Controlled Assembly of Fluorophores inside a Nanoliposome
Abstract
1. Introduction
2. Results and Discussions
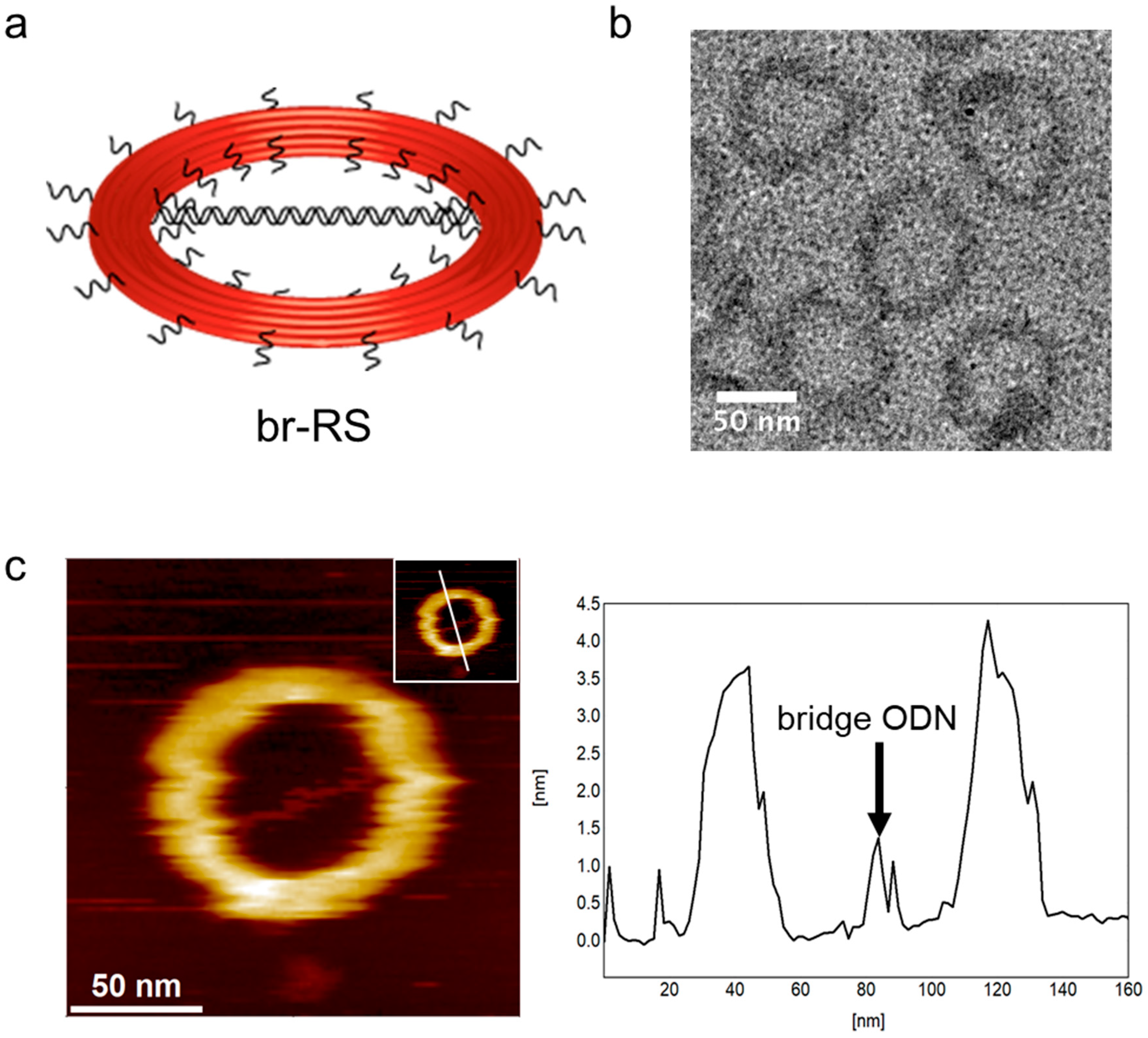
2.1. Design of a Ring-Shaped DNA Origami Skeleton with a Bridged DNA Platform
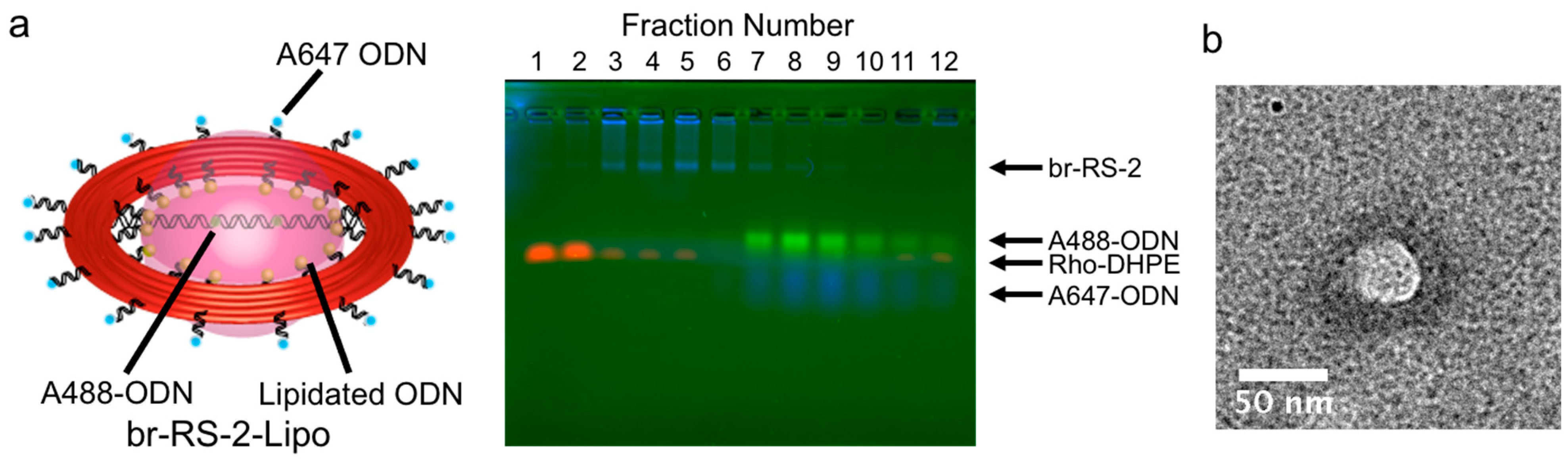
2.2. Construction of a Nanoliposome Guided by the DNA Origami Ring Skeleton to Encapsulate the Bridged DNA Platform
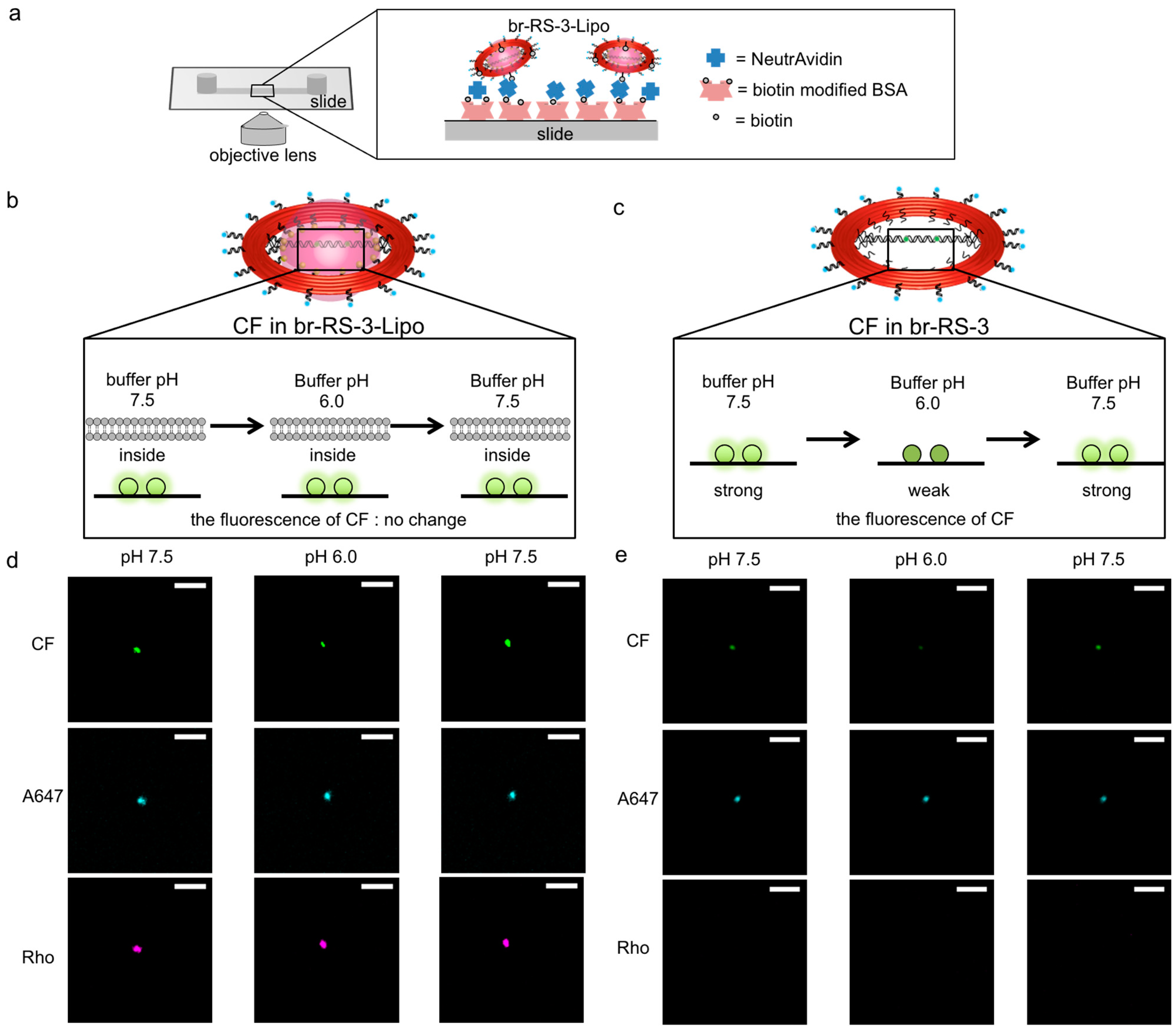
2.3. Controlled Assembly of Fluorophores on the Bridged DNA Platform Encapsulated in the Nanoliposome
3. Materials and Methods
3.1. Materials
3.2. Preparation of the DNA Scaffold
3.3. Preparation of Liposome on DNA Nanostructure
3.4. Synthesis of Lipidated DNA
3.5. Agarose Gel Electrophoresis
3.6. AFM Imaging
3.7. TEM Imaging
3.8. Fluorescent Microscopy
3.9. Fluorescence Measurement
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Agapakis, C.M.; Boyle, P.M.; Silver, P.A. Natural Strategies for the Spatial Optimization of Metabolism in Synthetic Biology. Nat. Chem. Biol. 2012, 8, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Bonacci, W.; Teng, P.K.; Afonso, B.; Niederholtmeyer, H.; Grob, P.; Silver, P.A.; Savage, D.F. Modularity of a Carbon-Fixing Protein Organelle. Proc. Natl. Acad. Sci. USA 2012, 109, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.H.; Silver, P.A. Designing biological compartmentalization. Trends Cell Biol. 2012, 22, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Brasch, M.; Putri, R.M.; de Ruiter, M.V.; Luque, D.; Koay, M.S.; Castón, J.R.; Cornelissen, J.J.L.M. Assembling Enzymatic Cascade Pathways inside Virus-Based Nanocages Using Dual-Tasking Nucleic Acid Tags. J. Am. Chem. Soc. 2017, 139, 1512–1519. [Google Scholar] [CrossRef] [PubMed]
- Walde, P.; Ichikawa, S. Enzymes inside Lipid Vesicles: Preparation, Reactivity and Applications. Biomol. Eng. 2001, 18, 143–177. [Google Scholar] [CrossRef] [PubMed]
