Computational Studies to Understand the Neuroprotective Mechanism of Action Basil Compounds
Abstract
:1. Introduction
2. Results and Discussion
2.1. Design of Ligand Library
2.2. Target Selection
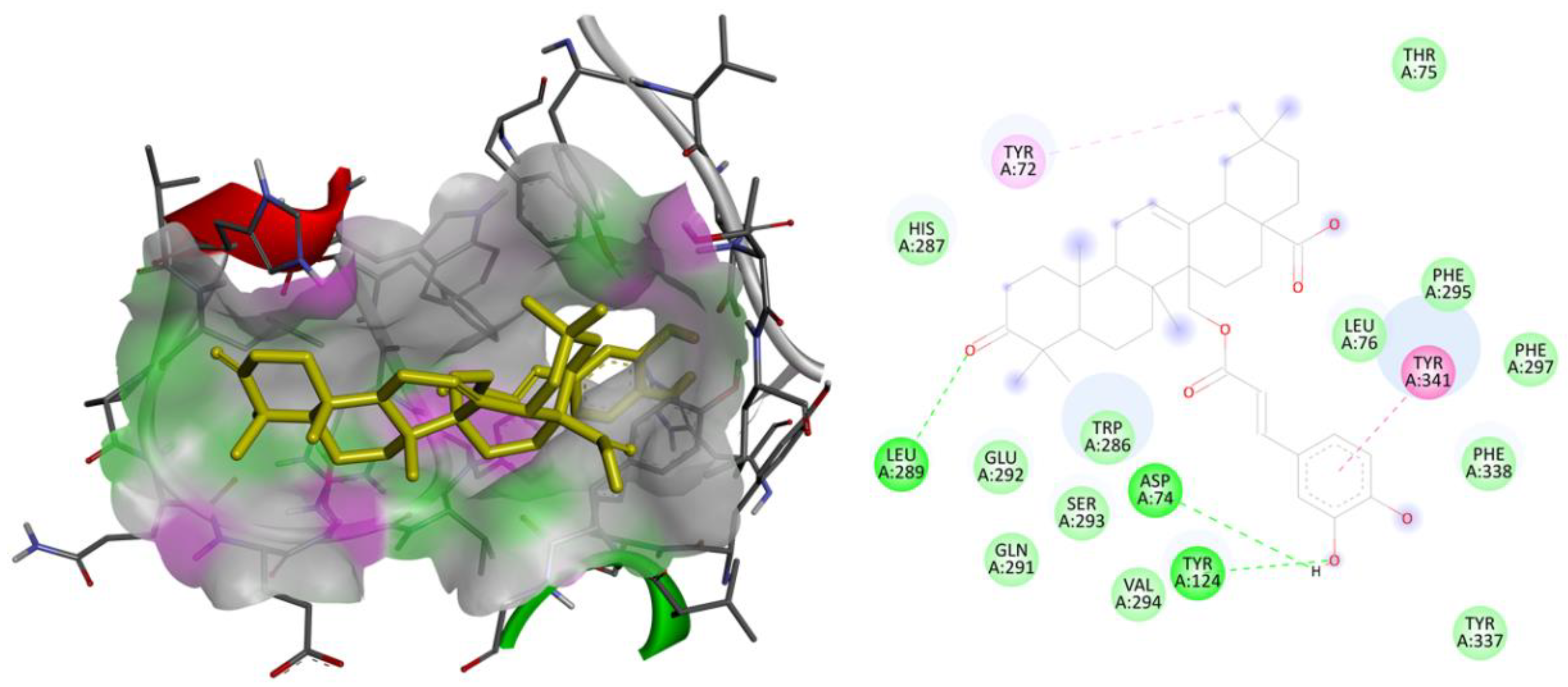
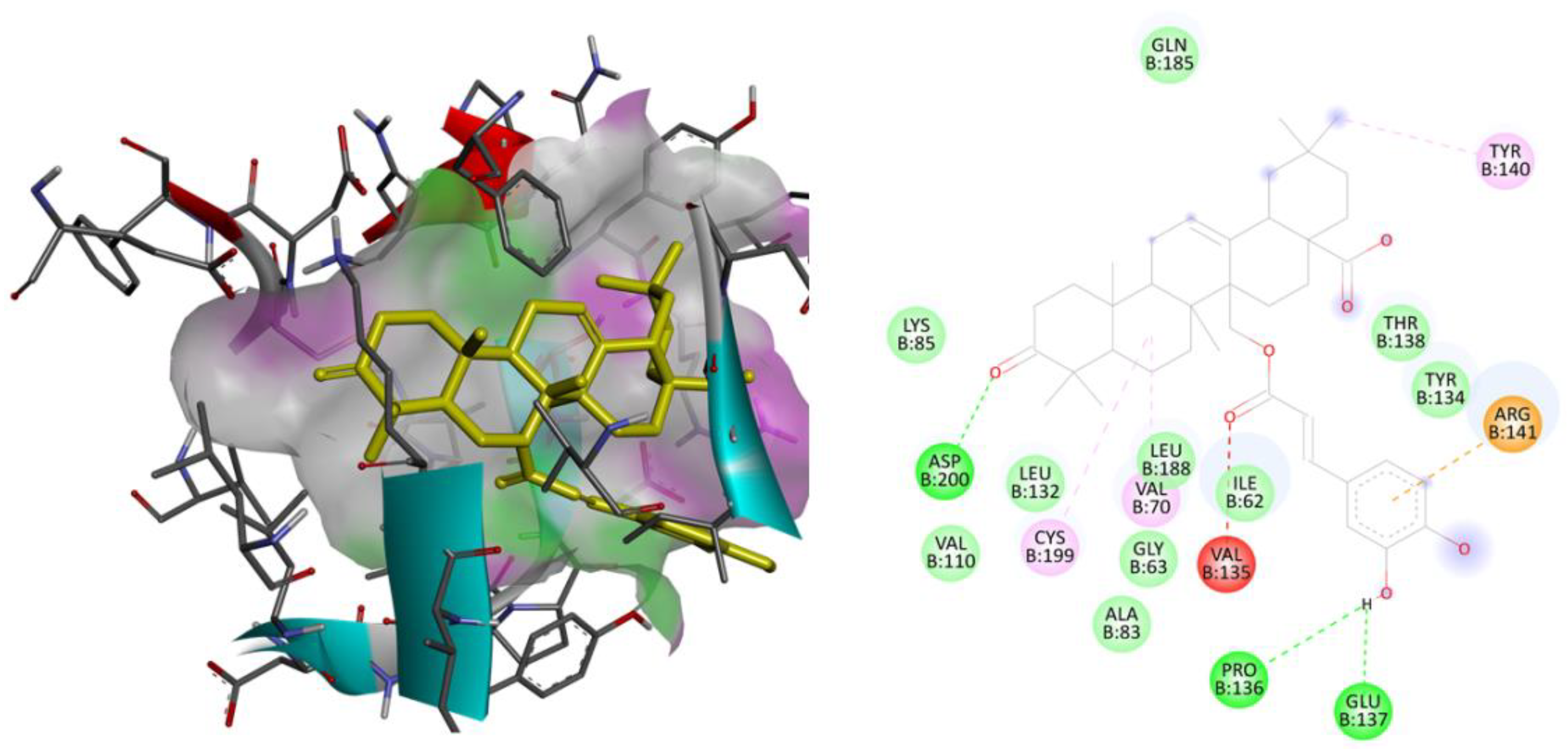
2.3. Molecular Docking Studies
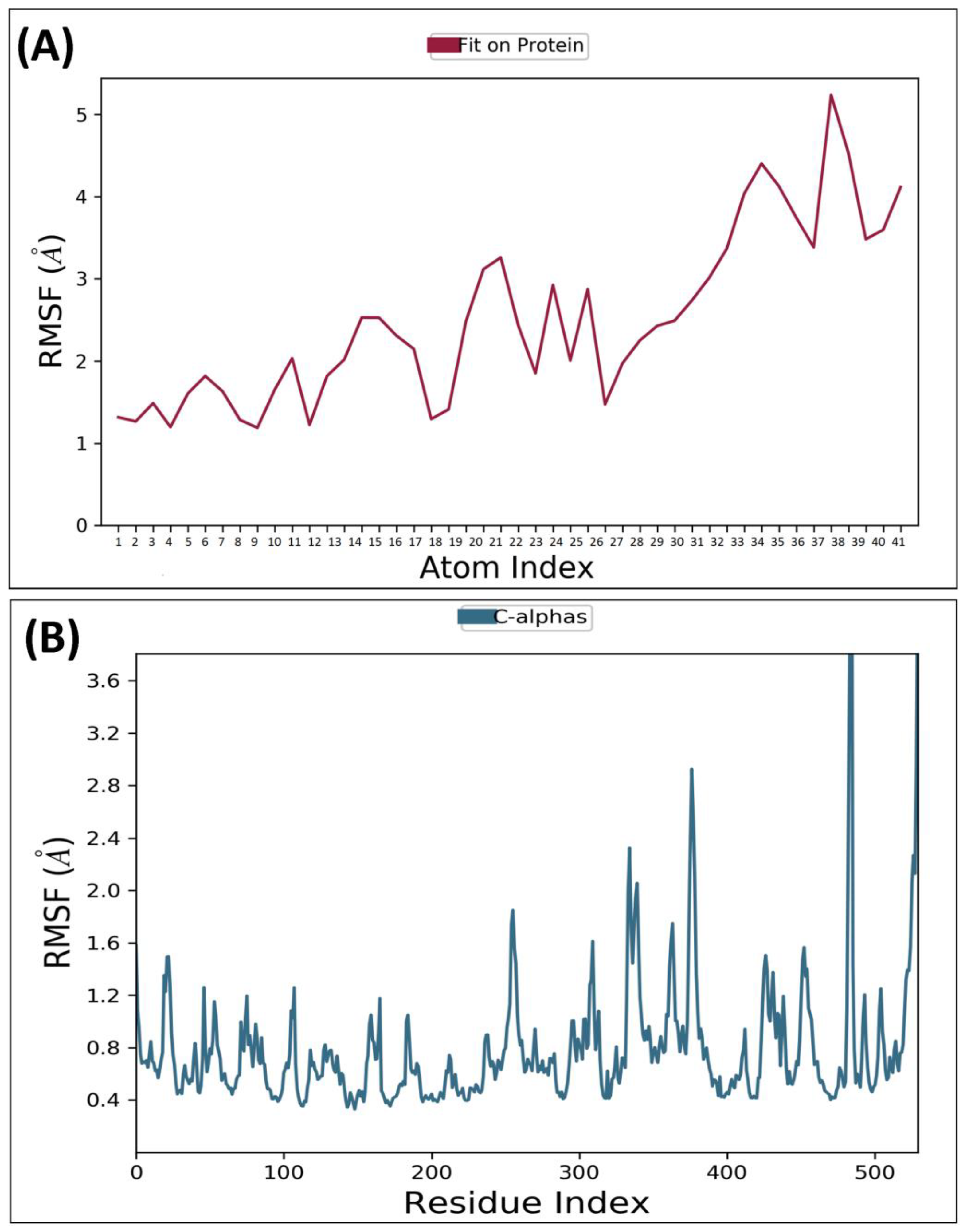
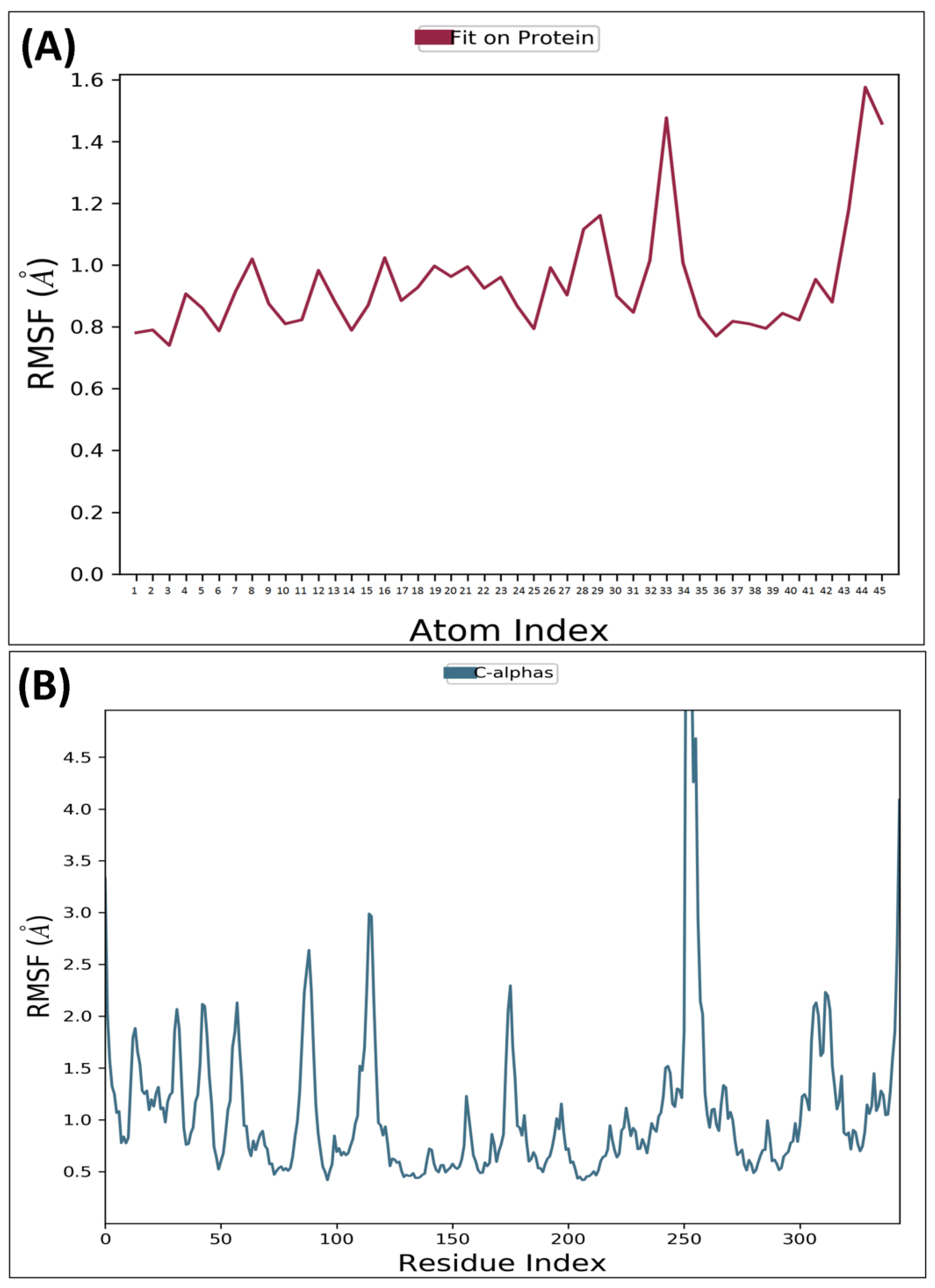
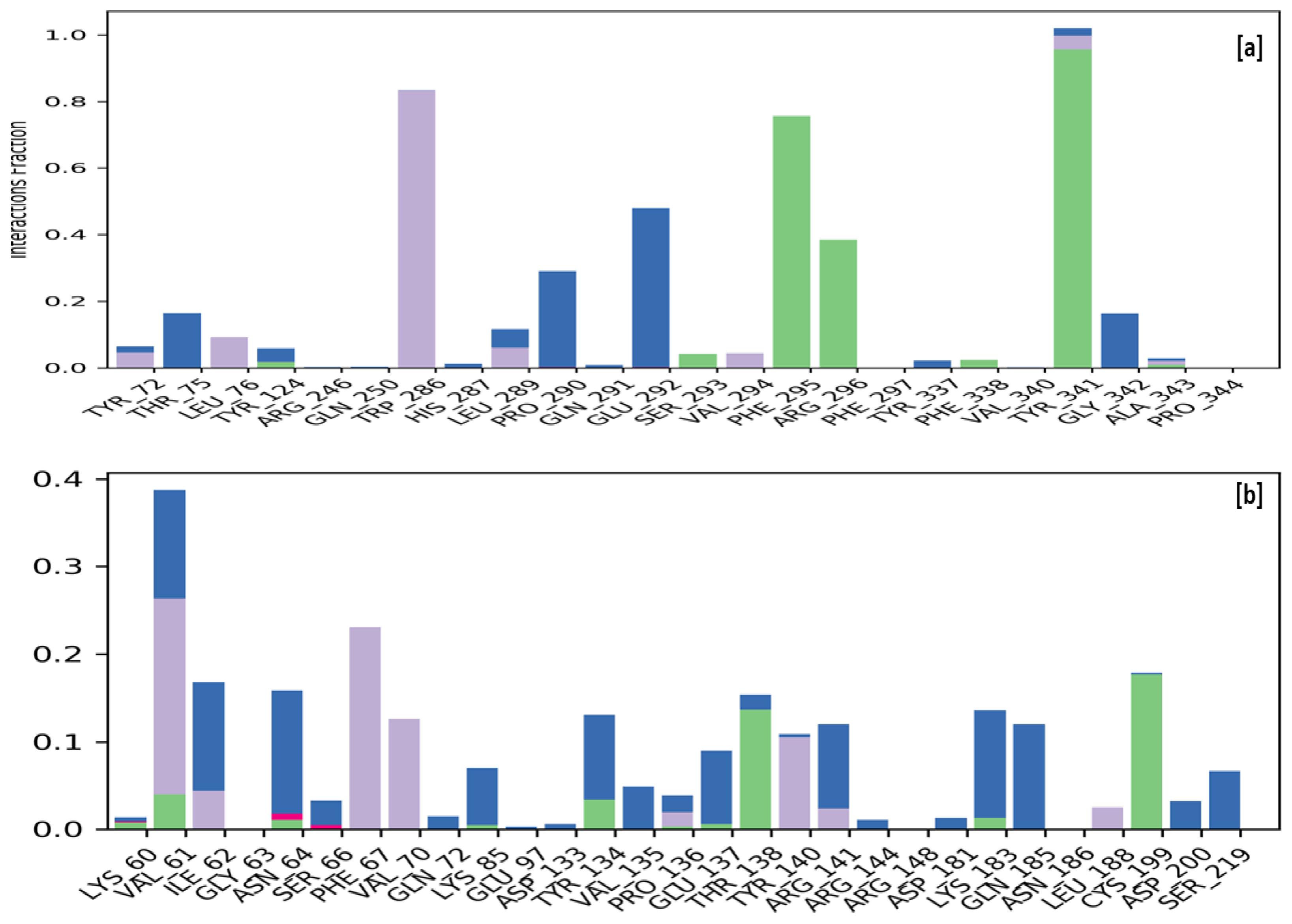
2.4. Molecular Dynamic Simulation
3. Materials and Methodology
3.1. Ligand Library Preparation
3.2. Macromolecular Target Selection and Preparation
3.3. Molecular Docking Studies
3.4. Molecular Dynamics Simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bright, F.; Werry, E.L.; Dobson-Stone, C.; Piguet, O.; Ittner, L.M.; Halliday, G.M.; Hodges, J.R.; Kiernan, M.C.; Loy, C.T.; Kassiou, M.; et al. Neuroinflammation in Frontotemporal Dementia. Nat. Rev. Neurol. 2019, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Tisher, A.; Salardini, A. A Comprehensive Update on Treatment of Dementia. Semin. Neurol. 2019, 39, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Magierski, R.; Sobow, T.; Schwertner, E.; Religa, D. Pharmacotherapy of Behavioral and Psychological Symptoms of Dementia: State of the Art and Future Progress. Front. Pharmacol. 2020, 11, 1168. [Google Scholar] [CrossRef] [PubMed]
- Van Hall, G.; Strømstad, M.; Rasmussen, P.; Jans, Ø.; Zaar, M.; Gam, C.; Quistorff, B.; Secher, N.H.; Nielsen, H.B. Pathogenesis of Dementia. Int. J. Mol. Sci. 2022, 24, 543. [Google Scholar] [CrossRef]
- Jang, J.E.; Eom, J.-I.; Jeung, H.-K.; Cheong, J.-W.; Lee, J.Y.; Seok, J.; Yoo, K.; Min, H. AMPK–ULK1-Mediated Autophagy Confers Resistance to BET Inhibitor JQ1 in Acute Myeloid Leukemia Stem Cells. Clin. Cancer Res. 2017, 23, 2781–2794. [Google Scholar] [CrossRef]
- Sarnik, J.; Popławski, T.; Tokarz, P. BET Proteins as Attractive Targets for Cancer Therapeutics. Int. J. Mol. Sci. 2021, 22, 11102. [Google Scholar] [CrossRef]
