) [17], Elaeocarpunone (5) [18], Dehydrovomifoliol (6) [19], 3-Hydroxy-5α, 6α-epoxy-β-ionone (7) [20], Grasshopper ketone (8) [21], (6R, 7E, 9R)-9-Hydroxy-4,7-megastigmadien-3-one (9) [17] and 4,5-Dihydroblumenol A (10) [22], respectively, based on the NMR data and literature comparison.
2.2. Antifeedant and Growth-Inhibitory Effects
It is well known that sesquiterpenes are naturally defensive compounds in plants and have significant antifeedant and growth-inhibitory effects on herbivorous insects [8]. From this, we speculated that the norsesquiterpenes (1–10) may be the potential defense substances for the latex of E. dentata against herbivorous insects. Field investigations found that H. armigera (a model herbivorous insect for evaluating chemical defense function) [23,24] could feed on a small amount of E. dentata. At the same time, E. dentata secretes white latex from its damaged area. These phenomena suggest that the latex of E. dentata has a defense mechanism against H. armigera. However, the secondary metabolites from the latex of E. dentata and their defense mechanisms against H. armigera are still unknown. Therefore, we studied their chemical defense functions (antifeedant and growth). As shown in Table 2, norsesquiterpenes (1–10) had antifeedant and growth-inhibitory effects with varying levels (100, 50, 25 μg/mL). In general, compounds 1–3 exhibited a stronger chemical defense function (antifeedant and growth-inhibitory effects) compared with other compounds (4–10). Among these, compounds 1 and 2 exhibited significant antifeedant effects at 100 μg/mL (85.16 ± 7.44% and 80.62 ± 6.55%, respectively), which was even comparable to neem oil (92.28 ± 7.11%). Moreover, compounds 1 and 2 also showed potent growth-inhibitory effects at 100 μg/mL (74.28 ± 8.35% and 78.11 ± 6.26%, respectively), and interestingly, they did not cause the death of H. armigera compared with the positive control.

Table 2.
Antifeedant and growth-inhibitory effects of compounds 1–10 on H. armigera.
2.3. Detoxifying Enzymes Effects
In the process of adapting to external stress, insects could produce a class of detoxifying enzymes that could metabolize a large number of foreign substances. AchE, CarE, GST, and MFO are important detoxifying enzymes in insects, playing an important role in metabolizing toxic compounds and maintaining normal physiological activities [13,14]. Therefore, the action mechanisms of active norsesquiterpenes (1–3) were revealed by detoxifying enzyme activities. The effects of compounds 1–3 on AchE, CarE, GST, and MFO activities of H. armigera at different times (0, 6, 12, 24, 48 h) are shown in Figure 4. In general, after treatment with different compounds (1–3), the AchE and GST activities of H. armigera decreased with the extension of time, MFO activities of H. armigera increased with the extension of time, and CarE activities of H. armigera increased first and then decreased with the extension of time. Among these, the changes in AchE, CarE, and MFO activities in each treatment group were not significantly different from those in the blank control. It is worth noting that GST activities significantly decreased by 35.54%, 18.41%, 41.25%, and 56.79% under compound 1 treatment; 13.99%, 35.60%, 54.16%, and 44.15% under compound 2 treatment; and 15.49%, 33.63%, 33.12%, and 31.42% under compound 3 treatment compared with the blank controls at 0–6, 6–12, 12–24 and 24–48 h, respectively.
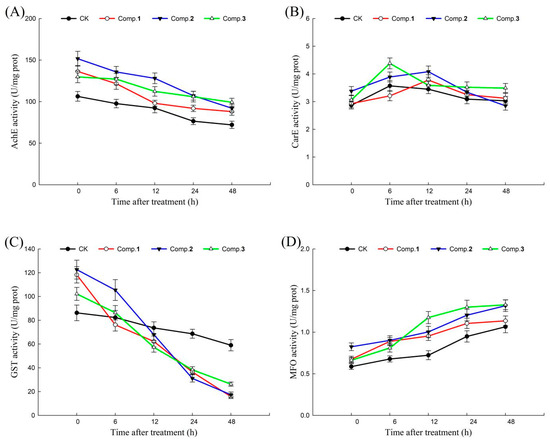
Figure 4.