- Klermund, L.; Poschenrieder, S.T.; Castiglione, K. Biocatalysis in Polymersomes: Improving Multienzyme Cascades with Incompatible Reaction Steps by Compartmentalization. ACS Catal. 2017, 7, 3900–3904. [Google Scholar] [CrossRef]
- Peters, R.J.R.W.; Marguet, M.; Marais, S.; Fraaije, M.W.; van Hest, J.C.M.; Lecommandoux, S. Cascade Reactions in Multicompartmentalized Polymersomes. Angew. Chem. Int. Ed. 2014, 53, 146–150. [Google Scholar] [CrossRef]
- Dubey, N.C.; Tripathi, B.P. Nature inspired multienzyme immobilization: Strategies and concepts. ACS Appl. Bio Mater. 2021, 4, 1077–1114. [Google Scholar] [CrossRef]
- Pinheiro, A.V.; Han, D.; Shih, W.M.; Yan, H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 2011, 6, 763–772. [Google Scholar] [CrossRef]
- Lin, C.; Jungmann, R.; Leifer, A.M.; Li, C.; Levner, D.; Church, G.M.; Shih, W.M.; Yin, P. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA. Nat. Chem. 2012, 4, 832–839. [Google Scholar] [CrossRef]
- Woehrstein, J.B.; Strauss, M.T.; Ong, L.L.; Wei, B.; Zhang, D.Y.; Jungmann, R.; Yin, P. Sub–100-nm metafluorophores with digitally tunable optical properties self-assembled from DNA. Sci. Adv. 2017, 3, e1602128. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Deng, Z.; Yan, H.; Cabrini, S.; Zuckermann, R.N.; Boker, J. Gold Nanoparticle Self-Similar Chain Structure Organized by DNA Origami. J. Am. Chem. Soc. 2010, 132, 3248–3249. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Chao, J.; Xiao, S.; Seeman, N.C. A proximity-based programmable DNA nanoscale assembly line. Nature 2010, 465, 202–205. [Google Scholar] [CrossRef]
- Fu, J.; Liu, M.; Liu, Y.; Woodbury, N.W.; Yao, H. Interenzyme Substrate Diffusion for an Enzyme Cascade Organized on Spatially Addressable DNA Nanostructures. J. Am. Chem. Soc. 2012, 134, 5516–5519. [Google Scholar] [CrossRef]