- Zlokovic, B.V. Vascular Disorder in Alzheimer’s Disease: Role in Pathogenesis of Dementia and Therapeutic Targets. Adv. Drug Deliv. Rev. 2002, 54, 1553–1559. [Google Scholar] [CrossRef]
- Kalaria, R.N. The Pathology and Pathophysiology of Vascular Dementia. Neuropharmacology 2018, 134, 226–239. [Google Scholar] [CrossRef]
- Gackowski, D.; Rozalski, R.; Siomek, A.; Dziaman, T.; Nicpon, K.; Klimarczyk, M.; Araszkiewicz, A.; Olinski, R. Oxidative Stress and Oxidative DNA Damage Is Characteristic for Mixed Alzheimer Disease/Vascular Dementia. J. Neurol. Sci. 2008, 266, 57–62. [Google Scholar] [CrossRef]
- Sharma, B.K.; Sharma, A. Future Prospect of Nanotechnology in Development of Anti-Ageing Formulations. Int. J. Pharm. Pharm. Sci. 2012, 4, 57–66. [Google Scholar]
- Behl, T.; Kaur, G.; Sehgal, A.; Bhardwaj, S.; Singh, S.; Buhas, C.; Judea-Pusta, C.; Uivarosan, D.; Munteanu, M.A.; Bungau, S. Multifaceted Role of Matrix Metalloproteinases in Neurodegenerative Diseases: Pathophysiological and Therapeutic Perspectives. Int. J. Mol. Sci. 2021, 22, 1413. [Google Scholar] [CrossRef] [PubMed]
- Marwat, S.K.; Khan, M.S.; Ghulam, S.; Anwar, N.; Mustafa, G.; Usman, K. Phytochemical constituents and pharmacological activities of sweet Basil-Ocimum basilicum L. (Lamiaceae). Asian J. Chem. 2011, 23, 3773–3782. [Google Scholar]
- Schwarz, S.; Froelich, L.; Burns, A. Pharmacological Treatment of Dementia. Curr. Opin. Psychiatry 2012, 25, 542–550. [Google Scholar] [CrossRef] [PubMed]
- Santaguida, P.; Raina, P.; Booker, L.; Patterson, C.; Baldassarre, F.; Cowan, D.; Gauld, M.; Levine, M.; Unsal, A. Pharmacological Treatment of Dementia: Summary. In AHRQ Evidence Report Summaries; Agency for Healthcare Research and Quality (US): Rockville, MD, USA, 2004; p. 97. [Google Scholar]
- Thapa, K.; Khan, H.; Sharma, U.; Grewal, A.K.; Singh, T.G. Poly (ADP-Ribose) Polymerase-1 as a Promising Drug Target for Neurodegenerative Diseases. Life Sci. 2021, 267, 118975. [Google Scholar] [CrossRef]