The effects of compounds 1–3 (100 μg/mL) on the detoxifying enzymes of H. armigera: (A) the effects of compounds 1–3 on AchE; (B) the effects of compounds 1–3 on CarE; (C) the effects of compounds 1–3 on MFO; and (D) the effects of compounds 1–3 on GST. CK = Control check.
2.4. Molecular Docking Analyses
In recent years, molecular docking analysis has become an important technique for exploring interactions between molecules (such as enzymes and compounds) [15]. The binding energies of compounds 1 and 2 with GST of H. armigera (−75.8056 and −82.0594 kcal/mol) were lower than those of compound 3 (−60.6084 kcal/mol) (Table S2). As shown in Figure 5, carbonyl groups of compounds 1–3 interacted with neighboring amino acids via hydrogen bonds, indicating that the carbonyl groups of norsesquiterpenes could play an important role in GST activities. In addition, compound 1 formed four hydrogen bonds with amino acids ARG68 (distance 2.73 Å), GLU66 (distance 1.96 Å), SER67 (distance 2.07 Å), and VAL54 (distance 2.17 Å) in GST. Compound 2 formed five hydrogen bonds with amino acids ARG68 (distance 2.50 Å), GLU66 (distance 2.39 Å), PPO55 (distance 2.51 Å), SER67 (distance 2.08 Å), and TRP65 (distance 2.01 Å) in GST. Compound 3 formed three hydrogen bonds with amino acids ARG68 (distance 2.64 Å), GLU66 (distance 1.93 Å), and SER67 (distance 2.64 Å) in GST.
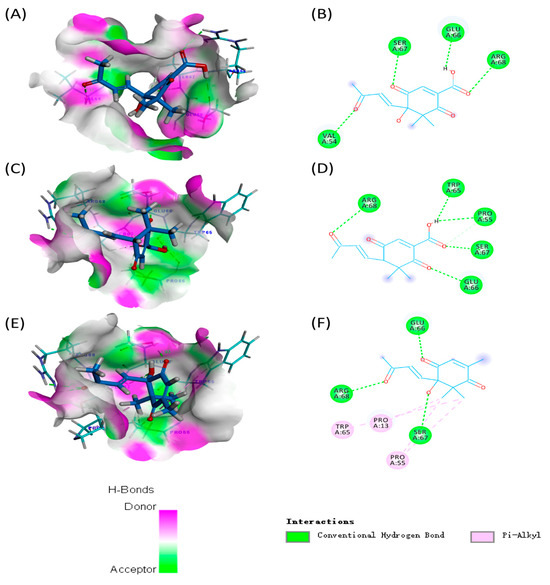
Figure 5.
Molecular docking analyses of compounds 1–3 with GST: (A) 3D diagram of the interactions between compound 1 and the active pocket of GST; (B) 2D diagram of the interactions between compound 1 and amino acids of GST; (C) 3D diagram of the interactions between compound 2 and the active pocket of GST; (D) 2D diagram of the interactions between compound 2 and amino acids of GST; (E) 3D diagram of the interactions between compound 3 and the active pocket of GST; and (F) 2D diagram of the interactions between compound 3 and amino acids of GST.