- Sacca, B.; Meyer, R.; Erkelenz, M.; Kiko, K.; Arndt, A.; Schroeder, H.; Rabe, K.S.; Niemeyer, C.M. Orthogonal Protein Decoration of DNA Origami. Angew. Chem. Int. Ed. 2010, 49, 9378–9383. [Google Scholar] [CrossRef]
- Zhang, F.; Nangreave, J.; Liu, Y.; Yan, H. Structural DNA Nanotechnology: State of the Art and Future Perspective. J. Am. Chem. Soc. 2014, 136, 11198–11211. [Google Scholar] [CrossRef]
- Sacca, B.; Niemeyer, C.M. DNA Origami: The Art of Folding DNA. Angew. Chem. Int. Ed. 2012, 51, 58–66. [Google Scholar] [CrossRef]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Fu, J.; Dhakal, S.; Alexander Johnson-Buck, A.; Liu, M.; Zhang, T.; Woodbury, N.W.; Liu, Y.; Walter, N.G.; Yan, H. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat. Commun. 2016, 7, 10619. [Google Scholar] [CrossRef]
- Grossi, G.; Dalgaard, E.J.M.; Kjems, J.; Andersen, E.S. Control of enzyme reactions by a reconfigurable DNA nanovault. Nat. Commun. 2017, 8, 992. [Google Scholar] [CrossRef]
- Lin, P.; Dinh, H.; Nakata, E.; Morii, T. Conditional dependence of enzyme cascade reaction efficiency on the inter-enzyme distance. Chem. Commun. 2021, 57, 11197–11200. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, A.; Nakata, E.; Nakano, S.; Morii, T. Nucleic-Acid-Templated Enzyme Cascades. ChemBioChem 2017, 18, 696–716. [Google Scholar] [CrossRef] [PubMed]
- Kong, G.; Xiong, M.; Liu, L.; Hu, L.; Meng, H.-M.; Ke, G.; Zhang, X.-B.; Tan, W. DNA origami-based protein networks: From basic construction to emerging applications. Chem. Soc. Rev. 2021, 50, 1846–1873. [Google Scholar] [CrossRef]
- Lin, C.; Perrault, S.D.; Kwak, M.; Graf, F.; Shih, W.M. Purification of DNA-origami nanostructures by rate-zonal centrifugation. Nucleic Acids Res. 2013, 41, e40. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, J.; Shigematsu, H.; Xu, W.; Shih, W.M.; Rothman, J.E.; Lin, C. Self-assembly of size-controlled liposomes on DNA nanotemplates. Nat. Chem. 2016, 8, 476–483. [Google Scholar] [CrossRef]