- Oladeji, O.S.; Adelowo, F.E.; Ayodele, D.T.; Odelade, K.A. Phytochemistry and pharmacological activities of Cymbopogon citratus: A review. Sci. Afr. 2019, 6, e00137. [Google Scholar] [CrossRef]
- Dhama, K.; Sharun, K.; Gugjoo, M.B.; Tiwari, R.; Alagawany, M.; Iqbal Yatoo, M.; Thakur, P.; Iqbal, H.M.; Chaicumpa, W.; Michalak, I.; et al. A comprehensive review on chemical profile and pharmacological activities of Ocimum basilicum. Food Rev. Int. 2023, 39, 119–147. [Google Scholar] [CrossRef]
- Škrovánková, S.; Mišurcová, L.; Machů, L. Antioxidant activity and protecting health effects of common medicinal plants. Adv. Food Nutr. Res. 2012, 67, 75–139. [Google Scholar] [CrossRef]
- Bhatia, S.; Singh, M.; Singh, T.; Singh, V. Scrutinizing the Therapeutic Potential of PROTACs in the Management of Alzheimer’s Disease. Neurochem. Res. 2023, 48, 13–25. [Google Scholar] [CrossRef]
- Edo, G.I.; Samuel, P.O.; Ossai, S.; Nwachukwu, S.C.; Okolie, M.C.; Oghenegueke, O.; Asaah, E.U.; Akpoghelie, P.O.; Ugbune, U.; Owheruo, J.O.; et al. Phytochemistry and pharmacological compounds present in scent leaf: A review. Food Chem. Adv. 2023, 3, 100300. [Google Scholar] [CrossRef]
- Shahrajabian, M.H.; Sun, W.; Cheng, Q. Chemical Components and Pharmacological Benefits of Basil (Ocimum basilicum): A Review. Int. J. Food Prop. 2020, 23, 1961–1970. [Google Scholar] [CrossRef]
- Hussain, A.I.; Anwar, F.; Hussain Sherazi, S.T.; Przybylski, R. Chemical Composition, Antioxidant and Antimicrobial Activities of Basil (Ocimum basilicum) Essential Oils Depends on Seasonal Variations. Food Chem. 2008, 108, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Jayasinghe, C.; Goto, N.; Aoki, T.; Wada, S. Phenolics Composition and Antioxidant Activity of Sweet Basil (Ocimum basilicum, L.). J. Agric. Food Chem. 2003, 51, 4442–4449. [Google Scholar] [CrossRef]