3. Discussion
Recently, researchers have paid more attention to the chemical relationships between invasive plants and native herbivorous insects [25,26]. This will help researchers to better reveal the invasion mechanism of alien plants from the perspective of chemical defense, and provides a new idea for the development and utilization of invasive plants. In our study, three new norsesquiterpenes (1–3), together with seven known analogues (4–10), were isolated from the latex of E. dentata and their structures were determined via HR-ESIMS verifications, NMR analyses, and ECD calculations. Among these, compounds 4, 8, and 9 were obtained from the Euphorbia genus for the first time. Moreover, this was also the first systematic study of the chemical composition of this plant. All norsesquiterpenes (1–10) showed chemical defense functions in H. armigera with varying levels (100, 50, 25 μg/mL), especially compounds 1 and 2. These results indicate that the norsesquiterpenes (1–3) from the latex of E. dentata could function as chemical defense substances against H. armigera, which may help E. dentata to gain a competitive advantage over other plants, as well as supporting theory for the defensive functions of sesquiterpenes against herbivorous insects [8]. By analyzing the structure–activity relationships of norsesquiterpenes (1–10), the presence of one carboxyl group at the position of C-3 may have a positive effect on chemical defense function. However, more studies of norsesquiterpene analogues are needed to confirm this.
In addition, in the detoxifying enzyme activities, the GST activities significantly decreased under treatment with compounds 1–3 compared with a blank control with an extension of time. This indicated that GST may be a key target of H. armigera for compounds 1–3 to exert their chemical defense functions. GST is an important detoxifying enzyme in insects, which could catalyze the binding of harmful substances and reduce glutathione, thereby increasing the water solubility of harmful substances and excreting them [13]. Norsesquiterpenes (1–3) can significantly inhibit the GST activity of H. armigera, which indicates that compounds 1–3 may block the excretion of harmful substances, resulting in the inhibition of the feeding and growth of H. armigera. On this basis, we explored the binding sites and modes of action of GST, as well as compounds 1–3, using molecular docking analyses. The binding energies of compounds 1 and 2 with GST of H. armigera were lower than those of compound 3, indicating that compounds 1 and 2 bound to GST more stably than compound 3. This was essentially consistent with the above results (antifeedant and growth-inhibitory effects of compounds 1–3). It is well-known that hydrogen bonds play a more important role than other forces in interactions between molecules [27]. Compounds 1 and 2 formed more hydrogen bonds with amino acids than compound 3, which not only verified that more hydrogen bonds led to a more stable binding between enzyme and compound, but also indicated that the common amino acids (ARG68, GLU66, and SER67) could be key active sites of GST interacting with compounds 1–3.
4. Materials and Methods
4.1. General
Column chromatography was carried out using silica gel (Qingdao Marine, Qingdao, China) and Sephadex LH-20 (GE Healthcare, Chicago, IL, USA). Preparative HPLC was carried out using a 1220 system (Agilent, Santa Clara, CA, USA) equipped with a 5C18-MS-II column (COSMOSIL, Tokyo, Japan). UV spectra were carried out using a 241 spectrophotometer (Perkin Elmer, Waltham, MA, USA). HR-ESIMS spectra were carried out using a 6545 Q-TOF instrument (Agilent, USA). NMR spectra were carried out using an AV-600 instrument (Bruker, Saarbrucken, Germany). ECD spectra were carried out using a MOS-450 instrument (Bio-Logic, Seyssinet Pariset, France).
4.2. Plant Material
E. dentata was identified by Professor Bo Qu and its voucher specimen (NO. ZW-2020-0073) and was kept in Shenyang Agricultural University. The latex of E. dentata (aerial part) was collected from Beijing Botanical Garden, China (40°01′ E, 116°21′ N) in August 2020.
4.3. Insect Material
H. armigera was purchased from Henan Keyun Bio-Pesticide Co., Ltd. (Henan, China) and identified by Associate Professor Lu Jiang (Shenyang Agricultural University).