- Han, J.; Burgess, K. Fluorescent Indicators for Intracellular pH. Chem. Rev. 2010, 110, 2709–2728. [Google Scholar] [CrossRef] [PubMed]
- Nakata, E.; Hirose, H.; Gerelbaatar, K.; Arafiles, J.V.V.; Zhang, Z.; Futaki, S.; Morii, T. A facile combinatorial approach to construct a ratiometric fluorescent sensor: Application for the real-time sensing of cellular pH changes. Chem. Sci. 2021, 12, 8231–8240. [Google Scholar] [CrossRef]
- Rajendran, A.; Krishnamurthy, K.; Giridasappa, A.; Nakata, E.; Morii, T. Stabilization and structural changes of 2D DNA origami by enzymatic ligation. Nucleic Acids Res. 2021, 49, 7884–7900. [Google Scholar] [CrossRef]
- Aime, S.; Sforzi, J.; Palagi, L. Liposome-Based Bioassays. Biology 2020, 9, 202. [Google Scholar] [CrossRef]
- Perrault, S.D.; Shih, W.M. Virus-Inspired Membrane Encapsulation of DNA Nanostructures To Achieve In Vivo Stability. ACS Nano 2014, 8, 5132–5140. [Google Scholar] [CrossRef]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA Origami: Scaffolds for Creating Higher Order Structures. Chem. Rev. 2017, 117, 12584–12640. [Google Scholar] [CrossRef] [PubMed]
- Nakata, E.; Dinh, H.; Ngo, T.A.; Saimura, M.; Morii, T. A modular zinc finger adaptor accelerates the covalent linkage of proteins at specific locations on DNA nanoscaffolds. Chem. Commun. 2015, 51, 1016–1019. [Google Scholar] [CrossRef] [PubMed]
- Ngo, T.A.; Nakata, E.; Saimura, M.; Morii, T. Spatially Organized Enzymes Drive Cofactor-Coupled Cascade Reactions. J. Am. Chem. Soc. 2016, 138, 3012–3021. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.M.; Nakata, E.; Saimura, M.; Dinh, H.; Morii, T. Design of Modular Protein-Tags for the Orthogonal Covalent Bond Formation at Specific DNA Sequences. J. Am. Chem. Soc. 2017, 139, 8487–8496. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.M.; Nakata, E.; Zhang, Z.; Saimura, M.; Dinh, H.; Morii, T. Rational design of a DNA sequence-specific modular protein tag by tuning the alkylation kinetics. Chem. Sci. 2019, 10, 9315–9325. [Google Scholar] [CrossRef]
- Ngo, T.A.; Dinh, H.; Nguyen, T.M.; Liew, F.-F.; Nakata, E.; Morii, T. Protein adaptors assemble functional proteins on DNA scaffolds. Chem. Commun. 2019, 55, 12428–12446. [Google Scholar] [CrossRef]
- Zhang, Z.; Nakata, E.; Dinh, H.; Saimura, M.; Rajendran, A.; Matsuda, K.; Morii, T. Tuning the Reactivity of a Substrate for SNAP-Tag Expands Its Application for Recognition-Driven DNA-Protein Conjugation. Chem. Eur. J. 2021, 27, 18118–18128. [Google Scholar] [CrossRef]

| Modules of br-RS or br-RS-Lipo | |||
|---|---|---|---|
| RS | Bridged DNA | Lipid | |
| br-RS-1 | A647 | none | none |
| br-RS-1-Lipo | NBD | ||
| br-RS-2 | A647 | A488 | none |
| br-RS-2-Lipo | Rhodamine (Rho) | ||
| br-RS-3 | A647 | Fluorescein (CF) | none |
| br-RS-3-Lipo | Rho | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konishi, H.; Nakata, E.; Komatsubara, F.; Morii, T. Controlled Assembly of Fluorophores inside a Nanoliposome. Molecules 2023, 28, 911. https://doi.org/10.3390/molecules28020911
Konishi H, Nakata E, Komatsubara F, Morii T. Controlled Assembly of Fluorophores inside a Nanoliposome. Molecules. 2023; 28(2):911. https://doi.org/10.3390/molecules28020911
Chicago/Turabian StyleKonishi, Hiroaki, Eiji Nakata, Futa Komatsubara, and Takashi Morii. 2023. "Controlled Assembly of Fluorophores inside a Nanoliposome" Molecules 28, no. 2: 911. https://doi.org/10.3390/molecules28020911
APA StyleKonishi, H., Nakata, E., Komatsubara, F., & Morii, T. (2023). Controlled Assembly of Fluorophores inside a Nanoliposome. Molecules, 28(2), 911. https://doi.org/10.3390/molecules28020911