- Sharma, P.; Singh, V.; Singh, M. N-methylpiperazinyl and piperdinylalkyl-O-chalcone derivatives as potential polyfunctional agents against Alzheimer’s disease: Design, synthesis and biological evaluation. Chem. Biol. Drug Des. 2023, 00, 121. [Google Scholar] [CrossRef]
- Sharma, V.K.; Singh, T.G.; Garg, N.; Dhiman, S.; Gupta, S.; Rahman, M.H.; Najda, A.; Walasek-Janusz, M.; Kamel, M.; Albadrani, G.M.; et al. Dysbiosis and Alzheimer’s Disease: A Role for Chronic Stress? Biomolecules 2021, 11, 678. [Google Scholar] [CrossRef]
- Farag, M.A.; Ezzat, S.M.; Salama, M.M.; Tadros, M.G. Anti-Acetylcholinesterase Potential and Metabolome Classification of 4 Ocimum Species as Determined via UPLC/QTOF/MS and Chemometric Tools. J. Pharm. Biomed. Anal. 2016, 125, 292–302. [Google Scholar] [CrossRef]
- Bhatia, S.; Singh, M.; Sharma, P.; Mujwar, S.; Singh, V.; Mishra, K.K.; Singh, T.G.; Singh, T.; Ahmad, S.F. Scaffold Morphing and In Silico Design of Potential BACE-1 (β-Secretase) Inhibitors: A Hope for a Newer Dawn in Anti-Alzheimer Therapeutics. Molecules 2023, 28, 6032. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.; Rawal, R.; Sharma, P.; Singh, T.; Singh, M.; Singh, V. Mitochondrial Dysfunction in Alzheimer’s Disease: Opportunities for Drug Development. Curr. Neuropharmacol. 2022, 20, 675–692. [Google Scholar] [CrossRef] [PubMed]
- Rosini, M.; Simoni, E.; Caporaso, R.; Minarini, A. Multitarget Strategies in Alzheimer’s Disease: Benefits and Challenges on the Road to Therapeutics. Future Med. Chem. 2016, 8, 697–711. [Google Scholar] [CrossRef]
- Russo, P.; Frustaci, A.; Del Bufalo, A.; Fini, M.; Cesario, A. Multitarget Drugs of Plants Origin Acting on Alzheimer’s Disease. Curr. Med. Chem. 2013, 20, 1686–1693. [Google Scholar] [CrossRef]
- Zheng, H.; Fridkin, M.; Youdim, M. From Single Target to Multitarget/Network Therapeutics in Alzheimer’s Therapy. Pharmaceuticals 2014, 7, 113–135. [Google Scholar] [CrossRef]