4.4. Extraction and Isolation
The latex of E. dentata (2 L) was suspended with 70% methanol (2 L) under an ultrasonic bath for 20 min and then centrifuged at 12,000 rpm for 10 min. The supernatant was merged and concentrated in vacuo. The concentrated extract (11 g) was subjected to a silica gel column (normal-phase, dichloromethane/methanol, 98:2–80:20, v/v) to give seven subfractions (Fr. A–Fr. F). Fr. B (110 mg) was subjected to a preparative HPLC (210 nm, methanol/H2O, 25:75, v/v, 5.0 mL/min) to give compounds 6 (9.1 mg, tR = 38.2 min) and 9 (13.5 mg, tR = 30.1 min), respectively. Fr. C (460 mg) was subjected to a Sephadex LH-20 column (methanol/H2O, 80:20, v/v) to give five subfractions (Fr. C-1–Fr. C-5). Fr. C-2 (61 mg) was subjected to a preparative HPLC (210 nm, methanol/H2O, 10:90, v/v, 5.0 mL/min) to give compounds 4 (13.4 mg, tR = 53.7 min). Fr. C-3 (125 mg) was subjected to a preparative HPLC (210 nm, acetonitrile/H2O, 10:90, v/v, 5.0 mL/min) to give compounds 5 (2.5 mg, tR = 77.3 min), 7 (9.5 mg, tR = 58.1 min), and 10 (6.2 mg, tR = 67.4 min), respectively. Fr. D (430 mg) was subjected to a Sephadex LH-20 column (methanol/H2O, 90:10, v/v) to give four subfractions (Fr. D-1–Fr. D-4). Fr. D-2 (83 mg) was subjected to a preparative HPLC (210 nm, acetonitrile/H2O, 10:90, v/v, 5.0 mL/min) to give compounds 2 (5.8 mg, tR = 37.5 min) and 8 (1.4 mg, tR = 48.0 min), respectively. Fr. E (246 mg) was subjected to a preparative HPLC (210 nm, methanol/H2O, 10:90, v/v, 5.0 mL/min) to give compounds 1 (7.5 mg, tR = 33.6 min) and 3 (8.4 mg, tR = 44.1 min), respectively.
4.5. Spectroscopic Data
Norsescycldione A (1): yellowish oil; ECD (methanol) λmax (Δε) 205 (+21.40), 215 (+20.10), 239 (−82.47), 254 (+52.85), 324 (+9.42) nm; HR-ESIMS at m/z 289.0628 [M + Na]+ (calculated for C13H14O6Na, 289.0688); 1H and 13C NMR data; see Table 1.
Norsescycldione B (2): yellowish oil; ECD (methanol) λmax (Δε) 201 (+31.26), 215 (+8.76), 242 (−34.99), 259 (+60.66), 310 (+12.12) nm; HR-ESIMS at m/z 273.0736 [M + Na]+ (calculated for C13H14O5Na, 273.0739); 1H and 13C NMR data; see Table 1.
Norsescycldione C (3): yellowish oil; ECD (methanol) λmax (Δε) 207 (+48.86), 241 (−105.20), 259 (+83.52), 302 (+5.96) nm; HR-ESIMS at m/z 259.0949 [M + Na]+ (calculated for C13H16O4Na, 259.0946); 1H and 13C NMR data; see Table 1.
(3S, 5R, 6S, 7E)-3,5,6-Trihydroxy-7-megastigmen-9-one (4): yellowish oil; 1H and 13C NMR data, see Figures S16 and S17.
Elaeocarpunone (5): yellowish oil; 1H and 13C NMR data; see Figures S18 and S19.
Dehydrovomifoliol (6): yellowish oil; 1H and 13C NMR data; see Figures S20 and S21.
Grasshopper ketone (8): yellowish oil; 1H and 13C NMR data, see Figures S24 and S25.
(6R, 7E, 9R)-9-Hydroxy-4,7-megastigmadien-3-one (9): yellowish oil; 1H and 13C NMR data; see Figures S26 and S27.
4,5-Dihydroblumenol A (10): yellowish oil; 1H and 13C NMR data; see Figures S28 and S29.
4.6. ECD Calculations
The ECD calculations were carried out by a previously reported method [25]. The conformational analyses of compounds 1–3 were performed using Spartan 14.0 software under the MMFF94 force field. The obtained conformations were optimized using Gaussian 09 software at the B3LYP/6-31G (d) level. Theoretical calculations were performed using TDDFT at the B3LYP/6-311+G (2d, p) level in methanol. The final ECD spectra were generated using SpecDis 1.60 software based on the Boltzmann weighting of each conformation.
4.7. Antifeedant and Growth-Inhibitory Assay
The antifeedant assay was carried out according to a previously reported method [26]. The leaf discs (1 cm in diameter from the leaves of Brassica chinensis) of the treatment group were painted with latex (50 µL) of E. dentata, methanol extract of latex (50 µL, 0.5 mg/mL), or methanol solution (20 µL) containing different concentrations of compounds (100, 50, and 25 μg/mL). The blank control group was painted with the same amount of methanol. The positive control group was painted with the same amount of commercial insecticide (neem oil). After natural drying, four discs (two treated and two blank controls) were placed in a Petri dish at a crosswise position. Two third-instar H. armigera (starved for 6 h) were placed in each Petri dish. When about 80% of the leaf discs (blank control) were consumed, the H. armigera was removed from the Petri dish, and the consumed area of the leaf discs was measured. Each assay was repeated at least five times. Antifeedant rate (%) was calculated as (AC − AT)/Ac × 100 (AC, the consumed area of leaf discs in the blank control group; AT, the consumed area of leaf discs in the treatment group).
The growth-inhibitory assay was carried out according to a previously reported method [28]. The leaf discs (1 cm in diameter from the leaves of B. chinensis) of the treatment group were painted with methanol solution (20 µL) containing 100 μg/mL compounds. The blank control group was painted with the same amount of methanol. The positive control group was painted with the same amount of commercial insecticide (neem oil). After natural drying, ten discs (ten treated or ten controls) were placed in a Petri dish. A weighed third-instar H. armigera (starved for 6 h) was placed in each Petri dish. The H. armigera were accurately weighed at 24, 48, and 72 h, respectively. Each assay was repeated at least five times. The growth-inhibitory rate (%) was calculated as (WC − WT)/Wc × 100 (WC, the gained weight of H. armigera in the blank control group; WT, the gained weight of H. armigera in the treatment group).
The leaf discs were pretreated as the growth-inhibitory assay above. After natural drying, ten discs (ten treated or ten controls) were placed in a Petri dish. A third-instar H. armigera (starved for 6 h) was placed in each Petri dish. The activities of detoxifying enzymes were measured at 0, 6, 12, 24, and 48 h, respectively. The extraction of enzymes was carried out according to a previously reported method [29]. Third-instar H. armigera with similar sizes were weighed, and nine volumes of normal saline were added. The obtained homogenate was then centrifuged at 12,000 rpm for 10 min, and the supernatant was used for the follow-up enzyme solution.
4.8. Detoxifying Enzymes Assay
The AchE assay was carried out according to Ellman’s method [30]. The enzyme solution (100 μL) was mixed with acetylthiocholine iodide (0.075 M, 100 μL). The mixture was incubated at 30 °C for 15 min and terminated with 5, 5′-dithiobis-(2-nitrobenzoic acid) (0.01 M, 50 μL). The absorbance was detected at 405 nm. The CarE assay was carried out according to Asperen’s method [31]. Enzyme solution (100 μL) was mixed with α-naphthyl acetate (0.03 M, 100 μL). The mixture was incubated at 30 °C for 15 min and terminated with diazo blue lauryl sulphate (0.01 M, 50 μL). The absorbance was detected at 600 nm. The GST assay was carried out according to Habig’s method [32]. The enzyme solution (100 μL) was mixed with 1-chloro-2, 4-dinitrobenzene (0.001 M, 100 μL) and glutathione (0.001 M, 100 μL). The absorbance was detected at 340 nm. The MFO assay was carried out according to Shang’s method [33]. The enzyme solution (100 μL) was mixed with paranitroanisole (0.05 M, 100 μL). The mixture was incubated at 30 °C for 15 min and terminated with HCl (0.01 M, 50 μL). The absorbance was detected at 400 nm. Each enzyme assay was repeated at least five times. One unit of enzyme activity was expressed as a change of 0.001 absorbance per milligram of protein per minute.