- Xia, W. γ-Secretase and Its Modulators: Twenty Years and Beyond. Neurosci. Lett. 2019, 701, 162–169. [Google Scholar] [CrossRef]
- Drenth, H.; Zuidema, S.U.; Krijnen, W.P.; Bautmans, I.; Van Der Schans, C.; Hobbelen, H. Association between Advanced Glycation End-Products and Functional Performance in Alzheimer’s Disease and Mixed Dementia. Int. Psychogeriatr. 2017, 29, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef] [PubMed]
- RCSB PDB-4EY7: Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Donepezil. Available online: https://www.rcsb.org/structure/4ey7 (accessed on 21 September 2023).
- Cheung, J.; Rudolph, M.J.; Burshteyn, F.; Cassidy, M.S.; Gary, E.N.; Love, J.; Franklin, M.C.; Height, J.J. Structures of Human Acetylcholinesterase in Complex with Pharmacologically Important Ligands. J. Med. Chem. 2012, 55, 10282–10286. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Ezzat, S.M.; Salama, M.M.; Tadros, M.G.; Serya, R.A. Anti-acetylcholinesterase activity of essential oils and their major constituents from four Ocimum species. Z. Naturforschung C 2016, 71, 393–402. [Google Scholar] [CrossRef]
- Sharma, P.; Singh, M.; Singh, V.; Singh, T.G.; Singh, T.; Ahmad, S.F. Recent Development of Novel Aminoethyl-Substituted Chalcones as Potential Drug Candidates for the Treatment of Alzheimer’s Disease. Molecules 2023, 28, 6579. [Google Scholar] [CrossRef]
- Xiao, S.; Tian, Z.; Wang, Y.; Si, L.; Zhang, L.; Zhou, D. Recent progress in the antiviral activity and mechanism study of pentacyclic triterpenoids and their derivatives. Med. Res. Rev. 2018, 38, 951–976. [Google Scholar] [CrossRef]
- Bonini, S.A.; Premoli, M.; Tambaro, S.; Kumar, A.; Maccarinelli, G.; Memo, M.; Mastinu, A. Cannabis sativa: A comprehensive ethnopharmacological review of a medicinal plant with a long history. J. Ethnopharmacol. 2018, 227, 300–315. [Google Scholar] [CrossRef]