4.9. Molecular Docking Analyses
The molecular docking analyses were carried out according to a previously reported method [26]. The 3D structure of the enzyme (GST of H. armigera) was constructed via homology modeling using the EasyModeller 4.0 software. The GST sequence of H. armigera was obtained from the NCBI database in FASTA format (GenBank NO. BK40535.1). The optimal template (PDB code: 3VK9) was searched using the BLAST server based on E value, sequence identity, and query coverage (Table S1). The 3D structures of compounds 1–3 were optimized by the ChemDraw-3D 14.0 software to minimize energy. The Molegro Virtual Docker 4.0 software was carried out for further docking analyses. An active pocket (radius = 15.0) centered on x (39.90), y (33.15), and z (39.55) was formed based on the binding sites of the original ligand (glycerol) and enzyme (3VK9).
5. Conclusions
In conclusion, three new norsesquiterpenes (1–3), together with seven known analogues (4–10), were isolated from the latex of E. dentata using multiple column chromatography and preparative HPLC. Additionally, their structures were identified using HR-ESIMS verifications, NMR analyses, and ECD calculations. All norsesquiterpenes (1–10) showed chemical defense functions on H. armigera with varying levels (100, 50, 25 μg/mL), especially compounds 1 and 2 (antifeedant effects: 85.16 ± 7.44% and 80.62 ± 6.55% at 100 μg/mL, growth-inhibitory effects: 74.28 ± 8.35% and 78.11 ± 6.26% at 100 μg/mL, respectively). In addition, the action mechanisms of active compounds (1–3) were revealed via detoxifying enzymes (AchE, CarE, GST and MFO) activities and corresponding molecular docking analyses. GST activities significantly decreased under treatment with compounds 1–3 compared with the blank control with the extension of time. The common amino acids (ARG68, GLU66, and SER67) could be key active sites of GST interacting with compounds 1–3. These results will help researchers to better reveal the secret effects of latex from E. dentata against herbivorous insects from the perspective of chemical defense, providing new ideas for the development and utilization of the latex of E. dentata.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28237681/s1, Figures S1–S29: NMR spectrum of compounds 1–10. Figure S30: The superimposed 3D structures of GST target and template. Figure S31: Antifeedant effects of the latex of E. dentata on H. armigera. Figure S32: Antifeedant effects of the methanol extract of latex on H. armigera. Figure S33: The flowchart of extraction and isolation of the latex of E. dentata. Table S1: The optimal template for 3D structure modelling of GST. Table S2: Moldock scores of the compounds (1–3) with GST of H. armigera.
Author Contributions
Conceptualization, Z.Y. and Z.L.; methodology, T.A.; software, T.A.; validation, D.C.; formal analysis, D.C.; investigation, X.H.; resources, Y.Z.; data curation, X.H.; writing—original draft preparation, Z.L.; writing—review and editing, Z.Y.; visualization, Y.Z.; supervision, D.C; project administration, Z.Y.; funding acquisition, Z.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All details and data can be found in the text.