- Houghton, P.J.; Howes, M.J. Natural products and derivatives affecting neurotransmission relevant to Alzheimer’s and Parkinson’s disease. Neurosignals 2005, 14, 6–22. [Google Scholar] [CrossRef]
- Sharma, R.; Kuca, K.; Nepovimova, E.; Kabra, A.; Rao, M.M.; Prajapati, P.K. Traditional Ayurvedic and herbal remedies for Alzheimer’s disease: From bench to bedside. Expert. Rev. Neurother. 2019, 19, 360–374. [Google Scholar] [CrossRef]
- Sharma, R.; Garg, N.; Verma, D.; Rathi, P.; Sharma, V.; Kuca, K.; Prajapati, P.K. Indian medicinal plants as drug leads in neurodegenerative disorders. In Nutraceuticals in Brain Health and Beyond; Academic Press: Cambridge, MA, USA, 2021; pp. 31–45. [Google Scholar] [CrossRef]
- Walczak-Nowicka, Ł.J.; Herbet, M. Acetylcholinesterase Inhibitors in the Treatment of Neurodegenerative Diseases and the Role of Acetylcholinesterase in Their Pathogenesis. Int. J. Mol. Sci. 2021, 22, 9290. [Google Scholar] [CrossRef] [PubMed]
- Moss, D.E. Improving Anti-Neurodegenerative Benefits of Acetylcholinesterase Inhibitors in Alzheimer’s Disease: Are Irreversible Inhibitors the Future? Int. J. Mol. Sci. 2020, 21, 3438. [Google Scholar] [CrossRef] [PubMed]
- Lauretti, E.; Dincer, O.; Praticò, D. Glycogen Synthase Kinase−3 Signaling in Alzheimer’s Disease. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2020, 1867, 118664. [Google Scholar] [CrossRef]
- dos Santos Picanco, L.C.; Ozela, P.F.; de Fatima de Brito Brito, M.; Pinheiro, A.A.; Padilha, E.C.; Braga, F.S.; de Paula da Silva, C.H.T.; dos Santos, C.B.R.; Rosa, J.M.C.; da Silva Hage-Melim, L.I. Alzheimer’s Disease: A Review from the Pathophysiology to Diagnosis, New Perspectives for Pharmacological Treatment. Curr. Med. Chem. 2016, 25, 3141–3159. [Google Scholar] [CrossRef]