Acknowledgments
The authors are grateful to Shenyang Pharmaceutical University for the ECD, NMR, and molecular docking analyses.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Futuyma, D.J.; Agrawal, A.A. Macroevolution and the biological diversity of plants and herbivores. Proc. Natl. Acad. Sci. USA 2009, 106, 18054–18061. [Google Scholar] [CrossRef] [PubMed]
- Konno, K.; One, H.; Nakamura, M.; Tateishi, K.; Hirayama, C.; Tamura, Y.; Hattori, M.; Koyama, A.; Kohno, K. Mulberry latex rich in antidiabetic sugar-mimic alkaloids forces dieting on caterpillars. Proc. Natl. Acad. Sci. USA 2006, 103, 1337–1341. [Google Scholar] [CrossRef] [PubMed]
- Salomé, A.; Luis, F.; Klinkhamer, P.G.L.; Choi, Y.H. Plant latex, from ecological interests to bioactive chemical resources. Phytochemistry 2019, 85, 856–868. [Google Scholar]
- Agrawal, A.A.; Konno, K. Latex: A model for understanding mechanisms, ecology, and evolution of plant defense against herbivory. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 311–331. [Google Scholar] [CrossRef]
- Ramos, M.V.; Demarco, D.; Souza, I.C.; Freitas, C.D.T. Laticifers, latex, and their role in plant defense. Trends Plant Sci. 2019, 24, 553–567. [Google Scholar] [CrossRef]
- Shi, Q.W.; Su, X.H.; Kiyota, H. Chemical and pharmacological research of the plants in genus Euphorbia. Chem. Rev. 2008, 108, 4295–4327. [Google Scholar] [CrossRef] [PubMed]
- Betancur-Galvis, L.; Palomares, E.; Marco, J.A.; Estornell, E. Tigliane diterpenes from the latex of Euphorbia obtusifolia with inhibitory activity on the mammalian mitochondrial respiratory chain. J. Ethnopharmacol. 2003, 85, 279–282. [Google Scholar] [CrossRef]
- Ashour, M.; Wink, M.; Gershenzon, J. Biochemistry of terpenoids: Monoterpenes, sesquiterpenes and diterpenes. Ann. Plant Rev. 2011, 40, 258–303. [Google Scholar]
- Barina, Z.; Shevera, M.; Sirbu, C.; Pinke, G. Current distribution and spreading of Euphorbia davidii (E. dentata agg.) in Europe. Cent. Eur. J. Biol. 2013, 8, 87–95. [Google Scholar] [CrossRef]
- Riaz, S.; Johnson, J.B.; Ahmad, M.; Fitt, G.P.; Naiker, M. A review on biological interactions and management of the cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). J. Appl. Entomol. 2021, 145, 467–498. [Google Scholar] [CrossRef]
- Tay, W.T.; Soria, M.F.; Walsh, T.; Thomazoni, D.; Silvie, P.; Behere, G.T.; Anderson, C.; Downes, S. A brave new world for an old world pest: Helicoverpa armigera (Lepidoptera: Noctuidae) in Brazil. PLoS ONE 2013, 8, e80134. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.R.; Trujillo, D.; Bernardi, O.; Rodrigues, J.C.V.; Bailey, W.D.; Gilligan, T.M.; Carrillo, D. Comparative toxicity of Helicoverpa armigera and Helicoverpa zea (Lepidoptera: Noctuidae) to selected insecticides. Insects 2020, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Piao, X.M.; Zhang, L.X.; Song, S.Y.; Xu, Y.H. Ginsenosides from the stems and leaves of Panax ginseng show antifeedant activity against Plutella xylostella (Linnaeus). Ind. Crop. Prod. 2018, 124, 412–417. [Google Scholar] [CrossRef]
- Wu, G.; Miyata, T. Susceptibilities to methamidophos and enzymatic characteristics in 18 species of pest insects and their natural enemies in crucifer vegetable crops. Pestic. Biochem. Phys. 2005, 82, 79–93. [Google Scholar] [CrossRef]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- Macias, F.A.; Varela, R.M.; Torres, A.; Oliva, R.M.; Molinillo, J.M.G. Bioactive norsesquiterpenes from Helianthus annuus with potential allelopathic activity. Phytochemistry 1998, 48, 631–636. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, D.G.; Yeon, S.W.; Kwon, H.S.; Ko, J.H.; Shin, D.J.; Park, H.S.; Kim, Y.S.; Bang, M.H.; Baek, N.I. Isolation of megastigmane sesquiterpenes from the silkworm (Bombyx mori L.) droppings and their promotion activity on HO-1 and SIRT1. Arch. Pharm. Res. 2011, 34, 533–542. [Google Scholar] [CrossRef]