- Jorissen, E.; De Strooper, B. γ-Secretase and the Intramembrane Proteolysis of Notch. Curr. Top. Dev. Biol. 2010, 92, 201–230. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, T.; Soares, G.A.B.E.; Chopra, H.; Rahman, M.M.; Hasan, Z.; Swain, S.S.; Cavalu, S. Applications of Phyto-Nanotechnology for the Treatment of Neurodegenerative Disorders. Materials 2022, 15, 804. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Behl, T.; Chadha, S. Synthesis of Physically Crosslinked PVA/Chitosan Loaded Silver Nanoparticles Hydrogels with Tunable Mechanical Properties and Antibacterial Effects. Int. J. Biol. Macromol. 2020, 149, 1262–1274. [Google Scholar] [CrossRef] [PubMed]
- Chandramowlishwaran, P.; Vijay, A.; Abraham, D.; Li, G.; Mwangi, S.M.; Srinivasan, S. Role of Sirtuins in Modulating Neurodegeneration of the Enteric Nervous System and Central Nervous System. Front. Neurosci. 2020, 14, 614331. [Google Scholar] [CrossRef]
- Anderson, K.A.; Madsen, A.S.; Olsen, C.A.; Hirschey, M.D. Metabolic Control by Sirtuins and Other Enzymes That Sense NAD+, NADH, or Their Ratio. Biochim. Biophys. Acta Bioenerg. 2017, 1858, 991–998. [Google Scholar] [CrossRef]
- Bai, X.C.; Rajendra, E.; Yang, G.; Shi, Y.; Scheres, S.H.W. Sampling the Conformational Space of the Catalytic Subunit of Human G-Secretase. Elife 2015, 4, e11182. [Google Scholar] [CrossRef]
- RCSB PDB—5FN2: Cryo-EM Structure of Gamma Secretase in Complex with a Drug DAPT. Available online: https://www.rcsb.org/structure/5FN2 (accessed on 27 July 2023).
- Demuro, S.; Sauvey, C.; Tripathi, S.K.; Di Martino, R.M.C.; Shi, D.; Ortega, J.A.; Russo, D.; Balboni, B.; Giabbai, B.; Storici, P.; et al. ARN25068, a Versatile Starting Point towards Triple GSK−3β/FYN/DYRK1A Inhibitors to Tackle Tau-Related Neurological Disorders. Eur. J. Med. Chem. 2022, 229, 114054. [Google Scholar] [CrossRef] [PubMed]
- RCSB PDB—7OY5: Crystal Structure of GSK3Beta in Complex with ARN25068. Available online: https://www.rcsb.org/structure/7OY5 (accessed on 27 July 2023).
- RCSB PDB—5Y5N: Crystal Structure of Human Sirtuin 2 in Complex with a Selective Inhibitor. Available online: https://www.rcsb.org/structure/5Y5N (accessed on 27 July 2023).