- Hossen, K.; Asato, Y.; Teruya, T.; Kato-Noguchi, H. Identification of four allelopathic compounds including a novel compound from Elaeocarpus floribundus Blume and determination of their allelopathic activity. J. Environ. Manag. 2023, 326, 116728. [Google Scholar] [CrossRef]
- Yuan, Z.G.; Zheng, X.W.; Zhao, Y.; Liu, Y.; Zhou, S.X.; Wei, C.X.; Hu, Y.X.; Shao, H. Phytotoxic compounds isolated from leaves of the invasive weed Xanthium spinosum. Molecules 2018, 23, 2840. [Google Scholar] [CrossRef]
- Kim, I.; Chin, Y.W.; Lim, S.W.; Kim, Y.C.; Kim, J. Norisoprenoids and hepatoprotective flavone glycosides from the aerial parts of Beta vulgaris var. cicla. Arch. Pharm. Res. 2004, 27, 600–603. [Google Scholar] [CrossRef]
- Kuang, H.X.; Yang, B.Y.; Xia, Y.G.; Feng, W.S. Chemical constituents from the flower of Datura metel L. Arch. Pharm. Res. 2008, 31, 1094–1097. [Google Scholar] [CrossRef]
- De Marino, S.; Borbone, N.; Zollo, N.; Ianaro, A.; Di Meglio, P.; Iorizzi, M. Megastigmane and phenolic components from Laurus nobilis L. leaves and their inhibitory effects on nitric oxide production. J. Agric. Food Chem. 2004, 52, 7525–7531. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.; Liu, Y.; Xiao, C.J.; Jing, S.X.; Luo, S.H.; Li, S.H. Chemical profile and defensive function of the latex of Euphorbia peplus. Phytochemistry 2017, 136, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.H.; Hua, J.; Niu, X.M.; Liu, Y.; Li, C.H.; Zhou, Y.Y.; Jing, S.X.; Zhao, X.; Li, S.H. Defense sesterterpenoid lactones from Leucosceptrum canum. Phytochemistry 2013, 86, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.X.; Wang, M.Q.; Tian, M.X.; Yuan, L.L.; Yu, B.M.; Qu, B.; An, T.; Feng, Y.L. Pyrrole alkaloids from Solanum rostratum and their chemical defense function against Henosepilachna vigintioctomaculata. Fitoterapia 2021, 155, 105031. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Tian, J.L.; An, T.; Lyu, F.N.; Jia, P.F.; Zhou, M.J.; Liu, Z.X.; Feng, Y.L. Secondary metabolites from Solanum rostratum and their antifeedant defense mechanisms against Helicoverpa armigera. J. Agric. Food Chem. 2020, 68, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, S.K.; Desiraju, G.R. Strong and weak hydrogen bonds in theprotein-ligand interface. Proteins 2007, 67, 128–141. [Google Scholar] [CrossRef]
- Zhou, Y.Y.; Luo, S.H.; Yi, T.S.; Li, C.H.; Luo, Q.; Hua, J.; Liu, Y.; Li, S.H. Secondary metabolites from Glycine soja and their growth inhibitory effect against Spodoptera litura. J. Agric. Food Chem. 2011, 59, 6004–6010. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Ren, L.L.; Chen, F.; Feng, Y.Q.; Luo, Y.Q. Antifeedant activity of Ginkgo biloba secondary metabolites against Hyphantria cunea Larvae: Mechanisms and application. PLoS ONE 2016, 11, e0155682. [Google Scholar] [CrossRef] [PubMed]
- Ellman, G.L.; Courtney, K.; Andres, V.; Featherstone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Asperen, K. A study of housefly esterases by means of a sensitive colorimetric method. J. Insect Physiol. 1962, 8, 401–416. [Google Scholar] [CrossRef]
- Habig, W.H.; Jakoby, W.B. Assays for differentiation of glutathione S-transferases. Methods Enzymol. 1981, 77, 398–405. [Google Scholar] [PubMed]
- Shang, C.C.; Soderlund, D.M. Monooxygenase activity of tobacco budworm (Heliothis virescens F.) larvae: Tissue distribution and optimal assay conditions for the gut activity. Comp. Biochem. Phys. B 1984, 79, 407–411. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).