- Mellini, P.; Itoh, Y.; Tsumoto, H.; Li, Y.; Suzuki, M.; Tokuda, N.; Kakizawa, T.; Miura, Y.; Takeuchi, J.; Lahtela-Kakkonen, M.; et al. Potent Mechanism-Based Sirtuin–2-Selective Inhibition by an in Situ-Generated Occupant of the Substrate-Binding Site, “Selectivity Pocket” and NAD+-Binding Site. Chem. Sci. 2017, 8, 6400–6408. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Upadhyay, P.K.; Mishra, P. Analgesic, Anti-Inflammatory Activity and Docking Study of 2-(Substituted Phenyl)−3-(Naphthalen−1-Yl)Thiazolidin−4-Ones. J. Indian Chem. Soc. 2020, 97, 39–46. [Google Scholar]
- Gupta, S.M.; Behera, A.; Jain, N.K.; Kumar, D.; Tripathi, A.; Tripathi, S.M.; Mujwar, S.; Patra, J.; Negi, A. Indene-Derived Hydrazides Targeting Acetylcholinesterase Enzyme in Alzheimer’s: Design, Synthesis, and Biological Evaluation. Pharmaceutics 2023, 15, 94. [Google Scholar] [CrossRef]
- Sharma, P.; Singh, M. An ongoing journey of chalcone analogues as single and multi-target ligands in the field of Alzheimer’s disease: A review with structural aspects. Life Sci. 2023, 320, 121568. [Google Scholar] [CrossRef]
- Er-rajy, M.; El fadili, M.; Mujwar, S.; Zarougui, S.; Elhallaoui, M. Design of Novel Anti-Cancer Drugs Targeting TRKs Inhibitors Based 3D QSAR, Molecular Docking and Molecular Dynamics Simulation. J. Biomol. Struct. Dyn. 2023, 1–14. [Google Scholar] [CrossRef]
- Cousins, K.R. ChemDraw Ultra 9.0. CambridgeSoft, 100 CambridgePark Drive, Cambridge, MA 02140. www.Cambridgesoft.Com. See Web Site for Pricing Options. J. Am. Chem. Soc. 2005, 127, 4115–4116. [Google Scholar] [CrossRef]

| Ligand | AChE (4ey7) | Gamma-Secretase (5fn2) | GSK3B (7oy5) | Sirtuin–2 (5y5n) |
|---|---|---|---|---|
| 18HODA | −5.09 | −2.28 | −4.37 | −7.06 |
| 2 | −6.17 | −2.74 | −4.30 | −7.56 |
| Apigenin | −8.15 | −5.72 | −7.58 | −9.33 |
| Caffeic Acid | −4.63 | −4.04 | −5.18 | −5.91 |
| Caffosyl Gluco | −6.61 | −3.64 | −5.30 | −8.05 |
| Caftaric Acid | −4.52 | −2.85 | −3.65 | −4.63 |
| Chichoric Acid | −5.22 | −2.43 | −5.30 | −6.66 |
| DHDM Flavone | −8.52 | −5.48 | −7.45 | −10.17 |
| DH Palmitic Acid | −4.04 | −1.91 | −3.33 | −5.95 |
| E Arabinoside | −8.64 | −5.36 | −7.43 | −7.61 |
| Ellagic Acid Pentoside | −9.31 | −5.78 | −7.70 | −8.47 |
| Feruloyl TA | −4.27 | −1.95 | −4.20 | −5.45 |
| Ferusoyl Glucosidase | −6.39 | −3.40 | −4.45 | −7.53 |
| GQ Apioside | −6.97 | −2.70 | −4.97 | +35.96 |
| HOD Dienoic Acid | −5.32 | −2.58 | −4.16 | −8.49 |
| Isomeli A | −5.93 | −3.27 | −4.85 | −8.78 |
| Isoquercetin | −8.71 | −4.80 | −6.93 | −7.77 |
| KoGluco | −6.82 | −3.30 | −5.32 | +1.83 |
| Linolenic Acid | −5.47 | −3.04 | −4.00 | −8.26 |
| Lithospermica A | −5.32 | −4.38 | −6.77 | −6.47 |
| Myricerone caffeoyl ester | −7.54 | −8.85 | −10.01 | +17.06 |
| Octa DA | −5.41 | −2.49 | −4.10 | −7.21 |
| Octadeca TA | −6.13 | −2.71 | −4.96 | −8.52 |
| Olenoliec Acid | −7.79 | −7.96 | −9.40 | +10.64 |
| Palmitic Acid | −5.33 | −2.30 | −3.81 | −6.72 |
| Quercetin 3oA | −6.10 | −3.72 | −4.86 | +0.23 |
| QuercetinDG | −6.40 | −3.21 | −5.02 | −1.42 |
| Rosmar Acid 3 Gluco | −6.42 | −1.70 | −3.56 | −4.42 |
| Rosmarinic Acid | −6.27 | −4.69 | −6.29 | −8.63 |
| Rutin | −5.99 | −4.22 | −4.72 | −3.08 |
| Salicylic Acid Gluco | −4.53 | −3.22 | −3.96 | −6.42 |
| Salicylic AoG | −5.70 | −3.57 | −5.01 | −6.43 |
| Salvianolic Acid | −8.75 | −5.68 | −7.06 | −11.50 |
| Savialinic Acid | −8.78 | −3.93 | −5.87 | −10.61 |
| Saviolonic AA | −6.58 | −4.29 | −6.68 | −9.25 |
| Savialonic AC | −8.27 | −6.00 | −6.80 | −10.75 |
| THOctadecadienoic Acid | −4.22 | −1.99 | −3.55 | −6.91 |
| Vicenin2 | −8.23 | −3.48 | −5.33 | +39.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, V.; Mujwar, S.; Singh, M.; Singh, T.; Ahmad, S.F. Computational Studies to Understand the Neuroprotective Mechanism of Action Basil Compounds. Molecules 2023, 28, 7005. https://doi.org/10.3390/molecules28207005
Singh V, Mujwar S, Singh M, Singh T, Ahmad SF. Computational Studies to Understand the Neuroprotective Mechanism of Action Basil Compounds. Molecules. 2023; 28(20):7005. https://doi.org/10.3390/molecules28207005
Chicago/Turabian StyleSingh, Varinder, Somdutt Mujwar, Manjinder Singh, Tanveer Singh, and Sheikh F. Ahmad. 2023. "Computational Studies to Understand the Neuroprotective Mechanism of Action Basil Compounds" Molecules 28, no. 20: 7005. https://doi.org/10.3390/molecules28207005
APA StyleSingh, V., Mujwar, S., Singh, M., Singh, T., & Ahmad, S. F. (2023). Computational Studies to Understand the Neuroprotective Mechanism of Action Basil Compounds. Molecules, 28(20), 7005. https://doi.org/10.3390/molecules28207005






